Abstract
A reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay targeting the nucleocapsid phosphoprotein gene of infectious bronchitis virus (IBV) was developed. The detection limits for the IBV RT-LAMP assay were 101 50% egg infection dose (EID50) per 50 μl of titrated viruses and no cross-reaction of IBV RT-LAMP was found when tested with other viruses including Newcastle disease virus (NDV), avian reovirus (ARV), and infectious laryngotrachietis virus (ILTV) due to their mismatch with IBV RT-LAMP primers. A total of 187 clinical tissues samples (88 blood, 62 kidney and 37 lung) were evaluated and compared to conventional RT-PCR. The sensitivity of RT-LAMP and RT-PCR assays for detecting IBV RNA in clinical specimens was 99.5% and 98.4%, respectively. These findings showed that the RT-LAMP assay has potential usefulness for rapid and sensitive diagnosis in outbreak of IBV.
Keywords: Infectious bronchitis virus (IBV), Detection, Reverse transcription loop-mediated isothermal amplification (RT-LAMP)
Infectious bronchitis virus (IBV) is a major cause of disease in domestic fowl and causes an acute, highly contagious disease of the respiratory tracts and sometimes also urogential tracts [1]. Current diagnostic assays for IBV include virus isolation in embryonating eggs or tracheal organ cultures, cell culture immunoassays and molecular assays that detect the viral RNA [2]. Virus isolation is generally considered the gold standard, but it is expensive and time consuming because several passages may be required to detect the virus. Immunoassays routinely use IBV-specific monoclonal antibodies to detect the virus in direct or indirect fluorescent antibody and enzyme-linked immunosorbent assay formats. Although the immunoassays are more rapid and simpler than virus isolation, which tend to lack specificity and sensitivity and none detect all strains or types of IBV [3], [4], [5]. Molecular assays for the detection of IBV are used commonly because they provide highly specific and sensitive results and can detect viral RNA directly from clinical samples or from viral isolates in a laboratory host system. Although RT-PCR and real-time RT-PCR methods are applied widely in the laboratories [6], [7], [8], [9], [10], [11], [12], [13], RT-LAMP need more simple device such as water bath or heat block and has high efficiency because there is no time loss for thermal change due to its isothermal reaction.
The loop-mediated isothermal amplification (LAMP), which employs a DNA polymerase and a set of four specially designed primers including a pair of outer primers and inner primers that recognize a total of six distinct sequences on the target DNA, is a novel nucleic acid amplification method. An inner primer containing sequences of the sense and antisense strands of the target DNA initiates LAMP reaction (Fig. 1 ). The final products are stem-loop DNAs with several inverted repeats of the target and cauliflower-like structures with multiple loops formed by annealing between alternately inverted repeats of the target in the same strand. Because LAMP recognizes the target by six distinct sequences initially and by four distinct sequences afterwards, it is expected to amplify the target sequence with high selectivity. The advantage of the LAMP assay is simple and easy to perform once the appropriate primers and a regular laboratory water bath or heat block are prepared for reaction. LAMP is also applicable to RNA upon use of reverse transcriptase (RTase) together with DNA polymerase [14].
Fig. 1.
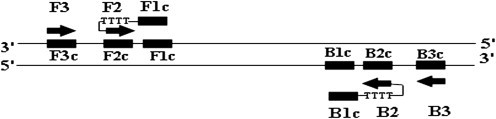
Primer design of RT-LAMP. Six distinct regions are designated on the target of infectious bronchitis virus. F2c, sequence complementary to F2; F3c, sequence complementary to F3; B2c, sequence complementary to B2; B3c, sequence complementary to B3. Two inner primers (FIP and BIP) and outer primers (F3 and B3) are used in RT-LAMP method. FIP (BIP) is a hybrid primer consisting of the F1c (B1c) sequence, a linker of TTTT and the F2 (B2) sequence.
Reverse transcription LAMP (RT-LAMP) has been applied successfully for the detection of influenza A virus, severe acute respiratory syndrome coronavirus and Newcastle disease virus [15], [16], [17], [18]. In the present study, the reference strain IBV01, IBV02 and IBV03 were from Chinese Veterinary Microorganism Conservation Center (CVMCC), the strains of IBV01 and IBV02 were Massachusetts serotype and IBV03 was the T strain. IBV01 was used to standardize the IBV RT-LAMP assay. The vaccine strains of H120 and M41 were supplied by a local vaccine manufacturer. The other viruses, including Newcastle disease virus (NDV), avian reovirus (ARV), and infectious laryngotrachietis virus (ILTV) were also from CVMCC and used to examine the specificity of IBV RT-LAMP. The median egg infectious dose (EID50) of the IBV01 virus used in sensitivity tests was calculated according to the Reed and Muench formula. A series of the 10 times dilutions spanning from 1 to 104 EID50 per 50 μl of titrated viruses was used as template.
Four primers of FIP, BIP, F, and B for the RT-LAMP test were designed by targeting the conserved regions of N gene (Fig. 2 ). RT-LAMP was performed in 25 μl of a mixture containing 2 μl of the genomic RNA, 40 pmol (each) of primers FIP and BIP, 5 pmol (each) of primers F and B, 1 U of the THERMO-X reverse transcriptase (Invitrogen) and 8 U of Bst DNA polymerase (New England Biolabs, Ipswich, MA) with the corresponding buffer, respectively. Amplification was carried out at 64 °C for 15, 30, 45, 60, 75 min, respectively, and then terminated by incubation at 80 °C for 2 min. The electrophoresis analysis indicated that 45 min are enough for IBV RT-LAMP in the study. The products of the reaction were also inspected by the naked eye following the addition of 1 μl of SYBR Green I dye to the tube. All the strains tested by RT-LAMP were also identified by RT-PCR and sequenced. Each assay was conducted in triplicates. The details of primers (NF and NR) and condition for RT-PCR assay for the detection of IBV have been previously described [11], with minor modifications (Fig. 2). A perfect correlation was found for all of the IBV strains which were positive by the RT-LAMP and RT-PCR and no cross-reaction of IBV RT-LAMP was tested with the healthy blood without IBV infection and other chicken tissue of NDV, ARV and ILTV (Fig. 4).
Fig. 2.

Oligonucleotide primers used for RT-PCR and RT-LAMP amplification of infectious bronchitis virus (GenBank accession number EU889030). The underlined letters indicate the sequences of RT-LAMP primers. The primers of NF and NR were designed according to the method described previously by Zwaagstra et al. (1992), with minor modifications.
Fig. 4.
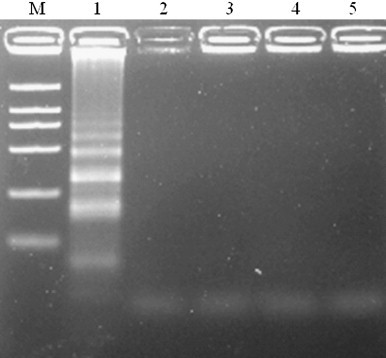
Agarose gel analysis of cross-reaction in the IBV RT-LAMP assay. Lane M, DNA Marker DL-2000; lane 1, IBV01; lane 2, NDV; lane 3, ARV; lane 4, ILTV; lane 5, negative control.
For further evaluation of RT-LAMP assay with clinical samples, 187 specimens of blood, kidney and lung tissue were obtained from IBV-infected chicken and proved to be positive by real-time PCR and serological test (Table 1 ). The result of RT-LAMP was analyzed and compared with RT-PCR with all clinical specimens. The general detection rates of IBV RT-LAMP and RT-PCR for above mentioned different clinical samples were 99.5% and 98.4%, respectively. In general, both assays showed higher sensitivity for blood and lung samples than kidney (Table 1). The results indicated that this diagnostic technique was reliable for the detection of IBV in blood, kidney and lung tissue samples. The preferred samples are the blood during the early stage of infection, which may have a higher predictive value of monitoring an outbreak.
Table 1.
Comparative evaluation of RT-LAMP assay with RT-PCR for 187 clinical samples.
| Specimen type | No. of positive samples | No. (%) of positive samples for assay |
|
|---|---|---|---|
| RT-PCR | RT-LAMP | ||
| Blood | 88 | 88 (100) | 88 (100) |
| Kidney | 62 | 60 (96.8) | 61 (98.3) |
| Lung | 37 | 36 (97.3) | 37 (100) |
| Total | 187 | 184 (98.4) | 186 (99.5) |
The result indicated the detection limit of IBV RT-LAMP was 10 EID50 per 50 μl with titrated viruses (Fig. 3 ). In addition, the reaction time of RT-LAMP method is 45 min, which is more rapid than conventional RT-PCR, and the reaction only needs a laboratory water bath. Another useful feature was RT-LAMP products could be directly observed by the addition of dyes, as the original orange color of SYBR green I would turn to be green in the positive reaction mixture. From a practical point of view, RT-LAMP is more suitable as a routine diagnostic tool, especially in clinics in which complicated equipment such as thermal cycling machines and electrophoresis apparatus are not available. In addition, RT-LAMP has a potential for field diagnosis. Nonetheless, the reliability of this assay should be further evaluated by large-scale investigation.
Fig. 3.
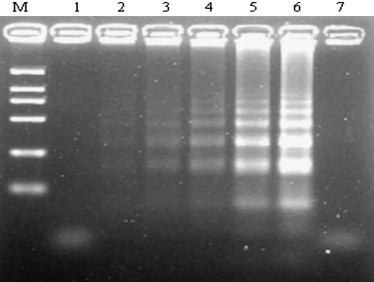
Sensitivity of RT-LAMP determined by agarose gel electrophoresis of RT-LAMP products from spiked with 10-fold serial dilution of IBV. Lines M, DNA marker DL-2000; Lanes 1–6, different IBV 50% egg infectious dose of RT-LAMP (10−1, 100, 101,102, 103, 104 EID50 per 50 μl of titrated viruses); lane 7, negative control.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 30700597). This work was also supported in part by grants from the National Key Technologies R&D Program of China (No. 2008BADB4B05) and the Natural Science Foundation of Gansu Province, China (No. 0803RJZA050).
References
- 1.King D.J., Cavanagh D. Infectious bronchitis. In: Calnek B.W., Barnes H.J., Beard C.W., Reid W.M., Yoder H.W. Jr., editors. Disease of Poultry. 9th ed. Iowa State University Press; Ames, Iowa: 1991. pp. 471–484. [Google Scholar]
- 2.Gelb J., Jr., Jackwood M.W. Infectious bronchitis. In: Swayne D.E., Glisson J.R., Jackwood M.W., Pearson J.E., Reed W.M., editors. A laboratory manual for the isolation and identification of avian pathogens. 4th ed. American Association of Avian Pathologists; Kennett Square, PA: 1998. pp. 169–174. [Google Scholar]
- 3.Karaca K., Naqi S. A monoclonal antibody blocking ELISA to detect serotype-specific infectious bronchitis virus antibodies. Vet Microbiol. 1993;34:249–257. doi: 10.1016/0378-1135(93)90015-y. [DOI] [PubMed] [Google Scholar]
- 4.Karaca K., Naqi S., Gelb J., Jr. Production and characterization of monoclonal antibodies to three infectious bronchitis virus serotypes. Avian Dis. 1992;36:903–915. [PubMed] [Google Scholar]
- 5.Naqi S.A., Karaca K., Bauman B. Amonoclonal antibody-based antigen capture enzyme-linked immunosorbent assay for identification of infectious bronchitis virus serotypes. Avian Pathol. 1993;22:555–564. doi: 10.1080/03079459308418943. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh D., David P.J., Cook J.K.A., Li D., Kant A., Koch G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- 7.Jackwood M.W., Yousef N.M., Hilt D.A. Further development and use of a molecular serotype identification test for infectious bronchitis virus. Avian Dis. 1997;41(1):105–110. [PubMed] [Google Scholar]
- 8.Keeler C.L., Jr., Reed K.L., Nix W.A., Gelb J., Jr. Serotype identification of avian infectious bronchitis virus by RT-PCR of the peplomer (S-1) gene. Avian Dis. 1998;42(2):275–284. [PubMed] [Google Scholar]
- 9.Kingham B.F., Keeler C.L., Jr., Nix W.A., Ladman B.S., Gelb J., Jr. Identification of avian infectious bronchitis virus by direct automated cycle sequencing of the S-1 gene. Avian Dis. 2000;44(2):325–335. [PubMed] [Google Scholar]
- 10.Kwon H.M., Jackwood M.W., Gelb J. Differentiation of infectious bronchitis virus serotypes using polymerase chain-reaction and restriction fragment- length-polymorphism analysis. Avian Dis. 1993;37(1):194–202. [PubMed] [Google Scholar]
- 11.Zwaagstra K.A., van der Zeijst B.A., Kusters J.G. Rapid detection and identification of avian infectious bronchitis virus. J Clin Microbiol. 1992;30:79–84. doi: 10.1128/jcm.30.1.79-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H.J., Lee L.H., Shih W.L., Lin M.Y., Liao M.H. Detection of infectious bronchitis virus by multiplex polymerase chain reaction and sequence analysis. J Virol Methods. 2003;109:31–37. doi: 10.1016/s0166-0934(03)00041-7. [DOI] [PubMed] [Google Scholar]
- 13.Callison S.A., Hilt D.A., Boynton T.O., Sample B.F., Robison R., Swayne D.E. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J Virol Methods. 2006;138:60–65. doi: 10.1016/j.jviromet.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H.T., Zhang J., Sun D.H., Ma L.N., Liu X.T., Cai X.P. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J Virol Methods. 2008;151:200–203. doi: 10.1016/j.jviromet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;43:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pham H.M., Nakajima C., Ohashi K., Onuma M. Loop-mediated isothermal amplification for rapid detection of Newcastle disease virus. J Clin Microbiol. 2005;43:1646–1650. doi: 10.1128/JCM.43.4.1646-1650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poon L.L.M., Leung C.S.W., Chan K.H., Lee J.H.C., Yuen K.Y., Guan Y. Detection of Human influenza A viruses by loop-mediated isothermal amplification. J Clin Microbiol. 2005;43:427–430. doi: 10.1128/JCM.43.1.427-430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


