Abstract
Human adenovirus type 14 (HAdV-B14) was first reported in 1955 from the Netherlands and since then had been associated with outbreaks of febrile respiratory illness (FRI). In China, sporadic HAdV-B14 infections were first identified in 2010, in Guangzhou and Beijing. In 2012, an outbreak of FRI occurred in Beijing and the etiological agent was determined to be HAdV-B14. We present a complete HAdV-B14 genome sequence isolated from this recent FRI outbreak. Virus in 30 throat swab samples was detected using polymerase chain reaction assays, and confirmed by sequencing of the fiber, hexon and penton genes. Comparative genomics and phylogenetic analysis showed that the newly isolated HAdV-B14 (HAdV-B14 CHN) shared highest sequence homology with a 2006 isolate from the United States and clustered closely with other HAdV-B14 strains. It is expected that data from the present study will help in devising better protocols for virus surveillance, and in developing preventative measures.
Keywords: Human adenovirus, Respiratory disease, Genome analysis, Bioinformatics
Highlights
-
•
Isolation of HAdV-B14 was performed from the outbreak of febrile respiratory illness.
-
•
Full genome sequence of HAdV-B14 CHN strain has been reported.
-
•
Several nucleotide substitutions were reported in the HAdV-B14 CHN genome.
1. Introduction
Adenoviruses (AdVs) are non-enveloped viruses with double-stranded, linear DNA genomes ranging from 26 to 45 kb. Human adenoviruses (HAdVs) belong to the genus Mastadenovirus, within the Adenoviridae family. Currently, there are seven HAdV species (A–G) with more than 65 serotypes. The various HAdV species are the etiological agents of several types of respiratory, ocular, and gastrointestinal diseases [1]. HAdV-C1, -C2, -C5, -B3, and -B7 cause upper respiratory tract infections, whereas HAdV-B3, -B7, -B21, and -E4 are associated with more severe infections of the lower respiratory tract [7], [8], [9].
HAdV-B14 was first isolated in 1955 from military recruits presenting with an acute respiratory disease in the Netherlands [2]. This particular AdV was also found to be associated with pharyngoconjunctival fever during outbreaks at schools in England in 1957 [3]. HAdV-B14 has occasionally been associated with febrile respiratory illness (FRI) in Eurasia, but has not resulted in fatalities [4], [5]. However, in 2006, an epidemic of HAdV-B14 infections suddenly re-emerged at a military camp in North America. This resulted in high incidences of FRI, and in some cases severe pneumonia and fatalities [6].
Large-scale epidemiological data related to HAdV infections is currently not available from China, however it is known that HAdVs are circulating among the population. Outbreaks associated with HAdV-3, -7, and -11 have been previously reported in China [7], with the first HAdV-B14 infection, designated GZ01 (Accession ID: JQ824845), reported in 2010 from Guangdong Province [8]. In 2011, another strain of HAdV-B14, designated BJ430 (Accession ID: JN032132), was isolated from a 6-month-old baby in Beijing [9]. In this study we aimed to sequence and characterize the entire genome of HAdV-B14 isolated from a 2012 FRI outbreak in Beijing.
2. Results and discussion
2.1. Genome features of HAdV-B14 CHN
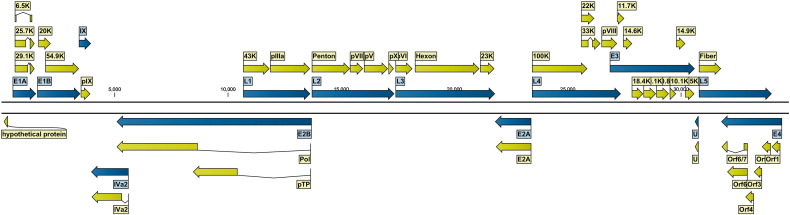
The isolate we sequenced was designated HAdV-B14 CHN and the genome found to have 34,760 bp. The CHN isolate had a G + C content of 48.8%, with base compositions of 26, 25, 24.4, and 24.4% for A, T, G and C, respectively. The G + C content was consistent with that of other type B HAdVs. The HAdV-B14 CHN genome organization and annotation details are graphically shown in Fig. 1 and described in Table 2. Detailed homology, and comparative sequence analysis of HAdV-B14 CHN genes and proteins were conducted with HAdV-B14 de Wit used as the reference strain.
Fig. 1.
Genome organization of HAdV-B14 CHN strain.
The genome is represented by two black horizontal lines marked at 5000 bp intervals. Protein encoding regions are shown as arrows indicating transcriptional orientation. Forward arrows (above the horizontal black line) show coding regions in the 5′ to 3′ direction and arrows pointing to the left (below the horizontal black line) show the coding regions encoded on the complementary strand. Genes are colored in blue and coding sequences are colored in light yellow.
Table 2.
Genome organization and coding sequences annotation from the HAdV-B14 CHN strain.
| Region | Nucleotide locality | Gene product | Product length | TATA box | ATG site | Stop site | Poly (A) signal |
|---|---|---|---|---|---|---|---|
| ITR | 1–137 | ||||||
| E1A | 587–1166, 1251–1459 | 29.1 kDa | 262 | 494–501 | 587 | 1457 | 1518–1523 |
| E1A | 587–1073, 1251–1459 | 25.7 kDa | 231 | 494–501 | 587 | 1457 | 1518–1523 |
| E1A | 587–658, 1251–1355 | 6.5 kDa | 58 | 494–501 | 587 | 1353–1355 | 1518–1523 |
| E1B | 1629–2171 | 20 kDa | 180 | 1575–1581 | 1629 | 2169–2171 | 3471–3476 |
| E1B | 1934–3418 | 54.9 kDa | 494 | 1575–1581 | 1934 | 3416–3418 | 3471–3476 |
| IVa2 | 3985c–5031c | IVa2 protein | 348 | ND | 5607c | 3985c–3987c | 3963c–3968c |
| E2A | 21833c–23389c | DNA binding protein | 518 | ND | 23387c | 21833c | 21791c–21796c |
| E2B | 5088c–8660c, 13643c–13651c | DNA polymerase | 1122 | 13693c–13698c | 13649c | 5088c | ND |
| E2B | 8459–10420, 13643–13651 | pTP | 656 | 13693c–13698c | ND | ND | ND |
| IX | 3498–3917 | 14.2 kDa | 139 | 3416–3421 | 3501 | 3918 | 3942–3947 |
| E4 | 31801c–32052c | ORF6/7 | 141 | 34452c–34459c | 32940c–32942c | 31795c–31797c | 31778c–31783c |
| E4 | 32043c–32942c | ORF6; 34 kDa | 299 | 34452c–34459c | 32940c–32942c | 32043c–32045c | 31778c–31783c |
| E4 | 32845c–33213c | ORF4; 13 kDa | 122 | 34452c–34459c | 33211c–33213c | 32845c–32847c | 31778c–31783c |
| E4 | 33222c–33575c | ORF3; 13.5 kDa | 117 | 34452c–34459c | 33573c–33575c | 33222c–33224c | 31778c–31783c |
| E4 | 33572c–33961c | ORF2; 14.3 kDa | 129 | 34452c–34459c | 33959c–33961c | 33572c–33574c | 31778c–31783c |
| E4 | 34004c–34381c | ORF1; 14.2 kDa | 125 | 34452c–34459c | 34379c–34381c | 34004c–34006c | 31778c–31783c |
| L1 | 10668–11828 | 43.9 kDa | 386 | ND | 10668–10670 | 11826–11828 | 13624–13629 |
| L1 | 11854–13617 | pIIIa; 65.6 kDa | 587 | ND | 11854–11856 | 13615–13617 | 13624–13629 |
| L2 | 13699–15375 | Penton | 558 | ND | 13699–13701 | 15373–15375 | 17335–17340 |
| L2 | 15380–15958 | pVII | 192 | ND | 15380–15382 | 15956–15958 | 17335–17340 |
| L2 | 16001–17056 | pV | 351 | ND | 16001–16003 | 17054–17056 | 17335–17340 |
| L2 | 17085–17315 | pX | 76 | ND | 17085–17087 | 17313–17315 | 17335–17340 |
| L3 | 17396–18136 | pVI | 246 | ND | 17396–17398 | 18134–18136 | 21779–21784 |
| L3 | 18252–21089 | Hexon | 945 | ND | 18252–18254 | 21087–21089 | 21779–21784 |
| L3 | 21126–21755 | 23 kDa | 209 | ND | 21126–21128 | 21753–21755 | 21779–21784 |
| L4 | 23420–25858 | 100 kDa | 812 | ND | 23420–23422 | 25856–25858 | 27333–27338 |
| L4 | 25590–26165 | 22 kDa | 191 | ND | 25590–25592 | 26163–26165 | 27333–27338 |
| L4 | 26047–26439 | 33 kDa | 130 | ND | 25590–25592 | 26437–26439 | 27333–27338 |
| L4 | 26489–27172 | pVIII | 227 | ND | 26489–26491 | 27170–27172 | 27333–27338 |
| E3 | 27172–27489 | 11.7 kDa | 105 | 26854–26859 | 27172–27174 | 27487–27489 | 30596–30601 |
| E3 | 27443–27838 | 14.6 kDa | 131 | 26854–26859 | 27443–27445 | 27836–27838 | 30596–30601 |
| E3 | 27823–28323 | 18.4 kDa | 166 | 26854–26859 | 27823–27825 | 28321–28323 | 30596–30601 |
| E3 | 28343–28888 | 20.1 kDa | 181 | 26854–26859 | 28343–28345 | 28886–28888 | 30596–30601 |
| E3 | 28906–29457 | 20.8 kDa | 183 | 26854–26859 | 28906–28908 | 29455–29457 | 30596–30601 |
| E3 | 29501–29776 | 10.1 kDa | 91 | 26854–26859 | 29501–29503 | 29774–29776 | 30596–30601 |
| E3 | 29781–30185 | 14.9 kDa | 134 | 26854–26859 | 29781–29783 | 30183–30185 | 30596–30601 |
| E3 | 30178–30585 | 15 kDa | 135 | 26854–26859 | 30178–30180 | 30583–30585 | 30596–30601 |
| U | 30609c–30733c | U protein | 54 | ND | 30771c | 30609c | ND |
| ND | 102c–281c | Hypothetical protein | 141 | ND | 102c | 281c | ND |
| L5 | 30788–31759 | Fiber | 323 | ND | 30788–30790 | 31757–31759 | 31762–31767 |
| ITR | 34624c–34760c |
Coding sequences with spliced regions are indicated by double entries separated by “,” in the nucleotide locality column. Coding sequences transcribed from the complementary strand are designated by “c”. ND; not determined. p: protein.
2.2. Molecular analysis of HAdV-B14 CHN
2.2.1. Inverted terminal repeats (ITRs)
The ITRs are regions located at the 5′ and 3′ termini of AdV genomes. The ITRs and flanking DNA contain several sites for the binding of viral proteins and cellular factors, possibly serving as origins of replication for AdV DNA via the strand displacement mechanism [10]. The 5′ and 3′ ITRs of HAdV-B14 CHN are 137 bp (Table 2) and similar to those for the de Wit strain. We observed two nucleotide substitutions in the 5′ ITR, (CHN)T68C(de Wit) and (CHN)C134G(de Wit), and one in the 3′ ITR, (CHN)A34702G(de Wit). Similar substitutions in the 5′ ITR have been reported in HAdV-B14 previously isolated from the United States [6]. The functional impact of these substitutions remains unclear. The extreme end of the HAdV-B14 CHN 5′ ITR contains a 10 nt motif (CATCATCAAT), and has been found to be conserved among other HAdV-B prototype strains. Another 10 nt conserved motif (ATAATATACC) within the 5′ ITR of HAdV is directly involved in the interaction of the terminal protein precursor (pTP) and DNA polymerase during viral DNA replication [11], and this was seen in HAdV-B14 CHN. Several cellular transcription factors, including nuclear factor 1 (NF1), nuclear factor III (NFIII), specificity protein 1 (Sp1), and activating transcription factors (ATFs) are known to enhance virus replication, and are important for efficient growth [12], [13]. The binding motifs for these factors are localized within ITRs of AdVs; to identify the binding motifs for these host transcription factors, we generated multiple ITR sequence alignments using various HAdV-B species (Table 1 ). Binding sites for NF1, NFIII, Sp1 and ATFs were identified at nucleotides 26–39 (TGGAATGGTGCCAA), 40–50 (ATGTAAATGA), 108–114 (GGGGCGG), and 131–136 (TGAGGT), respectively. The typical ATF motif (TGACGT) was found to be conserved among all representative sequences, however a nucleotide substitution, (CHN)G134C(de Wit), was noticed in the CHN strain when compared with the de Wit reference sequence.
Table 1.
GenBank accession numbers and details of adenoviruses genomes used in present study.
| S no | Viruses | Serotype | Accession IDs | Host | Collection date (year) | Country |
|---|---|---|---|---|---|---|
| 1 | HAdV-B11 (Slobitski) | B11 | AY163756 | Human | 1956 | USA |
| 2 | HAdV-B14p1 (303600) | B14 | FJ822614 | Human | 2006 | USA |
| 3 | HAdV-B14p1 (BJ430) | B14 | JN032132 | Human | 2011 | China |
| 4 | HAdV-B14 (CHN)# | B14 | JX892927.2 | Human | 2012 | China |
| 5 | HAdV-B14p (de Wit) | B14 | AY803294 | Human | 1955 | Netherlands |
| 6 | HAdV-B14 (GZ01) | B14 | JQ824845 | Human | 2010 | China |
| 7 | HAdV-B16 (ch. 79) | B16 | AY601636 | Human | – | USA |
| 8 | HAdV-B21 (AV-1645) | B21 | AY601633 | Human | – | USA |
| 9 | HAdV-B3 (GB) | B3 | AY599834 | Human | 2004 | USA |
| 10 | HAdV-B3 (Guangzhou01) | B3 | DQ099432 | Human | 2005 | China |
| 11 | HAdV-B3 (Guangzhou02) | B3 | DQ105654 | Human | 2004 | China |
| 12 | HAdV-B3 (NHRC 1276) | B3 | AY599836 | Human | 1997 | USA |
| 13 | HAdV-B34 (Compton) | B34 | AY737797 | Human | 1972 | USA |
| 14 | HAdV-B35 (Holden) | B35 | AY128640 | Human | 1973 | USA |
| 15 | HAdV-B50 (Wan) | B50 | AY737798 | Human | – | USA |
| 16 | HAdV-B7 (Gomen) | B7 | AY594255 | Human | 1954 | USA |
| 17 | HAdV-B7 (NHRC 1315) | B7 | AY601634 | Human | 1997 | USA |
| 18 | HAdV-B7 (0901 HZ) | B7 | JF800905 | Human | 2009 | China |
| 19 | HAdV-B55 (QS-DLL) | B55 | FJ643676 | Human | 2006 | China |
| 20 | SAdV-21 | B21 | AC_000010 | Chimpanzee | – | – |
| 21 | HAdV-D9 | D9 | AJ854486 | Human | – | – |
| 22 | HAdV-E4 | E4 | AY594253 | Human | – | – |
| 23 | HAdV-G52 | G52 | DQ923122 | Human | – | – |
| 24 | HAdV-C1 | C1 | AF534906 | Human | – | – |
| 25 | HAdV-F40 | F40 | NC_001454 | Human | – | – |
| 26 | HAdV-A12 | A12 | AC_000005 | Human | – | – |
Dashes (–) indicate data not available. HAdV-B14 isolate sequenced in the present study is indicated by (#).
2.2.2. Virus-associated RNAs
Virus-associated RNAs (VA RNAs) in AdVs are short, non-coding RNAs transcribed by RNA polymerase III to form double-stranded (ds) RNA-like secondary structures [14], [15]. The number of VA RNA genes varies among different AdV species. HAdV-B7, HAdV-B3 and B1 sub-species contain two VA RNA genes [16], whereas B2 sub-species, such as HAdV-B14 and -B11, contain only one VA RNA gene [17]. We identified one VA RNA gene (161 bp), located between nucleotides 10452–10613 of the HAdV-B14 CHN genome. The VA RNA gene in the HAdV-B14 CHN genome exhibited 100% identity to that in HAdV-B14 de Wit.
2.2.3. Early genes
Early transcription units in HAdVs comprised the following components: E1 (E1A and E1B), E2 (E2A and E2B), E3, and E4. E1 and E4 are located at the ends of HAdV genomes and are the first portions of the genome transcribed during infection. This is followed by transcription of the delayed early units (IX, Iva2, and E2 late) and the major late transcriptional units [18].
2.2.3.1. E1A
E1A is the first gene expressed after adenoviral infection. Alternative splicing of the E1A RNA precursor leads to the formation of multiple E1A transcripts. These play important roles as transcriptional regulators within the host cell, modulating the expression of viral and cellular genes [19], [20], [21]. For HAdV-B14 CHN, three open reading frames (ORFs) were identified within the E1A region coding for proteins of 262, 231, and 58 amino acids. These had corresponding molecular weights of 29.1, 25.7 and 6.5 kDa, respectively (Table 2 ). The putative TATA box and polyadenylation (poly (A)) signal were identified at nucleotide positions 494 and 1518, respectively. Homology analysis of these three proteins showed that they had 100% identity to HAdV-B14 proteins isolated from Ireland in 2009 (Dublin strain), and from China in 2010 (BJ430 strain). Compared with the same proteins in the HAdV-B14 de Wit strain, there was 98% identity. The Rb protein binding motif (LHCYE) was identified in the 29.1 kDa and 25.7 kDa proteins at amino acids 115–119. A C-terminal binding protein (CtBP) interacting motif (PLDLS), was also identified near the C-terminus of the 29.1 kDa and 25.7 kDa proteins at amino acids 251–255 and 220–224, respectively.
2.2.3.2. E1B
E1B proteins facilitate replication of viral DNA by blocking the host apoptosis machinery. Two ORFs were identified in the E1B transcription unit of HAdV-B14 CHN, encoding proteins of 180 and 494 amino acids with corresponding molecular weights of 20 and 55 kDa, respectively. The TATA box and poly (A) signal were identified at nucleotides 1575 and 3471, respectively (Table 2). The 20 kDa protein showed high homology to the small t-antigen, an anti-apoptotic protein that blocks the mitochondrial apoptosis pathway by inactivating BAK and BAX [22]. The 55 kDa E1B protein showed homology to the large t-antigen, whose function is to inhibit cellular p53-mediated host defense mechanisms. p53 is a tumor suppressor protein, and mediates antiviral host cell responses by initiating cell cycle arrest upon infection. The 55 kDa protein directly binds to p53 and represses its function [20], [21]. An E3-ligase complex comprising the large t-antigen, the E4 ORF6 protein, and certain cellular cofactors degrades the p53 protein [33], [34]. A BC-Box binding motif, 177ALRPDKQYKI186, known to be essential for the stability of this complex, was also identified in the 55 kDa protein. A third protein from the E1B transcription unit has sometimes been reported in HAdVs, especially B- and C-types. However, the ORF encoding this protein was not seen in the HAdV-B14 CHN genome.
2.2.4. E2
The E2A and E2B transcription units encode three viral proteins: DNA binding protein (DBP); pTP; and DNA polymerase (DNA pol). These are all required for viral DNA replication, and are located on the complementary strand of the genome. The ORF for DBP is located within the E2A segment, while those for pTP and DNA pol are in the E2B segment (Table 2). For HAdV-B14 CHN, the DBP is 518 amino acids (58.2 kDa), while pTP and DNA pol were 656 (75 kDa) and 1193 (136 kDa) amino acids, respectively. The poly (A) signal for these transcripts was located at nucleotide position 21791c (Table 2). Homology analysis of HAdV-B14 CHN DBP, pTP and DNA pol showed 100, 99, and 99% identity respectively with those in HAdV-B14 de Wit. The DBP had two zinc-binding domains at amino acids 252–367 and 381–508. A bipartite nuclear localization sequence (NLS; 44PPKRN48 and 86PPKKKP91) that could facilitate entry of DBP into the nucleus was identified at the N-terminal domain. This NLS was identical to that previously reported in the DBP of HAdV-B11 [23]. For pTP, an NLS of 12 amino acids (367RLPVRRRRRVP378) was identified, along with two AdV protease cleavage sites (172MRGF↓G176 and 333MRGG↓V337) and a putative site (180MHGR-T184). The pTP NLS and cleavage sites were identical to those in HAdV-B11 [23].
2.2.5. E3
The protein encoded by the E3 transcription unit is not essential for viral growth and propagation, but assists in evasion and modulation of the host immune responses to infection [24], [25]. The TATA box was predicted to be at nucleotide position 26854. A single poly (A) signal was identified at nucleotide position 30596. We identified eight ORFs encoding putative proteins of 10.1, 12, 14.6, 14.7, 15.2, 18.6, 20.1, and 20.8 kDa (Table 2). The organization and number of ORFs in the HAdV-B14 CHN E3 transcription unit were found to be similar to those in HAdV-B14 de Wit. The sequence identity between E3 proteins of HAdV-B14 CHN and HAdV-B14 de Wit ranged 99–100%.
2.2.6. E4
In AdVs, the E4 transcription unit is located on the complementary strand and possesses several ORFs that encode proteins which regulate a variety of functions [26]. In this study, six ORFs were identified within the HAdV-B14 CHN E4 unit (Table 2). The TATA box and poly (A) signal were identified at nucleotides 34452c and 31788c, respectively. The E4 ORF1 encoded a 14.2 kDa protein with 125 amino acids and a dUTPase domain. The HAdV-B14 CHN E4 ORF1 had 100% identity with that from a North American HAdV-B14 strain (303600). We also found 99% identity with the E4 ORF1 of the de Wit strain, along with two substitutions; (CHN)S45Y(de Wit) and (CHN)F61L(de Wit). The E4 ORF2 protein was 129 amino acids corresponding to a molecular weight of 14.3 kDa (Table 2). It had 100% identity with the E4 ORF2 (GenBank Accession No. AFH58055) from HAdV-B14 BJ430 (Table 2). In comparison with the de Wit strain, there was 99% identity and a substitution; (CHN)E32Q(de Wit). A 13 kDa protein of 122 amino acids was encoded by E4 ORF4 and had 99% identity with the E4 ORF4 of the de Wit strain despite the presence of a substitution; (CHN)R72K(de Wit). In AdVs, E4 ORF3 and ORF6 proteins enhance the stability of late viral mRNAs and increase their level of export from the nucleus, thereby increasing viral mRNA accumulation in the cytoplasm [26]. Additionally, ORF6 proteins also combine with the E1B 55 kDa protein. This complex then binds to p53 and blocks apoptosis, whereas E4 ORF6/7 has been shown to regulate cellular E2F levels. Putative proteins that were 13.5, 34.7, and 15.9 kDa were encoded by E4 ORF3, ORF6 and ORF6/7, respectively in HAdV-B14 CHN. ORFs 3 and 4 were 100% identical to ORFs 3 and 4 of the de Wit strain, whereas ORF6/7 was 98% identical. We found three substitutions; (CHN)Q70H(de Wit), (CHN)A83D(de Wit), and (CHN)Y96S(de Wit), in HAdV-B14 CHN ORF6/7 when compared with the de Wit strain.
2.2.7. Intermediate genes
Two ORFs encoding intermediate proteins IX and IVa2 were identified. The IX protein (pIX) was composed of 139 amino acids (14.2 kDa) and exhibited 100% sequence identity with HAdV-B14 de Wit pIX. The pIX protein is a minor capsid protein and supports activation of the major late promoter (MLP) [27]. It is also a structural protein, and influences hexon–hexon interactions. The HAdV-B14 CHN strain IVa2 protein consists of 448 amino acids (50 kDa), with its ORF located on the complementary strand of the genome (Fig. 1 and Table 2). The IVa2 protein had 99% sequence identity with the HAdV-B14 de Wit IVa2 protein, and also contained a substitution; (CHN)H161Q(de Wit). Similar to pIX, IVa2 enhances activation of MLP by interacting with the L1 protein during packaging and assembly of viral DNA.
2.2.8. Late genes
The late genes of AdV are predominantly transcribed from the MLP, following initiation of DNA replication [18]. Several regulatory elements of MLP from HAdV-B11 [23], HAdV-B7 [16] and HAdV-C2 [28] have been well characterized. We identified homologs of essential regulatory elements in the HAdV-B14 CHN MLP. These include the inverted CAAT box (5858–5867 bp), upstream element (5878–5886 bp), TATA box (5909–5915 bp), and the MAZ/Sp1 binding site (5899–5908 bp). The initiator element (INR), which includes the transcription start site for the MLP [29], is located at nucleotides 5938–5944. Two downstream elements that recognize IVa2 and enhance transcription from the MLP after the onset of DNA replication were identified at nucleotides 6025–6035 (DE1) and 6040–6055 (DE2a and DE2b). Three tripartite leader (TPL) sequences were also identified in HAdV-B14 CHN: TPL1 (nucleotides 5940–5980); TPL2 (nucleotides 7000–7071); and TPL3 (nucleotides 9514–9600). Based on the location of poly (A) signals, the L5 transcription unit encodes the major adenoviral structural proteins and is further subdivided into regions L1–L5, with each region expressed as a distinct mRNA species.
2.2.8.1. L1
The L1 transcription unit comprises two ORFs encoding the 52/55K and IIIa proteins in different AdV species. For HAdV-B14 CHN, two proteins of 43.9 kDa (386 amino acids) and 65.6 kDa (587 amino acids) were identified, with a common poly (A) signal at nucleotide position 13624 (Table 2). The 43.9 kDa protein was a homolog of the 52/55K protein; it serves as a scaffold during the DNA encapsidation process and facilitates virion assembly [30], [31]. It also interacts with the IVa2 protein to facilitate viral DNA packaging. The second predicted protein within the L1 region IIIa protein homolog; this is a structural hexon-associated protein that extends from the exterior to the interior of the capsid. It is also a phosphoprotein, which is cleaved by the viral protease during virion assembly [32]. The consensus viral protease cleavage motif (LGGRG) was predicted at amino acids 567–571 in the HAdV-B14 CHN IIIa protein.
2.2.8.2. L2
Four ORFs were identified in the HAdV-B14 CHN L2 region. A common poly (A) signal for L2 proteins was situated at nucleotide 17335 (Table 2). The protein encoded by the first ORF was 558 amino acids (66.6 kDa), and designated the penton protein. This is one of the three major capsid proteins in AdVs responsible for facilitating virus internalization via interaction with host integrins. Two conserved motifs [Arg-Gly-Asp (RGD) and Leu-Asp-Val (LDV)] within the penton protein are responsible for this interaction with integrins. The RGD motif interacts with αvβ3 and αvβ5 types of integrins, whereas LDV interacts with α4β1 and α4β7 integrins [33]. For HAdV-B14 CHN, RGD and LDV motifs were identified in the penton protein at amino acids 338–340 and 297–299, respectively. The HAdV-B14 CHN penton was 100% identical to other penton proteins in HAdV-B14 strains previously isolated from North America and with two previously reported B14 strains from China (BJ430 and GZ01). We identified two substitutions, (CHN)S328F(de Wit) and (CHN)N362D(de Wit), in the penton protein when compared with HAdV-B14 de Wit. A conserved fiber-interacting domain (ESRLSNLLGIRKK) was also identified at amino acids 262–274. In addition to the penton protein, the L2 region of AdVs encodes three additional proteins (pVII, pV, and pX). These serve as core proteins and facilitate packaging of viral DNA within the capsid, by forming associations with the viral DNA through arginine and lysine-rich regions [34]. The ORF for pVII was identified at nucleotides 15380–15958 in HAdV-B14 CHN, and the protein comprised 192 amino acids (21.3 kDa). Proteins of 351 and 74 amino acids (40.1 and 8.5 kDa, respectively) were predicted for pV (nucleotides 16001–17056) and pX (nucleotides 17085–17315) in the HAdV-B14 CHN genome. For pVII, a single protease cleavage site (21MYGG↓A25) was found, while in pX two protease cleavage sites (25MLGR↓G29 and 43LRGG↓F47) were identified. Similar cleavage sites have also been reported in pVIII and pX proteins of HAdV-B11 and HAdV-C5 [23].
2.2.8.3. L3
Three ORFs corresponding to the pVI, hexon, and protease proteins were identified in the L3 region. A common poly (A) signal for these proteins was situated at nucleotide 21779 (Table 2). Protein pVI is associated with the transport of hexon molecules to the nucleus, and participates in the disruption of the endosomal membrane during virus infection; however, mature pVI is a minor capsid protein [35]. The HAdV-B14 CHN pVI protein comprises 246 amino acids (26.6 kDa) and was 100% identical to its homolog in the HAdV-B14 (303600) North American strain. We found 99% identity to the corresponding protein in the HAdV-B14 de Wit strain, because of an amino acid substitution; (CHN)D3N(de Wit). Two nuclear localization signals (KRPRP and KRRR) were identified at amino acids 132–136 and 241–244, respectively. Two endoprotease cleavage motifs (29LNGG↓A33 and 232IVGL↓G236) were also determined near the N and C termini of HAdV-B14 CHN pVI. The hexon protein is the major structural component of the adenovirus capsid, constituting about 63% of the virion mass. The epitopes located on the hexon protein are targets for neutralizing antibodies in vivo. These epitopes also assist cytotoxic T cells in recognizing the virus and provide the basis for classification of AdVs [36]. The second ORF predicted at nucleotides 18252–21089 in the L3 region encodes the hexon protein (945 amino acids, 106.9 kDa; Table 2). The HAdV-B14 CHN hexon protein was 100% identical to that in the HAdV-B14 (303600) North American isolate, and 99% identical to that in the de Wit strain. We identified a single amino acid substitution, (CHN)I497M(de Wit), in the HAdV-B14 CHN hexon protein when compared with the HAdV-B14 CHN de Wit hexon. The third protein encoded in the L3 region is the 23.9 kDa (209 amino acids) viral protease. The function of this protein is to cleave other viral proteins, thereby allowing for viral assembly and maturation [37].
2.2.8.4. L4
Four ORFs were identified in the HAdV-B14 CHN L4 region, and corresponded to the 100 kDa hexon-assembly associated protein (23420–25858 bp), a 22 kDa protein (25590–26125 bp), a 33 kDa protein comprising two exons (25590–25908, 26078–26439 bp), and the pVIII protein (26489–27172 bp). A common poly (A) signal for L4 proteins was predicted at nucleotide 27333 (Table 2). The 100 kDa protein is a nonstructural protein; it is required for translation of late viral mRNAs, and acts to inhibit translation of cellular mRNAs. This protein also serves as a scaffold for hexon attachment, thereby facilitating its folding into a trimer, and transport to the nucleus [38], [39]. The predicted 100 kDa protein for HAdV-B14 CHN comprised 812 amino acids, which actually corresponds to a molecular weight of 91 kDa. This protein is 100 and 99% identical to its homologs in the HAdV-B14 (303600) North American and de Wit strains, respectively. An amino acid substitution, (CHN)G343D(de Wit), was identified in the de Wit strain. The 22 kDa protein, in conjunction with IVa2, is now known to play a critical role in the recognition of the packaging domain of the AdV genome, and leads to viral DNA encapsidation [40]. A homolog of this protein was identified in the HAdV-B14 CHN L4 region, and comprised 191 amino acids (21.7 kDa). Similar to the L4 100 kDa protein, this protein was also 100% identical to that in the HAdV-B14 (303600) North American strain, and 99% identical to that in the de Wit strain. When compared with the HAdV-B14 de Wit strain, we found a substitution, (CHN)V173I(de Wit). The L4 33 kDa protein plays an indispensable role in virion assembly [41]. The homolog of the 33 kDa protein in HAdV-B14 CHN was 226 amino acids (25.4 kDa) and 100% identical to the L4 33 kDa protein in the de Wit strain. The fourth ORF in the HAdV-B14 CHN L4 region encodes pVIII (227 amino acids, 25 kDa; Table 2), which is a minor capsid protein, found on the interior of the capsid with three other hexon-associated proteins (pIIIa, pVI and protein IX) [42].
2.2.8.5. L5
A single ORF, encoding the fiber protein (323 amino acids, 35 kDa), was identified in the L5 region. The poly (A) signal was predicted at nucleotide 30788 (Table 2). The fiber protein is a major structural protein, and is considered to be a major determinant of tissue tropism. Structurally, the fiber protein can be divided into three parts: the N-terminal tail; a central shaft with repeated motifs; and a C-terminal globular knob. A hydrophobic motif (10FNPVYPYE17) was identified at the N-terminal end of the HAdV-B14 CHN fiber protein. This has been shown to mediate the interaction between the penton base and the fiber through hydrogen bonds and salt bridges. The globular C-terminal knob domain binds to the host cell receptor [43], [44].
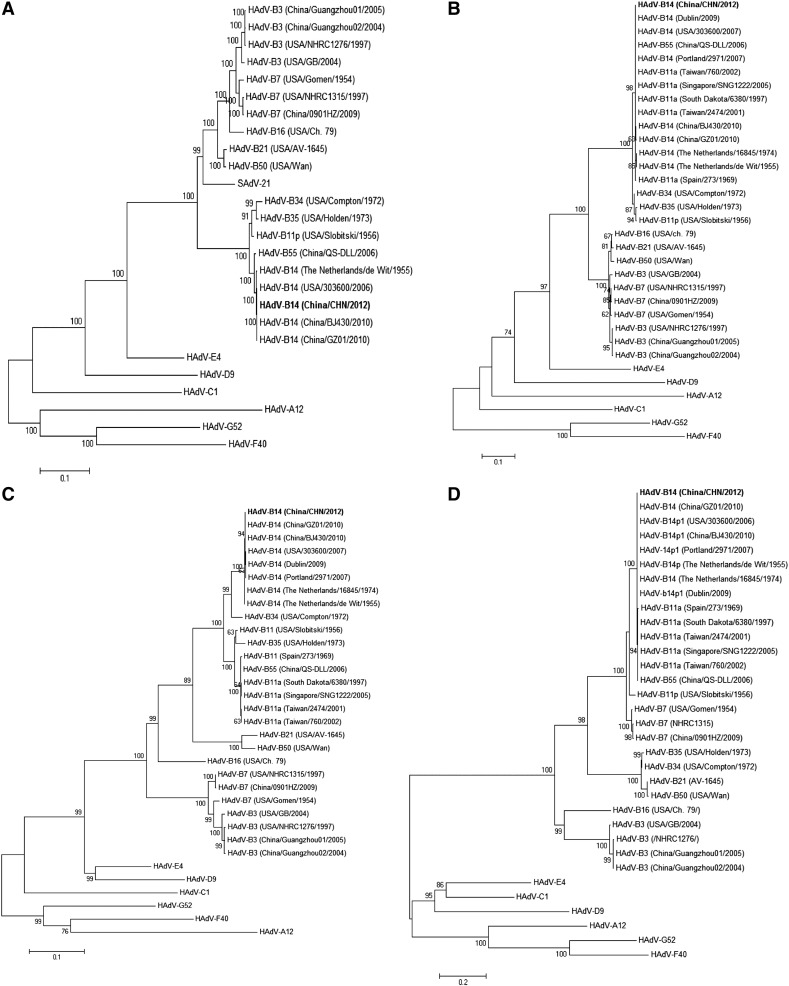
2.3. Recombination and phylogenetic analysis
Although HAdV-B14 CHN showed high homology to the reference HAdV-B14 de Wit strain, we wanted to determine if any potential recombination events occurred. Through recombination analysis we determined that HAdV-B14 CHN was closely related to HAdV-B14 de Wit, but is not a recombinant with respect to other recognized clades (data not shown). To determine the evolutionary relationship of HAdV-B14 CHN with other AdV, phylogenetic analyses of the entire genome and individual genes (E1A, fiber and hexon) were conducted. We found that HAdV-B14 CHN was a member of the B2 group of B-type AdVs, similar to HAdV-B14 strains previously isolated from China (Fig. 2A). The E1A, hexon and fiber genes also clustered with other members of the B2 group as shown in Fig. 2B, C and D respectively. Comparative sequence analysis at the nucleotide level indicated that HAdV-B14 CHN was 100% identical to HAdV-B14 (303600), and 99.9, 99.9, and 99.7% identical to HAdV-B14 (BJ430, China), HAdV-B14 (GZ01, China), and HAdV-B14 de Wit, respectively.
Fig. 2.
Phylogenetic analysis.
Maximum-likelihood phylogenetic trees were estimated using full-length (A) genomic sequences, (B) E1A gene sequences, (C) hexon gene sequences, and (D) fiber gene sequences of AdVs. The GenBank accession IDs and details of the full-length genomic sequences and E1A, hexon, and fiber genes sequences used in phylogenetic analysis are given in Table 1 and Table S1 respectively. Reference sequences representing different AdV genomes together with the newly sequenced HAdV-B14 CHN (bold format) were included in each tree construction dataset. Isolates were named using the following format: AdV type (sampling country/isolate name/isolation year). The percentage of trees in which the associated taxa clustered together is shown next to the branches. The reliability of the tree was assessed by bootstrap analysis with 1000 replications.
The majority of previous HAdV-B14 infections have resulted in severe outbreaks or epidemics; however, in China only sporadic infections have been identified. The first report of HAdV-B14 in China was from Guangzhou in 2010. The virus was isolated from a throat swab taken from a 17-month-old baby, who displayed symptoms of acute suppurative tonsillitis. This infection was regarded as unusual as these symptoms were not previously described for other HAdV-B14 infections [8]. Another report of a HAdV-B14 infection in a 6-month-old baby diagnosed with a bronchial pneumonia/acute respiratory tract infection (ARTI) came from Beijing Children's Hospital [9]. Based on presented symptoms, FRI or pneumonia was predominant in outbreaks caused by HAdV-B14 in the United States from 2006–2007. Although the two strains of HAdV-B14 isolated in China in 2010 were similar (≥ 99% nucleotide sequence identity) to those isolated in the United States from 2006–2007, infections due to Chinese isolates were mild [6].
In the present study, a Chinese isolate of HAdV-B14 associated with an outbreak of FRI has been subjected to genomic and bioinformatic analyses. While this manuscript was in preparation, another HAdV-B14-associated FRI outbreak that occurred in Gansu Province, China during 2001 was reported by Huang et al. [45]. This outbreak symptoms also differed from HAdV-B14-associated outbreaks reported in the United States. Sequences from Huang's study were not publically released in GenBank; however, according to their published report, they performed sequencing of fiber, hexon and E1A genes. Their results showed 99% identity to the fiber, hexon and E1A genes of HAdV-B14 strains reported for other Chinese isolates, and with the United States isolate associated with infections at military camps. Taken together, all these findings suggest that HAdV-B14 might have been circulating in China since, or even before, 2010. In summary, considering the prevalence of HAdV-B14 in China, close surveillance of circulating HAdVs in China is required to prevent future outbreaks, and respond to them once they occur.
3. Methods
3.1. Sample collection and ethics statement
A mild febrile outbreak was identified in Beijing in 2012. All cases were male, with a median age of 23 years (range, 19–33 years); of these cases, 16 (53%) reported fever, 12 (40%) reported coughing, and 26 (87%) reported a sore throat. Additionally, 2 of the 30 patients reported either vomiting or diarrhea, but none of the patients exhibited both symptoms. All cases required hospitalization. Throat swab specimens were collected from patients with acute respiratory disease. Informed consent was provided by patients and the study was approved by the Beijing Institute of Microbiology and Epidemiology Ethics Committee. Specimens were preserved in virus transport medium and stored at − 80 °C.
3.2. Determination of the etiological agent
Viral genomic DNA was extracted using a PureLink™ Viral RNA/DNA Kit (Life Technology, Grand Island, NY, USA) according to the manufacturer's instructions. Reverse-transcription polymerase chain reaction (RT-PCR) assays specific for the detection of human coronavirus and influenza virus types A and B were conducted as previously described. Normal PCR assays for the detection of HAdVs, and quantitative real-time RT-PCR (qPCR) assays for the detection of avian H5N1 influenza virus were conducted as previously described [17], [18], [19]. Throat swab samples were inoculated in A549 cells (ATCC CCL-185; Manassas, VA, USA) and maintained in 1640 medium (Life Technologies) supplemented with 2% fetal bovine serum (FBS; Life Technologies, USA) at 37 °C/5% CO2. Cultures were observed for any signs of cytopathic effect (CPE). If no CPE developed, inoculated cultures were harvested by freeze-thawing after 5 days, and passaged on fresh cell cultures over three serial passages. To confirm isolation of viruses, PCR detection and immunofluorescent staining of infected cells were done. Purified amplicons were directly sequenced on an ABI3730XL Sequencer using BigDye Terminator version 3.1 (Applied Biosystems).
3.3. Amplification of the HAdV-B14 genome
Primers (Table S1) were designed to cover the whole genome of HAdV-B14, using HAdV-B14 (BJ430) strain as the reference sequence. Neighboring amplicons overlapped around 100 bp. The PCRs were conducted in 50-μL volumes, and included 1 μL of high-fidelity Taq DNA polymerase (Transgene, Beijing, China). Thermal cycling conditions involved an initial denaturation step at 95 °C for 30 s, followed by 35 cycles of 95 °C for 20 s, 55 °C for 30 s, and 72 °C for 1 min, then a final extension step at 72 °C for 5 min after the 35th cycle. All reactions were conducted on a GeneAmp 9700 thermal cycler (Applied Biosystems). Amplicons were analyzed by electrophoresis on 1% (w/v) agarose gel stained with ethidium bromide. DNA fragments were extracted from gels and purified with a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA). Purified DNA was directly sequenced on an ABI3730XL Sequencer using BigDye Terminator version 3.1 (Applied Biosystems). Sequences were assembled using Vector NTI Advance 10 (Invitrogen, Carlsbad, CA, USA).
3.4. Genome annotation and sequence analysis
The full-length genome sequence of HAdV-B14 CHN strain was deposited into the GenBank database and assigned the accession number JX892927.2. Genome annotations and ORFs were completed using the Rapid Annotation using Subsystem Technology (RAST) server (http://rast.nmpdr.org/) [46] and National Center for Biotechnology Information (NCBI) ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). ORFs were further analyzed for nucleotide and protein sequence homology search by using blastn and blastp programs respectively in the BLAST software package (version 2.2.28) (http://www.ncbi.nlm.nih.gov/blast) [47]. Splice sites were identified by using on-line splice prediction software (http://www.fruitfly.org/seq_tools/splice.html) and the GENSCAN software (http://genes.mit.edu/GENSCAN.html). Sequence percent identities were calculated using tools from the EMBOSS package (http://www.ebi.ac.uk/Tools/emboss/).
3.5. Recombination analysis
Identification of potential recombinant events in the HAdV-B14 CHN genome was determined with the Recombination Detection Program (RDP) Beta (version 4.16) software suite (http://web.cbio.uct.ac.za/~darren/rdp.html) [48] that incorporates several phylogenetic-substitution and distance-based methods. Comparisons among similar sequences are unlikely to yield detectable signal of recombination and masking similar sequences increases the power of multiple tests. The P-value cut-off was set to 0.05 in all analyses and the Bonferroni correction was applied. The analyses settings were kept at defaults.
3.6. Phylogenetic analysis
The full genome sequence of HAdV-B14 CHN was subject to NCBI blastn analysis against previously reported AdV genomes. We retrieved 26 full-length genome sequences of different AdV species for phylogenetic analysis (Table 1). In addition, individual phylogenetic trees for the E1A, hexon and fiber genes were constructed. For each gene segment, the dataset comprised 33 full-length sequences (Table S2). The selected sequences were aligned using MUSCLE [49] within the Molecular Evolutionary Genetic Analysis (MEGA) (version 5.2) software suite (http://www.megasoftware.net/) [50]. Nucleotide substitution models were estimated using MEGA5.2 following the Akaike Information Criterion (AIC). The GTR + G + I, K2 + G + I, TN93 + G + I and HKY + G + I models were selected as best-fit models for the full genome, E1A, hexon, and fiber datasets, respectively. Phylogenetic trees were constructed using the maximum likelihood (ML) method by incorporating dataset specific nucleotide substitution models in MEGA5.2. The robustness of trees was assessed by bootstrap analysis of 1000 replicates, and is indicated as a percentage on each branch.
The following are the Supplementary data related to this article.
Primer sequences used for amplification and sequencing of HAdV-B14 CHN complete genome.
GenBank accession numbers and details of E1A, fiber and hexon genes from different adenoviruses used in the present study for phylogenetic analysis.
Conflicts of interest
None declared.
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (No. 81072350), the China Mega-Project on Major Drug Development (No. 2011ZX09401-023), the China Mega-Project on Infectious Disease Prevention (No. 2011ZX10004-001), and the State Key Laboratory of Pathogen and Biosecurity Program (No. SKLPBS1113).
Contributor Information
Wu-Chun Cao, Email: caowc@nic.bmi.ac.cn.
Yigang Tong, Email: tong62035@gmail.com.
References
- 1.Hierholzer J.C. Adenoviruses in the immunocompromised host. Clin. Microbiol. Rev. 1992;5:262–274. doi: 10.1128/cmr.5.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Der Veen J., Kok G. Isolation and typing of adenoviruses recovered from military recruits with acute respiratory disease in The Netherlands. Am. J. Hyg. 1957;65:119–129. doi: 10.1093/oxfordjournals.aje.a119860. [DOI] [PubMed] [Google Scholar]
- 3.Kendall E.J., Riddle R.W., Tuck H.A., Rodan K.S., Andrews B.E., McDonald J.C. Pharyngo-conjunctival fever; school outbreaks in England during the summer of 1955 associated with adenovirus types 3, 7, and 14. Br. Med. J. 1957;2:131–136. doi: 10.1136/bmj.2.5037.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mevzos L.M., Il'ina T.S., Makhmudov O.S., Zolotarskaia E.E., Dreizin R.S. An outbreak of acute respiratory infections among adults caused by adenovirus serotype 14. Vopr. Virusol. 1966;11:426–431. [PubMed] [Google Scholar]
- 5.Cooper R.J., Hallett R., Tullo A.B., Klapper P.E. The epidemiology of adenovirus infections in Greater Manchester, UK 1982–96. Epidemiol. Infect. 2000;125:333–345. doi: 10.1017/s0950268899004550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houng H.S., Gong H., Kajon A.E., Jones M.S., Kuschner R.A., Lyons A., Lott L., Lin K.H., Metzgar D. Genome sequences of human adenovirus 14 isolates from mild respiratory cases and a fatal pneumonia, isolated during 2006–2007 epidemics in North America. Respir. Res. 2010;11:116. doi: 10.1186/1465-9921-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu L., Liu Z., Li X., Qu J., Guan W., Liu Y., Song S., Yu X., Cao B. Severe community-acquired pneumonia caused by adenovirus type 11 in immunocompetent adults in Beijing. J. Clin. Virol. 2012;54:295–301. doi: 10.1016/j.jcv.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q., Seto D., Zhao S., Zhu L., Zhao W., Wan C. Genome sequence of the first human adenovirus type 14 isolated in China. J. Virol. 2012;86:7019–7020. doi: 10.1128/JVI.00814-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang L., An J., Xie Z., Dehghan S., Seto D., Xu W., Ji Y. Genome and bioinformatic analysis of a HAdV-B14p1 virus isolated from a baby with pneumonia in Beijing, China. PLoS One. 2013;8:e60345. doi: 10.1371/journal.pone.0060345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dan A., Elo P., Harrach B., Zadori Z., Benko M. Four new inverted terminal repeat sequences from bovine adenoviruses reveal striking differences in the length and content of the ITRs. Virus Genes. 2001;22:175–179. doi: 10.1023/a:1008125324346. [DOI] [PubMed] [Google Scholar]
- 11.Temperley S.M., Hay R.T. Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO J. 1992;11:761–768. doi: 10.1002/j.1460-2075.1992.tb05109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mul Y.M., Verrijzer C.P., van der Vliet P.C. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol. 1990;64:5510–5518. doi: 10.1128/jvi.64.11.5510-5518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatfield L., Hearing P. The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J. Virol. 1993;67:3931–3939. doi: 10.1128/jvi.67.7.3931-3939.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akusjarvi G., Mathews M.B., Andersson P., Vennstrom B., Pettersson U. Structure of genes for virus-associated RNAI and RNAII of adenovirus type 2. Proc. Natl. Acad. Sci. U. S. A. 1980;77:2424–2428. doi: 10.1073/pnas.77.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y., Mathews M.B. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J. Virol. 1996;70:5083–5099. doi: 10.1128/jvi.70.8.5083-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purkayastha A., Su J., Carlisle S., Tibbetts C., Seto D. Genomic and bioinformatics analysis of HAdV-7, a human adenovirus of species B1 that causes acute respiratory disease: implications for vector development in human gene therapy. Virology. 2005;332:114–129. doi: 10.1016/j.virol.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 17.Kidd A.H., Garwicz D., Oberg M. Human and simian adenoviruses: phylogenetic inferences from analysis of VA RNA genes. Virology. 1995;207:32–45. doi: 10.1006/viro.1995.1049. [DOI] [PubMed] [Google Scholar]
- 18.Shenk T.E. Lippincott - Raven Publishers; 1996. Adenoviridae: The Viruses and Their Replication. [Google Scholar]
- 19.Frisch S.M., Mymryk J.S. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 2002;3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- 20.Gallimore P.H., Turnell A.S. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene. 2001;20:7824–7835. doi: 10.1038/sj.onc.1204913. [DOI] [PubMed] [Google Scholar]
- 21.Berk A.J., Sharp P.A. Structure of the adenovirus 2 early mRNAs. Cell. 1978;14:695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- 22.Berk A.J. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 23.Mei Y.F., Skog J., Lindman K., Wadell G. Comparative analysis of the genome organization of human adenovirus 11, a member of the human adenovirus species B, and the commonly used human adenovirus 5 vector, a member of species C. J. Gen. Virol. 2003;84:2061–2071. doi: 10.1099/vir.0.19178-0. [DOI] [PubMed] [Google Scholar]
- 24.Horwitz M.S. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med. 2004;6(Suppl. 1):S172–S183. doi: 10.1002/jgm.495. [DOI] [PubMed] [Google Scholar]
- 25.Windheim M., Hilgendorf A., Burgert H.G. Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr. Top. Microbiol. Immunol. 2004;273:29–85. doi: 10.1007/978-3-662-05599-1_2. [DOI] [PubMed] [Google Scholar]
- 26.Leppard K.N. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J. Gen. Virol. 1997;78(Pt 9):2131–2138. doi: 10.1099/0022-1317-78-9-2131. [DOI] [PubMed] [Google Scholar]
- 27.Boulanger P., Lemay P., Blair G.E., Russell W.C. Characterization of adenovirus protein IX. J. Gen. Virol. 1979;44:783–800. doi: 10.1099/0022-1317-44-3-783. [DOI] [PubMed] [Google Scholar]
- 28.Young C.S. The structure and function of the adenovirus major late promoter. Curr. Top. Microbiol. Immunol. 2003;272:213–249. doi: 10.1007/978-3-662-05597-7_8. [DOI] [PubMed] [Google Scholar]
- 29.Hu S.L., Manley J.L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc. Natl. Acad. Sci. U. S. A. 1981;78:820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasson T.B., Soloway P.D., Ornelles D.A., Doerfler W., Shenk T. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 1989;63:3612–3621. doi: 10.1128/jvi.63.9.3612-3621.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gustin K.E., Imperiale M.J. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 1998;72:7860–7870. doi: 10.1128/jvi.72.10.7860-7870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuillel M., Milleville M., D'Halluin J.C. Expression of protein IIIa of human adenovirus type 2 in Escherichia coli. Gene. 1987;55:295–301. doi: 10.1016/0378-1119(87)90289-7. [DOI] [PubMed] [Google Scholar]
- 33.Wickham T.J., Mathias P., Cheresh D.A., Nemerow G.R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee P.K., Vayda M.E., Flint S.J. Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J. Mol. Biol. 1986;188:23–37. doi: 10.1016/0022-2836(86)90477-8. [DOI] [PubMed] [Google Scholar]
- 35.Wodrich H., Guan T., Cingolani G., Von Seggern D., Nemerow G., Gerace L. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 2003;22:6245–6255. doi: 10.1093/emboj/cdg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebner K., Pinsker W., Lion T. Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: phylogenetic, taxonomic, and clinical implications. J. Virol. 2005;79:12635–12642. doi: 10.1128/JVI.79.20.12635-12642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber J. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol. 1976;17:462–471. doi: 10.1128/jvi.17.2.462-471.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes B.W., Telling G.C., Myat M.M., Williams J.F., Flint S.J. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 1990;64:2732–2742. doi: 10.1128/jvi.64.6.2732-2742.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oosterom-Dragon E.A., Ginsberg H.S. Characterization of two temperature-sensitive mutants of type 5 adenovirus with mutations in the 100,000-dalton protein gene. J. Virol. 1981;40:491–500. doi: 10.1128/jvi.40.2.491-500.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostapchuk P., Anderson M.E., Chandrasekhar S., Hearing P. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 2006;80:6973–6981. doi: 10.1128/JVI.00123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finnen R.L., Biddle J.F., Flint J. Truncation of the human adenovirus type 5 L4 33-kDa protein: evidence for an essential role of the carboxy-terminus in the viral infectious cycle. Virology. 2001;289:388–399. doi: 10.1006/viro.2001.1130. [DOI] [PubMed] [Google Scholar]
- 42.Liu G.Q., Babiss L.E., Volkert F.C., Young C.S., Ginsberg H.S. A thermolabile mutant of adenovirus 5 resulting from a substitution mutation in the protein VIII gene. J. Virol. 1985;53:920–925. doi: 10.1128/jvi.53.3.920-925.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caillet-Boudin M.L. Complementary peptide sequences in partner proteins of the adenovirus capsid. J. Mol. Biol. 1989;208:195–198. doi: 10.1016/0022-2836(89)90095-8. [DOI] [PubMed] [Google Scholar]
- 44.Green N.M., Wrigley N.G., Russell W.C., Martin S.R., McLachlan A.D. Evidence for a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J. 1983;2:1357–1365. doi: 10.1002/j.1460-2075.1983.tb01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang G., Yu D., Zhu Z., Zhao H., Wang P., Gray G.C., Meng L., Xu W. 2013. Outbreak of Febrile Respiratory Illness Associated with Human Adenovirus Type 14p1 in Gansu Province. Influenza and Other Respiratory Viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Martin D.P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for amplification and sequencing of HAdV-B14 CHN complete genome.
GenBank accession numbers and details of E1A, fiber and hexon genes from different adenoviruses used in the present study for phylogenetic analysis.