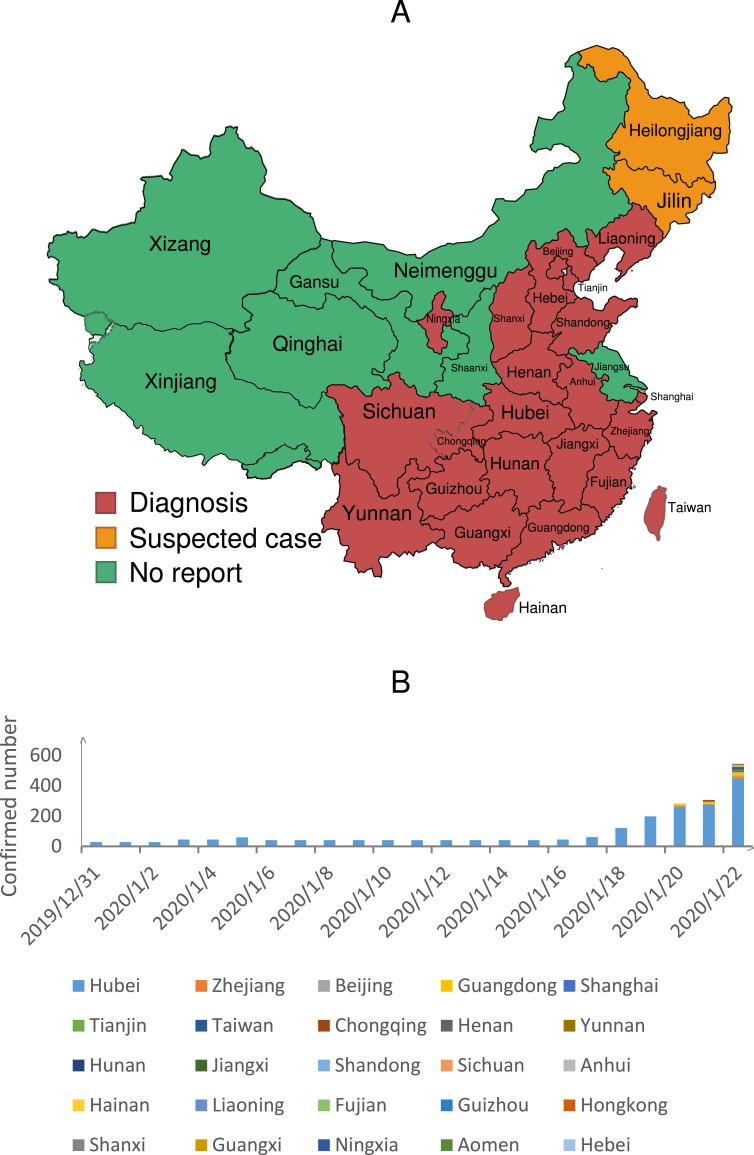
Recently, the emergence of African swine fever virus in China has raised great concern in this journal.1 , 2 The epidemic is not over yet, outbreak of a new SARS-like Coronavirus in Wuhan at the end of 2019 poses new public health challenges in China. Coronaviruses are a large family of viruses that cause respiratory illnesses. Although Coronaviruses (CoVs) have been known for decades, they did not raise great attention in human medicine until the outbreaks of Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS).3 SARS-CoV first emerged in November 2002 in Guangdong province of Southern China and then rapidly spread to 29 countries and regions, infecting over 8000 individuals with a death toll of nearly 800.3 Ten years after the SARS, MERS emerged in 2012, have caused 2494 human infections with 858 deaths (as of November 2019) and remains a disease of global, and particularly Middle Eastern, public health concern. In 2017, a novel HKU2-related bat coronavirus, swine acute diarrhea syndrome coronavirus (SADS-CoV), caused the death of 24,693 piglets,4 , 5 raising further concern about these coronavirus. In December 2019, a new coronavirus (2019-nCoV), which is about 70% similar to SARS-Cov, was discovered in the central Chinese city of Wuhan, with 545 cases in 25 provinces being diagnosed (Fig. 1 A, January 22, 2020). According to the World Health Organization (https://www.who.int), in addition to mainland China, this virus has also been detected in Japan, Thailand, Republic of Korea and the United States in travelers from Wuhan. The number of confirmed cases has been gradually increasing (Fig. 1B), which might be partially due to recent the establishment of relevant detection methods, but, at the same time, could signal that there could be a quick expansion of the epidemic.
Fig. 1.
The 2019-nCoV epidemic.
(A) Spread of 2019-nCoV in China. Provinces with 545 confirmed 2019-nCoV cases to date (1/22/2020 8:00 PM) are indicated in red (total 25). Provinces in yellow have suspected cases, while those in green have reported no cases. The epidemic is mainly distributed in central and southern China.
(B) Increasing number of cases. The number of confirmed cases by date are plotted, which shows a gradual increase over time.
The three basic elements required for an infectious disease epidemic are source of the infection, route of transmission, and susceptible hosts (humans). Eliminating the source of the infection and cutting off the transmission route are usually effective means to block the spread of an infectious diseases. The successful precedent of emergent CoV containment based on the elimination of the primary reservoir is SARS. Bats are suggested to be the reservoir hosts of SARS-CoVs.6 , 7 However, without an intermediary host, bat derived CoVs cannot directly infect humans. The Carnivora–intermediate amplifying host (civets) of SARS-CoVs was found.8 The quick control of the intermediate amplifying host by banning wild animal trade was the key factor in the effective control of SARS. Many of the 2019-nCoV infected people had either worked at or frequently visited a seafood market in Wuhan. Apart from fish, the market also sold other live wild animals – sparking concerns that the virus might have been transmitted from an animal to humans, just like SARS and MERS. The Wuhan CoVs cluster with SARS/SARS-like coronaviruses, and have about 70% overall genome sequence similarity. As this virus clusters with various bat Betacoronavirus, it is reasonable to deduce that bats are the native host for the 2019-nCov. However, its origin (source) remains unknown, that is, the animals that are the origin and amplifying hosts for this virus. This makes the elimination of this disease from the source difficult.
The control of the route of transmission is another effective means of epidemic control. Infections of healthcare workers and family clusters suggest that 2019-nCoV has the ability to spread from human to human. Although the seafood market has been closed, the number of confirmed cases continues to gradually increase (Fig. 1B). This further supports the conclusion that this virus can spread by human-to-human contact. Previous research has shown that coronaviruses usually have an ability to rapidly mutate.9 We cannot exclude the possibility that some 2019-nCoVs will mutate to become “super-spreaders”, as seen in SARS. Frequent disinfection of people-intensive places and animal trading markets should help stop the spread of the virus. Unfortunately, the outbreak has cast a shadow over the celebrations for the Lunar New Year, which falls on January 25. Millions of people in China travel over the course of the Spring Festival, both within the country and overseas. In addition, millions of college students (Wuhan has more than 1 million college students) will return to school after the winter vacation. This large-scale population migration could lead to further spread of this virus. Therefore, it is possible that this epidemic will be more serious after the Spring Festival. At present, the National Health Commission of China (http://www.nhc.gov.cn) has listed 2019-nCoV as Class B infectious disease and managed as Class A. Measures including the temperature monitoring of passengers at railway stations, airports, terminals and other transportation hubs have been carried out, which should slow the spread of the virus to a certain extent. However these disease control policies are far from enough, more strict methods to control population flow should be on the table.
It is increasingly recognized that a One Health approach at the human–animal–ecosystem interface is needed for effective investigation, prevention and control of any emerging zoonotic disease. In the context of emerging zoonoses, human and veterinary medicine must work together. These viruses emerge from animal trading markets, where veterinary workers need to be vigilant and concerned whether these viruses have the potential to spread between animals and the impact the livestock industry. Veterinary workers should also actively invest in epidemiological investigations to find the natural hosts of these viruses and develop corresponding detection methods. Considering that the original source of the 2019-nCoV is very likely related to a wild animal, additional strict laws to limit wildlife markets should be made, otherwise, more emerging zoonoses from wild animals will occurred in the near future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31822056), Fund for the key program and Creative Research Group of the Department of Education of Guangdong Province, and the 111 Project (D20008).
References
- 1.Shen X., Pu Z., Li Y., Yu S., Guo F., Luo T. Phylogeographic patterns of the African swine fever virus. J Infect. 2019;79(2):174–187. doi: 10.1016/j.jinf.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Li X., Xiao K., Zhang Z., Yang J., Wang R., Shen X. The recombination hot spots and genetic diversity of the genomes of African swine fever viruses. J Infect. 2020;80(1):121–142. doi: 10.1016/j.jinf.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):E59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong L., Li J., Zhou Q., Xu Z., Chen L., Zhang Y. A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg Infect Dis. 2017;23(9):1607. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H. Bats are natural reservoirs of SARS-Like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 8.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 9.Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.-.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]