Abstract
The interferon-induced double-strand RNA activated protein kinase (PKR) plays important roles in host defense against viral infection. Here we demonstrate the significant antiviral role of PKR against foot-and-mouth disease virus (FMDV) and report that FMDV infection inhibits PKR expression and activation in porcine kidney (PK-15) cells. The viral nonstructural protein 3 C proteinase (3Cpro) is identified to be responsible for this inhibition. However, it is independent of the well-known proteinase activity of 3Cpro or 3Cpro-induced shutoff of host protein synthesis. We show that 3Cpro induces PKR degradation by lysosomal pathway and no interaction is determined between 3Cpro and PKR. Together, our results indicate that PKR acts an important antiviral factor during FMDV infection, and FMDV has evolved a strategy to overcome PKR-mediated antiviral role by downregulation of PKR protein.
Keywords: Foot-and-mouth disease virus, PKR, 3Cpro, Lysosomes
Highlights
-
•
FMDV infection triggers PKR mRNA expression, while decreases PKR protein levels.
-
•
3Cpro was responsible for FMDV-induced inhibition of PKR expression and activation.
-
•
3Cpro-induced PKR reduction was independent of its proteinase activity.
-
•
3Cpro induces PKR degradation by lysosomal pathway.
1. Introduction
Foot-and-mouth disease virus (FMDV) is an RNA virus belonging to the genus Aphthovirus of family Picornaviridae. FMDV is the causative agent of foot-and-mouth disease (FMD) that causes highly contagious viral disease of cloven-hoofed animals (Grubman and Baxt, 2004). The outbreak of FMD often results in severe economic consequences due to its impact on trade and the slaughtering of large amounts of animals (Jamal and Belsham, 2013). The genome of FMDV is a positive-strand RNA of ~8.5 kb encoding a large polyprotein. The viral leader proteinase (Lpro), 2 A protein, and 3 C proteinase (3Cpro) subsequently cleaves the polyprotein to yield mature structural and nonstructural proteins (Steinberger et al., 2014). Lpro and 3Cro are multifunctional proteins. In addition to the vital roles in processing of the polyprotein precursor, it is well-known that Lpro and 3Cro are significantly involved in the viral antagonistic activity against host antiviral responses (Lawrence et al., 2012; Liu et al., 2015; Stenfeldt et al., 2016).
Lpro is widely known to promote virus propagation by disrupting the interferon (IFN) signaling pathway to suppress host innate immune responses (Liu et al., 2015; Steinberger and Skern, 2014). 3Cpro belongs to the family of chymotrypsin-like cysteine proteases, and plays an important role in FMDV pathogenesis (Curry et al., 2007). 3Cpro can suppress host antiviral responses by different mechanisms, such as, inducing cleavage of eukaryotic translation initiation factor 4G (eIF4G) to shut off host protein synthesis; cleaving host antiviral proteins and inhibition of IFN-α/β production (Du et al., 2014a; Lawrence et al., 2012; Liu et al., 2015). The significant roles of 3Cpro in viral pathogenesis make 3Cpro as an attractive target for the design of antiviral agents (Sweeney et al., 2007). The mechanisms about the 3Cpro-mediated antagonistic effects are further exploited to provide insights for uncovering the viral pathogenesis.
The interferon-induced double-strand RNA activated protein kinase (PKR) plays important roles in host defense against viral infection. PKR is a serine-threonine kinase that constitutively expresses in mammalian cells (Habjan et al., 2009; Nallagatla et al., 2011). As an antiviral factor, PKR can be induced by IFN treatment and is activated by binding of double-stranded RNA (dsRNA) or the PKR activator (PACT) protein. Activation results in PKR phosphorylation and subsequently phosphorylates the α subunit of the eIF2 translation initiation factor (eIF2α), blocks cellular and viral protein synthesis, thereby interfering with viral transcription or translation in infected cells (Nallagatla et al., 2011). To disrupt or antagonize antiviral effects of PKR, various viruses have evolved different strategies to inhibit PKR activation. Influenza A virus non-structural (NS1) protein interacts with PKR or directly binds the double-stranded RNA to block PKR activation and counteract PKR-mediated inhibition of viral replication (Bergmann et al., 2000, Li et al., 2006, Lu et al., 1995). Hepatitis C virus (HCV) internal ribosome entry site binds to PKR in competition with PKR ligands and prevents the phosphorylation and activation of PKR (Vyas et al., 2003). Our previous study found that FMDV infection leads to a significant decrease of PKR protein as infection progresses (Li et al., 2016b). It implies that PKR may suppress FMDV infection and the virus decreases PKR protein expression to block PKR activation and antagonize the antiviral response.
In the present study, we examined the FMDV modulation of PKR singling in porcine cells. Here we report that overexpression of PKR could significantly suppress FMDV replication. FMDV infection led to decreased levels of PKR protein in comparison with that in the mock-infected cells. The viral 3Cpro was determined to be responsible for this decrease of PKR. 3Cpro-induced PKR reduction was independent of its proteinase activity and the ability to cleave eIF4G.
2. Results
2.1. Dynamics of PKR expression in response to FMDV infection
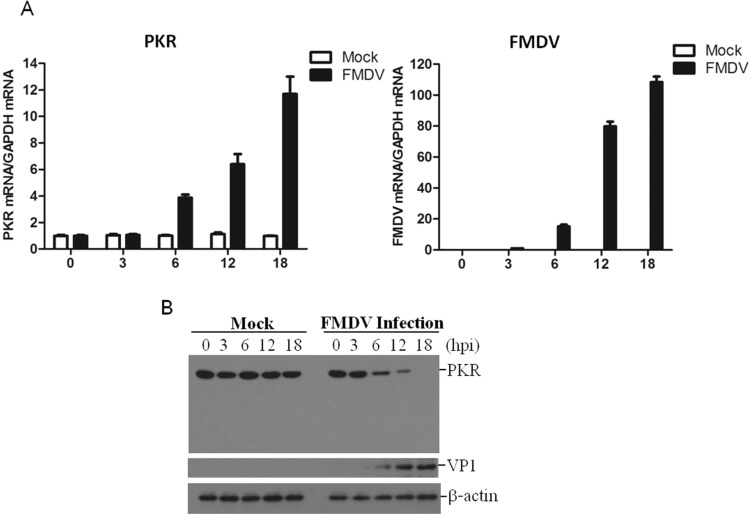
The addition of 2-aminopurine (2-AP), an inhibitor of dsRNA inducible protein kinase (PKR), increases the yield of FMDV in PK-15 cells (Chinsangaram et al., 2001). This indicates the important role of PKR in inhibition of FMDV replication. It was therefore necessary to investigate the state of PKR during FMDV infection. We first investigated the kinetics of PKR to determine the state of PKR in FMDV-infected cells. PK-15 cells were infected by FMDV at an MOI of 0.5, and the dynamics of PKR were examined. It showed that the mRNA level of PKR was significantly upregulated starting at 6 h post-infection (hpi), and gradually increased as the infection progressed; no significant changes in PKR mRNA expression levels were observed in the mock-infected cells ( Fig. 1A). To explore the correlation between PKR expression and FMDV replication, we also determined viral RNA levels. No viral RNA was detectable in mock-infected cells. In FMDV-infected cells, the viral RNA was detectable at 3 hpi, and gradually increased, similar to the increase of PKR (Fig. 1A). The state of PKR protein was also examined at different time points after FMDV infection, which showed that FMDV infection resulted in a significant loss of PKR protein. As shown in Fig. 1B, the amounts of PKR began to decrease at 6 hpi; and by 18 hpi, the amount of PKR almost could not be detected by western blotting. These results suggested that FMDV infection triggers PKR mRNA expression, while PKR protein levels are gradually reduced as infection progresses.
Fig. 1.
FMDV infection inhibits PKR protein expression. (A) PK-15 cells were mock-infected or infected with FMDV for 0, 3, 6, 12 or 18 h, the transcripts of FMDV and PKR were detected by qPCR. (B) The expression levels of viral and PKR proteins in PK-15 cells were detected by western blotting at the indicated time points post-infection.
2.2. Upregulation of PKR inhibits FMDV replication
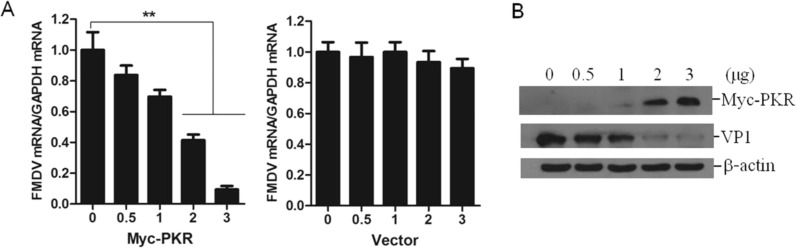
PKR plays an important role in inhibition of FMDV replication. FMDV infection reduced PKR expression. To explore whether upregulation of PKR suppresses FMDV replication, PK-15 cells were transfected with different amounts of empty vector plasmids or Myc-PKR expressing plasmids; at 12 h post-transfection (hpt), the cells were infected with 0.5 MOI of FMDV for 12 h. The viral RNA expression levels were determined and compared. The relative abundance of viral RNA was significantly reduced by overexpression of PKR, showing a dose-dependent manner ( Fig. 2A). No remarkable inhibitive effect was observed in the vector-transfected cells. This observation was further confirmed by western blotting analysis. The presence of VP1 was used as an indicator of FMDV replication. As shown in Fig. 2B, a remarkable decrease of VP1 levels was detected in PKR overexpressing cells, and it also showed a clear dose-dependent manner. These results determined that overexpression of PKR significantly suppresses FMDV infection in the virus-infected cells, which indicated that the presence of PKR in FMDV-infected cells is essential for suppressing virus replication.
Fig. 2.
High abundance of PKR inhibits FMDV replication. Upregulation of PKR suppresses FMDV replication in a dose-dependent manner in PK-15 cells. (A) PK-15 cells were transfected with increasing amounts of Myc-PKR or empty vector plasmids (0, 0.5, 1, 2 or 3 μg). The cells were infected with equal amounts of FMDV (0.5 MOI) at 12 hpt for 12 h. The expression of viral RNA was detected by qPCR; and (B) The expression of viral protein VP1 was detected by western blotting. **P<0.01.
2.3. FMDV infection inhibits PKR activation
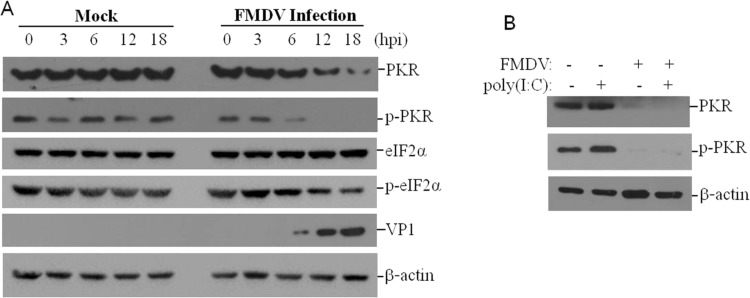
Poliovirus infection degrades PKR levels, while, the levels of phosphorylated PKR (p-PKR) significantly increased (Black et al., 1989). It suggests the decrease of total PKR levels not always results in a similar decrease of PKR phosphorylation levels. Therefore, we also investigated the state of PKR phosphorylation in FMDV-infected cells. The PK-15 cells were inoculated with 0.5 MOI of FMDV and harvested at 0, 3, 6, 12 or 18 hpi. Western blotting result showed that the level of p-PKR in FMDV-infected cells is significantly lower than that in the mock-infected cells ( Fig. 3A). To further confirm this observation, we also investigated the effect of FMDV infection on poly(I:C)-induced PKR phosphorylation. poly(I:C) is an artificial dsRNA that can significantly induce the activation of PKR. PK-15 cells were transfected with poly(I:C) for 6 h, and then infected with FMDV for 12 h, the p-PKR levels were then assessed by western blotting. Transfection of poly(I:C) significantly induced PKR phosphorylation in the mock-infected cells. However, in the FMDV-infected cells, PKR phosphorylation was significantly blocked (Fig. 3B). The phosphorylation of the PKR substrate eukaryotic translation initiation factor 2α (eIF-2α) in the FMDV-infected cells was also evaluated; the relative levels of the phosphorylated eIF-2α (p-eIF2α) were also reduced after FMDV infection, and the total eIF2α level had minimum change (Fig. 3A). These results indicate that FMDV infection blocks PKR activation.
Fig. 3.
FMDV inhibits PKR activation. (A) PK-15 cells were mock-infected or infected with FMDV (0.5 MOI) for 0, 3, 6, 12 or 18 h. The expression levels of PKR, p-PKR, eIF-2α and p-eIF-2α levels were detected by western blotting. (B) PK-15 cells were transfected with 1 μg of empty vector or poly(I:C); and the cells were mock-infected or infected with FMDV (0.5 MOI) for 12 h. The expression levels of PKR and p-PKR levels were detected by western blotting.
2.4. 3Cpro inhibits PKR protein expression and PKR activation
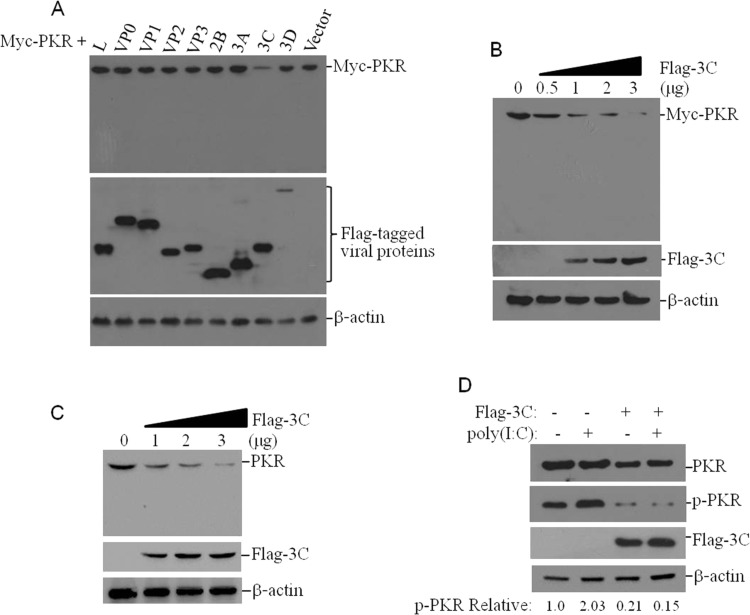
FMDV infection reduces PKR protein levels. To identify the viral proteins that are responsible for the decrease of PKR protein levels, various Flag-tagged viral proteins and Myc-tagged PKR expressing plasmids were co-transfected into HEK293T cells. The expression levels of Myc-PKR were measured by western blotting at 36 hpt. It was observed that 3Cpro significantly reduced PKR abundance ( Fig. 4A). The dose-dependent experiment was further performed, and the results confirmed 3Cpro-mediated suppressive effect on PKR expression (Fig. 4B). 3Cpro-induced decrease of endogenous PKR was also assessed by western blotting, and it showed that 3Cpro significantly reduced endogenous PKR protein levels in a dose-dependent manner (Fig. 4C), which was consistent with those observed above. The effect of 3Cpro on poly(I:C)-induced PKR phosphorylation was subsequently examined by Western blotting. It showed that 3Cpro also significantly suppressed poly(I:C)-induced PKR phosphorylation (Fig. 4D). These results suggest FMDV 3Cpro reduces PKR protein levels and suppresses PKR activation during FMDV infection.
Fig. 4.
3Cpro inhibits PKR protein expression and PKR activation. (A) HEK293T cells were co-transfected with 2 μg Myc-PKR plasmids and 2 μg vector plasmids or 2 μg various Flag-tagged viral proteins expressing plasmids. The cells were collected at 36 hpt. The expressions of Myc-PKR and Flag-tagged viral proteins were detected by western blotting. (B) HEK293T cells were co-transfected with 2 μg Myc-PKR plasmids and increasing amounts of Flag-3C plasmids (0, 0.5, 1, 2 or 3 μg) for 36 h. The expressions of Myc-PKR were detected by western blotting. The empty vector plasmids were used in the transfection process to ensure that the same amount of cells received the same amount of total plasmids. (C) PK-15 cells were transfected with increasing amounts of Flag-3C plasmids (0, 1, 2 or 3 μg) for 36 h. The empty vector plasmids were used in the transfection process to ensure that the same amount of cells received the same amount of total plasmids. The expressions of endogenous PKR were detected by western blotting. (D) PK-15 cells were transfected with vector plasmids or Flag-3C plasmids in the presence or absence of poly(I:C) (1 μg). The PKR and p-PKR levels were detected by western blotting. Relative level of p-PKR is indicated in folds below the images after normalization with β-actin.
2.5. 3Cpro-induced PKR reduction is independent of disruption of eIF4G
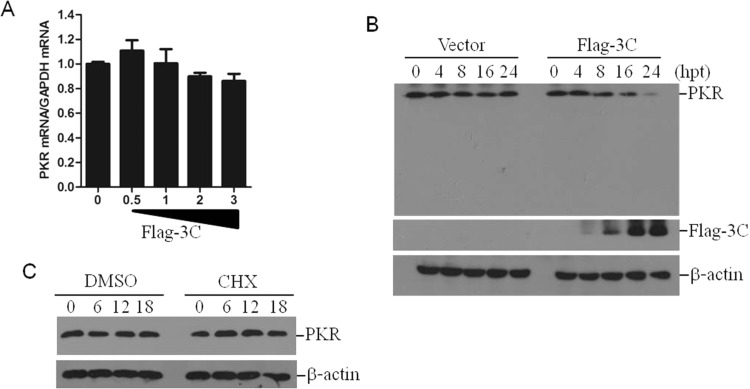
The abundance of PKR mRNA in the Flag-3C-transfected cells was subsequently evaluated to investigate if the reduction of PKR protein was the result of the specific decrease in mRNA expression. The expression of 3Cpro had no significant affection on PKR mRNA expression ( Fig. 5A). This indicates that 3Cpro-induced PKR reduction was not due to reduced synthesis of PKR mRNA. 3Cpro is a viral proteinase that can directly induce the cleavage of various host proteins. The kinetics of 3Cpro-induced reduction of PKR were subsequently examined to detect any PKR cleavage product(s). As shown in Fig. 5B, 3Cpro gradually induced the decrease of PKR as time progressed; however, no cleavage bands were observed. This suggests that 3Cpro does not induce the cleavage of PKR. It is well known that 3Cpro induces the cleavage of the eukaryotic cellular translation initiation factor 4 G (eIF4G) and results in a so-called “shut-off” of host cell protein synthesis. To determine whether the reduction of PKR was completely due to this shut-off effect, a translational inhibitor cycloheximide (CHX) was used to evaluate this correlation. The PK-15 cells were mock-treated or treated with CHX for 0, 6, 12 or 18 h, the PKR levels were detected by western blotting. The inhibition of host protein synthesis did not resulted in detectable reduction of PKR for 18 h (Fig. 5C). This indicates that FMDV- or 3Cpro-induced reduction of PKR protein levels is not related to the well-characterized blocking effect of 3Cpro on cellular translation (viral host cell shutoff).
Fig. 5.
3Cpro induces PKR reduction at posttranslational levels. (A) PK-15 cells were transfected with increasing amounts of Flag-3C plasmids (0, 0.5, 1, 2 or 3 μg) for 36 h. The empty vector plasmids were used in the transfection process to ensure that the same amount of cells received the same amount of total plasmids. The expressions of PKR mRNA levels were detected by qPCR method. (B) PK-15 cells were transfected with vector plasmids or Flag-3C plasmids. The cells were collected at 0, 4, 8, 16 or 24 hpt and subjected to western blotting analysis. The PKR protein levels were then detected and compared. (C) PK-15 cells were treated with DMSO (solvent control) or CHX (100 μg/mL) for 0, 6, 12 or 18 h, the expression of PKR was detected by western blotting.
2.6. 3Cpro does not interact with PKR and the catalytic triad active-site residues of 3Cpro are not essential for 3Cpro-induced PKR reduction
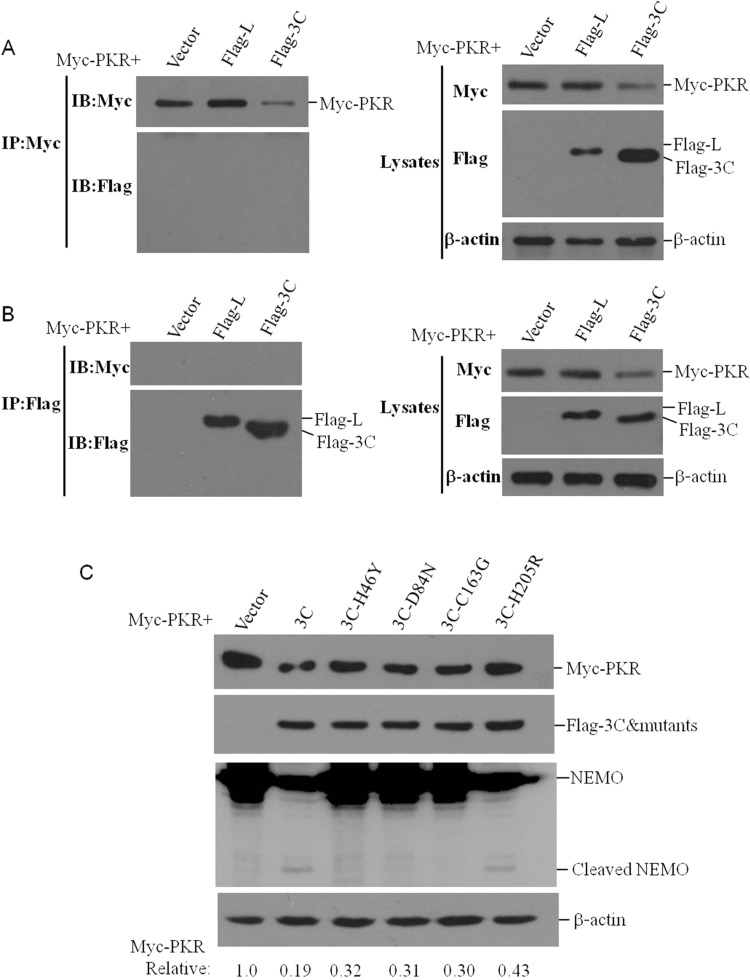
To investigate whether there is a direct interaction between 3Cpro and PKR, the co-immunoprecipitation assay was performed. HEK293T cells were co-transfected Myc-PKR plasmid and vector, Flag-L or Flag-3C plasmids. Lpro showed no effect on PKR expression (Fig. 4), therefore, was used as a control. The cells were collected at 36 hpt, and the cell lysates were immunoprecipitated with anti-Myc antibody. As shown in Fig. 6A, PKR did not precipitate 3 Cpro or Lpro. A reverse immunoprecipitation was also performed using Flag antibody, which showed that 3 Cpro or Lpro did not precipitate PKR (Fig. 6B). These results suggested that there was no direct interaction between PKR and 3Cpro. The catalytic triad of H46, D84, and C163 of FMDV 3Cpro have been determined as crucial sites that were essential for the enzyme activity of 3Cpro (Birtley et al., 2005, Du et al., 2014b, Grubman et al., 1995). To determine whether the enzyme activity was involved in 3Cpro-induced PKR reduction, we constructed 3 mutant plasmids that expressing 3Cpro mutant (Flag-3C-H46Y, Flag-3C-D84N and Flag-3C-C163G) which eliminated the protease activity of 3 Cpro. Another mutant plasmid Flag-3C-H205R that had no influence on the enzyme activity of 3 Cpro was constructed and used as a control. We co-transfected the vector plasmids, mutant plasmids or Flag-3C plasmids with Myc-PKR plasmids into the HEK293T cells, the expression levels of Myc-PKR were detected at 36 hpt. As shown in Fig. 6C, compared to the vector control, the wild-type 3 Cpro, 3C-H205R, 3C-H46Y, 3C-D84N and 3C-C163G all significantly repressed PKR expression. The cleavage of NEMO was used to evaluate the enzyme activity of 3Cpro and 3Cpro mutant. As expected, the mutation of H46Y, D84N and C163G indeed abolished the enzyme activity of 3Cpro (Fig. 6C). It indicated that the elimination of the protease activity of 3Cpro did not impair 3Cpro-induced reduction of PKR.
Fig. 6.
3Cpro does not interact with PKR and the protease activity is not essential for 3Cpro to induce PKR reduction. (A) HEK293T cells grown in the 10-cm dish were co-transfected with Myc-PKR (5 μg) and vector, Flag-L or Flag-3C plasmids (5 μg) for 36 h. The cell lysates were immunoprecipitated with mouse anti-Myc antibody and subjected to western blotting. (B) HEK293T cells grown in the 10-cm dish were co-transfected with Myc-PKR (5 μg) and vector, Flag-L or Flag-3C plasmids (5 μg) for 36 h. The cell lysates were immunoprecipitated with rabbit anti-Flag antibody and subjected to western blotting. (C) HEK293T cells were co-transfected with 2 μg of Myc-PKR plasmids and 3 μg of vector plasmids or various 3 C mutant plasmids for 36 h. The expression of Myc-PKR and host protein NEMO was detected by western blotting. Relative level of Myc-PKR was indicated in folds below the images after normalization with β-actin.
2.7. 3Cpro induces PKR reduction through lysosomal pathway
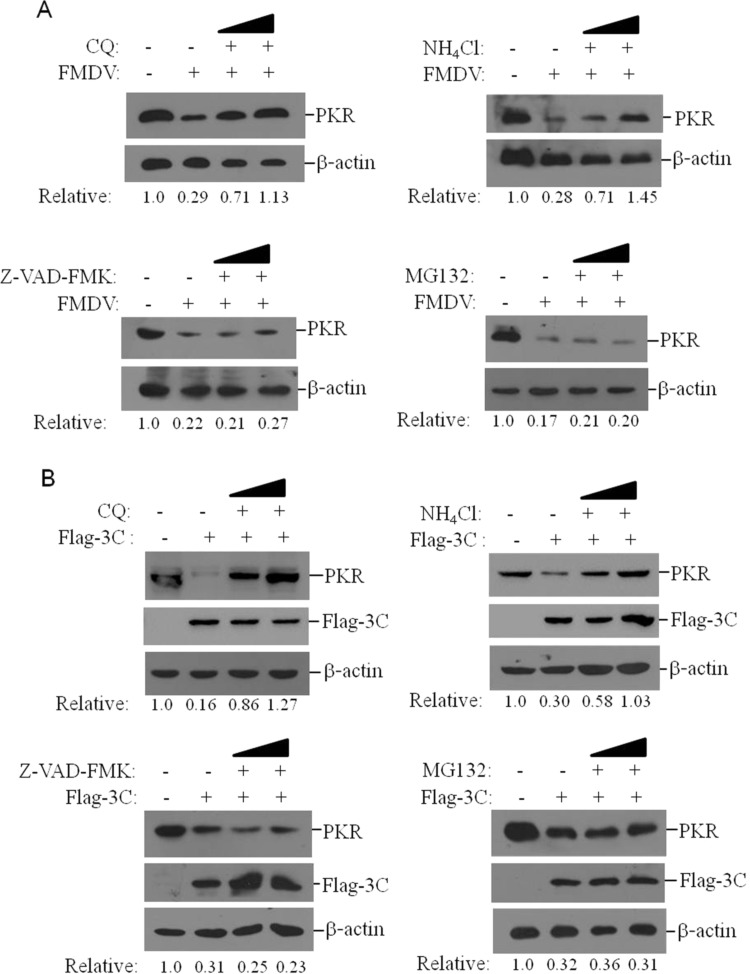
To investigate whether the proteasomes, lysosomes or caspase-dependent pathways are involved in FMDV- and 3Cpro-induced decrease of PKR, the proteasome inhibitor MG132, lysosome inhibitor ammonium chloride (NH4Cl) and chloroquine diphosphate (CQ), and a general caspases inhibitor Z-VAD-FMK were used to examine the inhibitive effects. As show in Fig. 7A, incubation of CQ and NH4Cl significantly restored PKR levels in FMDV-infected cells. However, MG132 and Z-VAD-FMK had no such effect on PKR restoration. The effect of these inhibitors on 3 Cpro-induced decrease of PKR was also evaluated by western blotting, which indicated that incubation of CQ and NH4Cl also restored PKR levels in Flag-3C-tranfected cells, and MG132 and Z-VAD-FMK had no such effect (Fig. 7B). Taken together, these results suggest that FMDV 3Cpro induces PKR degradation through the lysosome pathway.
Fig. 7.
FMDV and 3Cpro induces PKR reduction through lysosomal pathway. (A) PK-15 cells were mock-infected or infected with FMDV (0.5 MOI) in the absence or presence of CQ (50 or 100 μM), NH4Cl (10 or 20 mM), Z-VAD-FMK (10 or 50 μM) or MG132 (2 or 20 μM). The expression of PKR was detected by western blotting at 12 hpi. (B) PK-15 cells were transfected with 2 μg vector plasmids or 2 μg Flag-3C plasmids in the absence or presence of CQ (50 or 100 μM), NH4Cl (10 or 20 mM), Z-VAD-FMK (10 or 50 μM) or MG132 (2 or 20 μM). The expression of PKR was detected by western blotting at 36 hpt. Relative level of PKR was indicated in folds below the images after normalization with β-actin.
3. Discussion
FMDV is one of the most concerned animal virus due to its enormous potential to cause epidemics and economic losses (Kitching, 2005). FMDV infection strongly suppresses host innate antiviral responses by various strategies (Liu et al., 2015). 3Cpro, as an important viral proteinase, is responsible for inducing cleavage of several viral polyprotein and has the ability to antagonize host antiviral effects (Curry et al., 2007). For instance, 3Cpro specifically incises host translation initiation factor eIF4G, resulting in shut-off of host proteins synthesis (Belsham et al., 2000). 3Cpro also induces the nuclear factor kappa B (NF-κB) essential modulator (NEMO) and karyopherin α1 cleavage to impair innate antiviral response (Du et al., 2014b; Wang et al., 2012a). PKR, as a viral RNA sensor, plays important roles in host antiviral response (Balachandran et al., 2000; Dabo and Meurs, 2012). It has previously been shown that incubation of a PKR inhibitor, 2-AP, significantly increases the yield of FMDV in PK and EBK cells, suggesting an important antiviral role of PKR against FMDV (Chinsangaram et al., 2001). Our data here demonstrate that overexpression of PKR also suppresses FMDV, and FMDV infection degrades PKR to abate the antiviral action of PKR. This indicates that PKR functions as an antiviral factor during FMDV infection, and the virus has developed strategies to antagonize PKR-mediated antiviral action.
The importance of PKR in antiviral defense has been supported by various viruses, and PKR regulates diverse cellular pathways to perform its antiviral action (Balachandran et al., 2000, Garcia et al., 2006, Garcia et al., 2007, Nakamura et al., 2010). Meanwhile, most viruses have developed mechanisms to block or impair PKR-induced antiviral response by using delicate strategies; the well-known strategies include expression of dsRNA-binding proteins or small RNAs which compete with PKR for its ligand, interaction with PKR by viral proteins, expression of a decoy substrate, or dephosphorylation of eIF2α (Garcia et al., 2007, Habjan et al., 2009, Langland et al., 2006, Lussignol et al., 2013, Xiao et al., 2016). For example, the accessory protein 4a of Middle East respiratory coronavirus prevents PKR activation by sequestering its ligand, dsRNA (Rabouw et al., 2016). NS5A protein of HCV directly interacts with PKR to inhibit PKR dimerization and activation (Tan et al., 1998). To our knowledge, rift valley fever virus (RVFV), Mengovirus and poliovirus are the only identified viruses that possess the activity to degrade PKR. NSs protein of RVFV specifically induces PKR degradation via the proteasome pathway (Habjan et al., 2009); the responsible viral factor for Mengovirus- and poliovirus-induced PKR degradation has not been clarified (Black et al., 1989, Feng et al., 2012). We report here that FMDV infection induces PKR degradation and 3Cpro is the antagonistic factor responsible for this degradation. 3Cpro-induced cleavage of eIF4G blocks the expression of many host proteins (Belsham et al., 2000). However, we found that 3Cpro specifically reduces PKR protein levels after inhibition of cellular translational function, indicating that 3Cpro-induced reduction of PKR protein levels is not totally related to the well-characterized blocking effect of 3Cpro on cellular translation. Lpro also induces the cleavage of host eIF4G to inhibit host protein synthesis (Kirchweger et al., 1994); however, we observed that overexpression of Lpro had no detectable effect on PKR expression (Fig. 4A); it was similar to the observation in the cells treated with a translational inhibitor CHX (Fig. 5C), and the long half-life of PKR may result in this minimum change. This also explains why Lpro does not degrade PKR. The co-transfection of the plasmids expressing other FMDV proteins with the PKR expressing plasmids showed no significant affection on PKR protein levels. These results suggest that 3Cpro performs an important role in induction of PKR decrease in FMDV-infected cells.
Mengovirus infection induces significant decrease of total PKR protein levels (Dubois and Hovanessian, 1990). A recombinant mengovirus virus (mengo-Zn) which includes mutations in the Zn finger domain of the leader protein causes the decrease of total PKR levels similar as the wildtype Mengovirus in the virus-infected cells. However, the level of PKR phosphorylation in the mengo-Zn-infected cells is strongly induced even with reduced PKR levels (Rabouw et al., 2016). Poliovirus infection also triggers PKR phosphorylation, even though PKR is significantly degraded (Black et al., 1989). These indicate that the PKR phosphorylation status and PKR abundance are not always correlated with each other. Our data reveal that both FMDV infection and transfection of 3Cpro clearly suppress the PKR phosphorylation induced by poly(I:C). The state of PKR phosphorylation and PKR levels are similar during FMDV infection or transfection of 3Cpro. Therefore, we deem that the degradation of PKR may contribute to the decreased PKR phosphorylation in FMDV-infected cells.
Both Lpro and 3Cpro possess the proteinase activity, we observed that PKR could not be cleaved during FMDV infection or transfection of Lpro or 3Cpro (Fig. 1C and Fig. 4). It indicates that PKR degradation by FMDV 3Cpro occurs in a specific manner and not by the proteinase activity. The mutation of the catalytic sites of 3Cpro further confirmed this characteristic. No direct interaction was identified between PKR and 3Cpro, the lysosomal pathway was determined to be involved in 3Cpro-induced PKR reduction. Many RNA viruses induce degradation of host cell proteins to facilitate virus replication. Such as, in influenza A virus infection, the viral nonstructural protein NS1 induces lysosomal degradation of eukaryotic translation initiation factor 4B to promote virus replication (Wang et al., 2014); HIV Nef protein promotes lysosomal targeting of host CD4 receptor to facilitate release of newly produced viral particles (Amorim et al., 2014). How 3Cpro induces PKR degradation in the lysosomes without a direct interaction remains unknown. Experiments to further characterize the degradation mechanism are currently under way.
In conclusion, our study demonstrated the significant antiviral role of PKR during FMDV infection. It also showed a novel mechanism by which FMDV 3Cpro has evolved to induce a degradation of PKR by lysosomal pathway and counteract PKR-induced antiviral effect.
4. Materials and methods
4.1. Antibodies
The commercial antibodies against PKR, Flag and β-Actin were purchased from Sigma-Aldrich. Rabbit anti-phospho-PKR (Thr451) antibody was purchased from Millipore Corporation (Merck Millipore). Mouse anti-eIF2α monoclonal antibody and rabbit anti-phospho-eIF2α (Ser51) (D9G8) XP were purchased from Cell Signaling Technology (CST). Mouse anti-myc monoclonal antibody and HRP-conjugated goat anti-rabbit IgG antibody, as well as HRP-conjugated goat anti-mouse IgG antibody were purchased from Santa Cruz Biotechnology. An anti-VP1 polyclonal antibody was generated in our laboratory (unpublished data).
4.2. Viruses and cells
Porcine kidney (PK-15) cells and human embryonic kidney 293 T (HEK293T) cells were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The cells were maintained in Dulbecco's Modified Eagle Media (DMEM, Invitrogen), supplemented with 10% heated-inactivated fetal bovine serum (FBS) and then cultured at 37 °C in a humidified 5% CO2 incubator. FMDV strain O/BY/CHA/2010 (GenBank number: JN998085) was conserved by National Foot and Mouth Diseases Reference Laboratory, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences and used for virus infection in this study.
4.3. Plasmid constructions and transfection
A Myc-tagged PKR eukaryotic expression construct was generated through inserting full-length porcine PKR cDNA fragment into a pcDNATM3.1/myc-His A vector. Various pCAGGs plasmids expressing Flag-tagged viral protein were constructed from the cDNA of FMDV strain O/BY/CHA/2010 by standard reverse transcription (RT)-PCR by our lab previously as described (Li et al., 2016a). Specific mutations (H46Y, D84N, C163G, and H205R) were introduced to pCAGGs-3C by the site-directed mutagenesis PCR as previously described (Zhu et al., 2013). All the plasmids were verified by DNA sequencing analysis. The plasmids were transfected into the cells using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacture's protocol.
4.4. Viral infection
The viral infection experiment was carried out as previously described (Li et al., 2016b). Briefly, the cell cultures were washed with PBS for 3 times, and the virus was incubated for 1 h (h) in serum-free medium at the multiplicity of infection (MOI) of 0.5. The infected cells were then washed with PBS, and the supernatant was removed and then maintained in fresh medium supplemented with 1% FBS. The mock infection was carried out as the viral infection method using the serum-free medium instead of the virus for incubation. The samples were collected at different hours post-infection (hpi).
4.5. RNA extraction, reverse transcription, and real-time PCR
Total RNAs were isolated from the cells using the TRIzol® Reagent (Invitrogen) following the manufacturer's protocol. Reverse transcription (RT)-PCR was conducted to synthesize cDNAs. M-MLV reverse transcriptase (Promega) and random hexamer primers (TaKaRa) was used for cDNA amplification in RT-PCR. The generated cDNAs were used for quantitative real-time PCR (qPCR) assay. The cDNAs were quantified using SYBR Premix Ex Taq reagents (TaKaRa) and the Mx3005 P QPCR System (Agilent Technologies). Detection of house keeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also conducted for normalization of total input RNA. Relative transcript levels were calculated by 2−ΔΔCT (where CT is threshold cycle) method as previously described (Zhu et al., 2016). The primers used in this study were shown in Table 1.
Table 1.
The primers used in this study.
| Gene | Primers (5′→3′) | Purpose |
|---|---|---|
| PKR | Forward: GGAAGAAAACAAACACAGCTTGAA | qPCR |
| Reverse: CCAAATCCACCTGAGCCAATT | ||
| GAPDH | Forward: ACATGGCCTCCAAGGAGTAAGA | qPCR |
| Reverse: GATCGAGTTGGGGCTGTGACT | ||
| FMDV | Forward: CACTGGTGACAGGCTAAGG | qPCR |
| Reverse: CCCTTCTCAGATTCCGAGT | ||
| PKR | Forward: ATCTCGAGATGGCCAGTGGTCGTTCACCGTGTT | CDS amplification |
| Reverse: CGTGGTACCACATGTGTTTCGTTTCTTTTTCTCTGTGACG | ||
| 3C-H46Y | Forward: TACCTCGTGCCTCGTTATCTCTTCGCAGAGAAG | 3C mutagenesis |
| Reverse: CTTCTCTGCGAAGAGATAACGAGGCACGAGGTA | ||
| 3C-D84N3C-D84N | Forward: CAGGACATGCTCTCAAACGCCGCGCTCATGGTG | 3C mutagenesis |
| Reverse: CACCATGAGCGCGGCGTTTGAGAGCATGTCCTG | ||
| 3C-C163G | Forward: ACCAAGGCTGGCTACGGTGGAGGAGCCGTTCTC | 3C mutagenesis |
| Reverse: GAGAACGGCTCCTCCACCGTAGCCAGCCTTGGT | ||
| 3C-H205R | Forward: CTTAAAATGAAGGCACGCATTGACCCCGAACCA | 3C mutagenesis |
| Reverse: TGGTTCGGGGTCAATGCGTGCCTTCATTTTAAG |
4.6. Western blotting
The collected cell samples were lysed in the lysis buffer supplemented with phenylmethylsulfonyl fluoride (PMSF; Sigma). The protein-containing supernatants were analyzed by 10% sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto Immobilon-P membranes (Millipore). 5% skim milk in TBST was used to block the membranes. The membranes were incubated and probed with appropriate antibodies to generate antibody–antigen complexes. The generated complexes were visualized by enhanced chemiluminescence detection reagents (Thermo). Beta-actin was determined as a protein loading control.
4.7. Chemical inhibitors assay
The proteasome inhibitor MG132, the lysosome inhibitor chloroquine diphosphate (CQ) and NH4Cl, and caspase inhibitor benzyloxycarbony (Cbz)-l-Val-Ala-Asp (OMe)-fluoromethylketone (Z-VAD-FMK) were used to suppress various pathways involved in the protein degradation process. Monolayer PK-15 cells were mock-infected or infected with FMDV (0.5 MOI) in the absence or presence of CQ (50 μM or 100 μM), NH4Cl (10 mM or 20 mM), Z-VAD-FMK (10 μM or 50 μM) or MG132 (2 μM or 20 μM); the cells were collected at 12 hpi, and then subjected to western blotting analysis. As for transfection of Flag-3C expressing plasmids, the monolayer cells were transfected with 2 μg of Flag-3C plasmids or empty vector using Lipofectamine 3000 and maintained in the absence or presence of CQ (50 μM or 100 μM), NH4Cl (10 mM or 20 mM), Z-VAD-FMK (10 μM or 50 μM) or MG132 (2 μM or 20 μM) for 36 h. The expression levels of PKR and 3Cpro were then detected by western blotting. The cycloheximide (CHX) was used to block protein synthesis of cells and evaluate the half-life of PKR protein. The PK-15 cells were cultured in the presence or absence of 100 μg/mL of CHX for the indicated periods at 37 °C (Wang et al., 2012b). The collected cells were subjected to western blotting analysis.
4.8. Statistical analysis
All the experiments were repeated at least three times. The results were present as mean values ± standard error (mean ± SE) of three independent experiments. The Student's t-test was used to analyze the statistically significance (two- tailed Student's t-test). Statistical values of *P<0.05 was considered significant, and **P<0.01 was considered highly significant.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by grants from the Project Supported by National Science and Technology Ministry (2015BAD12B04), the Gansu Science Foundation for Distinguished Young Scholars (no. 1606RJDA313), and the Gansu Science Foundation for Young Scholars (1606RJYA280).
Contributor Information
Zixiang Zhu, Email: zhuzixiang@caas.cn.
Haixue Zheng, Email: haixuezheng@163.com.
References
- Amorim N.A., da Silva E.M.L., de Castro R.O., da Silva-Januario M.E., Mendonca L.M., Bonifacino J.S., da Costa L.J., daSilva L.L.P. Interaction of HIV-1 Nef Protein with the Host Protein Alix Promotes Lysosomal Targeting of CD4 Receptor. J. Biol. Chem. 2014;289:27744–27756. doi: 10.1074/jbc.M114.560193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S., Roberts P.C., Brown L.E., Truong H., Pattnaik A.K., Archer D.R., Barber G.N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- Belsham G.J., McInerney G.M., Ross-Smith N. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J. Virol. 2000;74:272–280. doi: 10.1128/jvi.74.1.272-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann M., Garcia-Sastre A., Carnero E., Pehamberger H., Wolff K., Palese P., Muster T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtley J.R., Knox S.R., Jaulent A.M., Brick P., Leatherbarrow R.J., Curry S. Crystal structure of foot-and-mouth disease virus 3C protease. J. Biol. Chem. 2005;280:11520–11527. doi: 10.1074/jbc.M413254200. [DOI] [PubMed] [Google Scholar]
- Black T.L., Safer B., Hovanessian A., Katze M.G. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J. Virol. 1989;63:2244–2251. doi: 10.1128/jvi.63.5.2244-2251.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsangaram J., Koster M., Grubman M.J. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J. Virol. 2001;75:5498–5503. doi: 10.1128/JVI.75.12.5498-5503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry S., Roqué-Rosell N., Zunszain P.A., Leatherbarrow R.J. Foot-and-mouth disease virus 3C protease: recent structural and functional insights into an antiviral target. Int. J. Biochem. Cell Biol. 2007;39:1–6. doi: 10.1016/j.biocel.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabo S., Meurs E.F. dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection. Virus.-Basel. 2012;4:2598–2635. doi: 10.3390/v4112598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Bi J., Liu J., Liu X., Wu X., Jiang P., Yoo D., Zhang Y., Wu J., Wan R., Zhao X., Guo L., Sun W., Cong X., Chen L., Wang J. 3Cpro of foot-and-mouth disease virus antagonizes the interferon signaling pathway by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2014;88:4908–4920. doi: 10.1128/JVI.03668-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y.J., Bi J.S., Liu J.Y., Liu X., Wu X.J., Jiang P., Yoo D.W., Zhang Y.G., Wu J.Q., Wan R.Z., Zhao X.M., Guo L.H., Sun W.B., Cong X.Y., Chen L., Wang J.B. 3C(pro) of Foot-and-Mouth Disease Virus Antagonizes the Interferon Signaling Pathway by Blocking STAT1/STAT2 Nuclear Translocation. J. Virol. 2014;88:4908–4920. doi: 10.1128/JVI.03668-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M.F., Hovanessian A.G. Modified subcellular localization of interferon-induced p68 kinase during encephalomyocarditis virus infection. Virology. 1990;179:591–598. doi: 10.1016/0042-6822(90)90126-c. [DOI] [PubMed] [Google Scholar]
- Feng Q., Hato S.V., Langereis M.A., Zoll J., Virgen-Slane R., Peisley A., Hur S., Semler B.L., van Rij R.P., van Kuppeveld F.J.M. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol. Biol. Rev. 2006;70(4):1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Meurs E.F., Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Grubman M.J., Baxt B. Foot-and-mouth disease. Clin. Microbiol. Rev. 2004;17:465–493. doi: 10.1128/CMR.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubman M.J., Zellner M., Bablanian G., Mason P.W., Piccone M.E. Identification of the active-site residues of the 3c proteinase of foot-and-mouth-disease virus. Virology. 1995;213:581–589. doi: 10.1006/viro.1995.0030. [DOI] [PubMed] [Google Scholar]
- Habjan M., Pichlmair A., Elliott R.M., Överby A.K., Glatter T., Gstaiger M., Superti-Furga G., Unger H., Weber F. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 2009;83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal S.M., Belsham G.J. Foot-and-mouth disease: past, present and future. Vet. Res. 2013;44:116. doi: 10.1186/1297-9716-44-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchweger R., Ziegler E., Lamphear B.J., Waters D., Liebig H.D., Sommergruber W., Sobrino F., Hohenadl C., Blaas D., Rhoads R.E. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eif-4 gamma. J. Virol. 1994;68:5677–5684. doi: 10.1128/jvi.68.9.5677-5684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitching R.P. Global epidemiology and prospects for control of foot-and-mouth disease. Curr. Top. Microbiol. Immunol. 2005;288:133–148. doi: 10.1007/3-540-27109-0_6. [DOI] [PubMed] [Google Scholar]
- Langland J.O., Cameron J.M., Heck M.C., Jancovich J.K., Jacobs B.L. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Lawrence P., Schafer E.A., Rieder E. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology. 2012;425:40–52. doi: 10.1016/j.virol.2011.12.019. [DOI] [PubMed] [Google Scholar]
- Li D., Yang W., Yang F., Liu H., Zhu Z., Lian K., Lei C., Li S., Liu X., Zheng H. The VP3 structural protein of foot-and-mouth disease virus inhibits the IFN-β signaling pathway. FASEB J. 2016 doi: 10.1096/fj.15-281410. (fj. 15-281410) [DOI] [PubMed] [Google Scholar]
- Li S., Min J.-Y., Krug R.M., Sen G.C. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Li W., Zhu Z., Cao W., Yang F., Zhang X., Li D., Zhang K., Li P., Mao R., Liu X., Zheng H. Esterase D enhances type I interferon signal transduction to suppress foot-and-mouth disease virus replication. Mol. Immunol. 2016;75:112–121. doi: 10.1016/j.molimm.2016.05.016. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhu Z., Zhang M., Zheng H. Multifunctional roles of leader protein of foot-and-mouth disease viruses in suppressing host antiviral responses. Vet. Res. 2015;46:127. doi: 10.1186/s13567-015-0273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Wambach M., Katze M.G., Krug R.M. Binding of the Influenza-Virus Ns1 Protein to Double-Stranded-Rna Inhibits the Activation of the Protein-Kinase That Phosphorylates the Elf-2 Translation Initiation-Factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- Lussignol M., Queval C., Bernet-Camard M.F., Cotte-Laffitte J., Beau I., Codogno P., Esclatine A. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J. Virol. 2013;87:859–871. doi: 10.1128/JVI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Furuhashi M., Li P., Cao H.M., Tuncman G., Sonenberg N., Gorgun C.Z., Hotamisligil G.S. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–U341. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla S.R., Toroney R., Bevilacqua P.C. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr. Opin. Struct. Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouw H.H., Langereis M.A., Knaap R.C., Dalebout T.J., Canton J., Sola I., Enjuanes L., Bredenbeek P.J., Kikkert M., de Groot R.J., van Kuppeveld F.J. Middle east respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog. 2016;12:e1005982. doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger J., Grishkovskaya I., Cencic R., Juliano L., Juliano M.A., Skern T. Foot-and-mouth disease virus leader proteinase: structural insights into the mechanism of intermolecular cleavage. Virology. 2014;468–470:397–408. doi: 10.1016/j.virol.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger J., Skern T. The leader proteinase of foot-and-mouth disease virus: structure-function relationships in a proteolytic virulence factor. Biol. Chem. 2014;395:1179–1185. doi: 10.1515/hsz-2014-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenfeldt C., Diaz-San Segundo F., de Los Santos T., Rodriguez L.L., Arzt J. The pathogenesis of foot-and-mouth disease in pigs. Front. Vet. Sci. 2016;3:41. doi: 10.3389/fvets.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T.R., Roque-Rosell N., Birtley J.R., Leatherbarrow R.J., Curry S. Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J. Virol. 2007;81:115–124. doi: 10.1128/JVI.01587-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.-L., Gale M.J., Katze M.G. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol. Cell. Biol. 1998;18:2431–2443. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas J., Elia A., Clemens M.J. Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA. Rna-a Publ. Rna Soc. 2003;9:858–870. doi: 10.1261/rna.5330503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Li K., Zhong H., Fan J., Ouyang C., Zhang H., Duan E., Luo R., Zhang Z., Liu X., Chen H., Xiao S. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J. Virol. 2012;86:9311–9322. doi: 10.1128/JVI.00722-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chi X.J., Wei H.T., Chen Y.H., Chen Z.L., Huang S.L., Chen J.L. Influenza A Virus-Induced Degradation of Eukaryotic Translation Initiation Factor 4B Contributes to Viral Replication by Suppressing IFITM3 Protein Expression. J. Virol. 2014;88:8375–8385. doi: 10.1128/JVI.00126-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.D., Deng X.F., Yan W.J., Zhu Z.X., Shen Y., Qiu Y.F., Shi Z.X., Shao D.H., Wei J.C., Xia X.Z., Ma Z.Y. Stabilization of p53 in Influenza A Virus-infected Cells Is Associated with Compromised MDM2-mediated Ubiquitination of p53. J. Biol. Chem. 2012;287:18366–18375. doi: 10.1074/jbc.M111.335422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.Q., Ma Z.X., Wang R., Yang L.P., Nan Y.C., Zhang Y.J. Downregulation of protein kinase PKR activation by porcine reproductive and respiratory syndrome virus at its early stage infection. Vet. Microbiol. 2016;187:1–7. doi: 10.1016/j.vetmic.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Wang G., Yang F., Cao W., Mao R., Du X., Zhang X., Li C., Li D., Zhang K. Foot-and-mouth disease virus viroporin 2B antagonizes RIG-I mediated antiviral effects by inhibition of its protein expression. J. Virol., JVI. 2016:01310–01316. doi: 10.1128/JVI.01310-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.X., Shi Z.X., Yan W.J., Wei J.C., Shao D.H., Deng X.F., Wang S.H., Li B.B., Tong G.Z., Ma Z.Y. Nonstructural Protein 1 of Influenza A Virus Interacts with Human Guanylate-Binding Protein 1 to Antagonize Antiviral Activity. PloS One. 2013;8(2):e0055920. doi: 10.1371/journal.pone.0055920. [DOI] [PMC free article] [PubMed] [Google Scholar]