Graphical abstract

Keywords: RNA structure and function, RNA interactions, Epitranscriptomics, Small molecules, RNA-based therapeutics, RNA in disease, Drug target
Abstract
The increasing appreciation for the crucial roles of RNAs in infectious and non-infectious human diseases makes them attractive therapeutic targets. Coding and non-coding RNAs frequently fold into complex conformations which, if effectively targeted, offer opportunities to therapeutically modulate numerous cellular processes, including those linked to undruggable protein targets. Despite the considerable skepticism as to whether RNAs can be targeted with small molecule therapeutics, overwhelming evidence suggests the challenges we are currently facing are not outside the realm of possibility. In this review, we highlight the most recent advances in molecular techniques that have sparked a revolution in understanding the RNA structure-to-function relationship. We bring attention to the application of these modern techniques to identify druggable RNA targets and to assess small molecule binding specificity. Finally, we discuss novel screening methodologies that support RNA drug discovery and present examples of therapeutically valuable RNA targets.
1. Introduction
Recent advances in the field of RNA biology has led to the discovery of a plethora of coding and non-coding (nc) transcripts with novel modes of action,1 new types of RNA epitranscriptomic modifications,2, 3 and an unforeseen diversity of mechanisms that lead to the production of RNAs.4 These discoveries prompted the parallel development of modern molecular techniques that provide in-depth insights to the RNA structure and function.5, 6, 7 Biophysical characterization of RNA motifs with X-ray crystallography,8 nuclear magnetic resonance,9 and cryo-electron microscopy10 has yielded a solid foundation for understanding the chemical and structural basis of RNA functions at atomic resolution. However, experimental requirements of these methodologies often restrict their throughput and suitability for studying conformationally heterogeneous RNAs. In addition, the isolation of RNA motifs from full-length transcript, or the removal of RNAs from their cellular context, may introduce biases in the final conformation. The development of RNA-centric deep-sequencing probing techniques opened up the possibility for the global assessment of RNA structures at a single nucleotide resolution, and in various biological contexts.11, 12, 13 In addition, techniques that address RNA interactions with effector molecules14, 15, 16, 17, 18 prompted the comprehension that a myriad of molecular pathways involve regulatory RNAs while also facilitating the development of high affinity ligands to interfere with these processes. These innovations created a new paradigm in the field of biomedicine, where RNA is viewed as a novel and largely unexplored drug target.
2. RNA as a drug target
The vast diversity of RNAs expanding beyond coding transcripts to several classes of ncRNAs that vary in length, biogenesis, polarity, and putative functions, increases the repertoire of druggable targets.19, 20 Here, specific RNA properties contribute to its attractive, yet challenging makeup as a target molecule. RNA can form complex three-dimensional structures through canonical Watson-Crick base pairing and complex tertiary interactions that are mediated by non-canonical bonds. Such structures can be as intricate and stable as those formed by proteins and can recognize small-molecule ligands, other nucleic acids, and/or proteins with high affinity and specificity.21, 22, 23, 24
At the same time, the highly dynamic conformation and repetitive character of its surface presents difficulties for drug design.25, 26 In addition, many putative small molecule-binding pockets in RNA are much more polar and solvent exposed than binding sites on proteins, complicating ligand design efforts. Compounding these issues, most target RNAs are expressed at low levels, with the exception of ribosomal (rRNA) and transfer (tRNA) RNAs, which constitute 80–90% and 10–15% of total cellular RNA, respectively.27 Other abundant RNAs, such as messenger (mRNA), small nuclear (snRNA), and small nucleolar (snoRNA) are present at levels that are about 1–2 orders of magnitude lower than rRNA and tRNA. Certain small RNAs, such as micro (miRNA) and piwi (piRNAs) can be present at very high levels; however, this appears to be cell type dependent. One should keep in mind that if the RNA has catalytic activity or if it acts as a scaffold to regulate chromosomal architecture, as it is the case for many long non-coding (lncRNAs), the RNA may not need to be present at very high levels to be able to perform its task. RNA stability is another detrimental aspect that needs to be considered. For example, long non-coding RNAs (lncRNA) have, on average, shorter half-life (median 3.5 h) than messenger RNAs (mRNAs, median 3.5 h).28 Yet, the intergenic, cis-antisense, as well as spliced lncRNAs with cytoplasmic localization tend to challenge that rule.
Efforts to therapeutically harness RNAs began as soon as they were discovered, even though their functionality was not well understood. Early RNA targeting approaches relied on the application of aminoglycosides - potent molecules that bind to bacterial rRNA through electrostatic interactions with, at best, modest selectivity and systemic toxicity.29, 30 Despite their rather unattractive physical, chemical, pharmacological and synthetic characteristics, they remain a pillar of antibacterial treatment. Studies of RNA riboswitch regulatory elements that bind diverse metabolites and regulate gene expression primarily in bacteria further showed that small metabolites can be recognized by RNA with remarkable specificity.31, 32 These discoveries were followed by the development of small molecule screens against functionally defined viral RNA motifs, such as Human immunodeficiency virus-1 (HIV-1) transactivating response element (TAR) RNA and its interacting trans-activator of transcription (Tat) protein,33 HIV-1 Rev response element (RRE) and Rev protein,34 and Hepatitis C virus (HCV) internal ribosome entry site (IRES).10, 35 These studies established that RNA is a legitimate target of small molecules and that ligands can indeed bind discrete binding pockets in RNA, cementing the applicability of small-molecule treatment to target RNAs.
As compared to antisense oligonucleotide-based (ASO) strategies, which pose significant limitations as pharmaceutical agents,36 small molecules offer the unique possibility to target particular RNA structural elements. The chemical and biophysical tunability of small molecules, in addition to their overall superior cellular and tissue permeability, make them an appealing alternative.37 In addition, by recognizing specific hallmarks of RNA fold, small molecules can be utilized as RNA structural probes to yield information on RNA conformation. For example, methidiumpropyl-EDTA (MPE) has been applied as a through-space cleavage reagent for structural interrogation of the folding transition of HIV-1 RRE2 substructures.35
Despite the spectacular advances in the RNA biology, experts in the field are still faced with a long list of considerations. Overall, three major components are required to support the development of efficacious RNA-based small molecule therapeutics: (1) the identification of therapeutically valuable RNA target, (2) the development of a screening method that will establish drug-like molecules against target RNA, and (3) the identification of RNA motifs that accommodate ligand binding with high affinity and specificity. In this review, we will discuss the modern RNA-centric techniques that can be harnessed to elucidate RNA structure-to-function relationship and their potential application to evaluate small molecule specificity. We will shed light on RNA properties e.g. stability, level of expression, structure and biological importance that need to be considered before a given transcript can be designated as druggable. Finally, we will present most recent developments in screening strategies that support identification of RNA-specific ligands, and we will highlight the newest discoveries in the field of therapeutically-valuable target RNAs.
3. Assessing RNA potential as a drug target by biochemical probing
As our knowledge of RNA biology and its importance in regulation of cellular processes increases, it is becoming clear that specific preponderant characteristics of RNA, such as its structure and conformational dynamics, may influence whether a given RNA target can be efficiently targeted with drugs.38 In the past, a wide range of chemicals and enzymes have been used to monitor RNA folding.39, 40 DMS is one of the most well-established chemicals used to study RNA structure. It is highly reactive with solvent accessible, unpaired residues but reliably unreactive with bases engaged in Watson-Crick interactions, thus nucleotides that are strongly protected or reactive to DMS can be inferred to be base-paired or unpaired, respectively. DMS treatment has been more recently coupled to a massively parallel sequencing readout (DMS-seq) by randomly fragmenting the pool of modified RNAs and size-selecting prior to 3′ ligation with a specific adapter oligo.41 Because DMS modifications at adenine and cytosine residues block reverse transcription (RT), sequencing of the fragments reveals the precise site of DMS modification, with the number of reads at each position providing a measure of relative reactivity of that site. DMS footprinting has proven to be a tremendously versatile method and has been applied to a large fraction of known structured RNAs42 as well as to probe the structure of modified RNAs and ligand binding to RNA.43
In recent years, a group of electrophiles, known as SHAPE reagents, have become widely used for high-throughput RNA structural probing.12 These compounds preferentially modify the backbone of RNA in structurally flexible single-stranded regions by reacting with the 2′-hydroxyl group on ribose and generating bulky 2′ O-adducts. The choice of electrophile can be tailored to specific experimental systems. For example, probes that react with RNA rapidly are well suited to obtain a structural “snap-shot” of target RNA,44, 45 while electrophiles with long half-life are suitable for studying RNA slower local motions, shown to have the greater potential to govern RNA folding, ligand recognition, and ribonucleoprotein assembly reactions.46 There is also a wide palette of cell-permeable probes that can be used to interrogate RNA folding in living cells, yielding information on how RNA structure changes in response to various environmental stimuli.11, 12, 47 The probe selection is followed with chemical adduct detection, which involves processing modified RNA samples and then performing reverse transcription (RT) to record probe modifications as either truncations (RT-stop) or mutations (RT-mutate) in the resultant cDNA sequences.
SHAPE and mutational profiling (SHAPE-MaP) is an example of RT-mutate methods that have recently been applied in living cells to resolve the secondary structure of lncRNAs with emerging roles in human diseases. The metastasis associated lung adenocarcinoma transcript 1 (MALAT1), a highly abundant nuclear transcript associated with metastasis in non-small cell lung cancer,48 polyadenylated nuclear (PAN) RNA, the key regulator of Kaposi’s sarcoma associate herpesvirus lytic reactivation,49, 50 and X-inactive specific transcript (XIST)51 a transcript that orchestrates mammalian X-chromosome inactivation (XCI), are just to name a few that led to increased appreciation of the functional capabilities of RNA structure. From the partition function of base pairing, a SHAPE-MaP data-informed Shannon entropy52 can be calculated, which yields information on the diversity of RNA conformation within a given region (Fig. 1 A). In general, regions that have both low SHAPE reactivities and low Shannon entropy are likely to exist in a single structural state. These have been found to be highly correlated with known functional RNA structures,52, 53 and as such they potentially represent valuable drug targets. In addition, SHAPE-MaP allows researchers to study the effects of small molecule binding on native RNA structure. The ΔSHAPE-MaP comparative analysis can yield information on not only whether a small molecule binds, but also which specific nucleotides constitute potential binding sites(s). The effectiveness of this approach has been proven on HIV-1 TAR and thienopyridine, a novel TAR-specific ligand.54 Here, however, the prior version of RNA probing method, namely SHAPE, was performed on in vitro transcribed HIV-1 5′ untranslated region (UTR) to validate the ligand specificity.
Fig. 1.
Structural and functional aspects of target RNAs for informed design of small molecule therapeutics. [A] SHAPE-MaP data-informed Shannon entropy yields information on RNA alternative conformation. Structurally well-defined motifs constitute valuable RNA targets. The ΔSHAPE-MaP comparative analysis can identify specific nucleotides that constitute ligand binding sites. [B] The interactions of target RNA with effector molecules expand the desirable structural space for ligand binding. [C] RNA epitranscriptomic modification, e.g. m6A affects RNA structure and RNA-binding sites.
Coupling systematic mutagenesis with high-throughput chemical mapping during two-dimensional mutate-and-map approach expands the information content of RNA chemical mapping.55, 56 Mutate-and-map seeks to determine not just chemical reactivities at each nucleotide but also how these reactivities are affected by systematic perturbations – nucleotide mutations, thus providing the insight into its tertiary conformation as well as validating its predicted structure. In practice, some mutations might not lead to the desired ‘release’ of the pairing partners, and some mutations might produce larger perturbations, such as the unfolding of an entire helix. Nevertheless, if even a subset of the probed mutations leads to precise release of interacting nucleotides, the base-pairing pattern of the RNA could potentially be read out from this extensive data set.
Still, the biochemical methods for RNA structure probing in living cells generate averaged reactivity profiles and have a limited capacity to capture the complexity of RNA structures that include long-range structures, pseudoknots, and alternative conformations. Psoralen analysis of RNA interactions and structures (PARIS) address these challenges by directly identifying base-paired helices and RNA-RNA interactions.16 In this method, cross-linking is performed with 4′-aminomethyl trioxsalen (AMT) that intercalates in RNA helices and, upon photo-activation, crosslinks the two strands, with a preference for staggered uridines.57 RNase and proteinase digestion followed by 2D gel electrophoresis ensure that the identified crosslinks are representing the directly base-paired RNA fragments. Proximity ligation of duplex RNA fragments, photo-reversal of crosslinks, and high throughput sequencing ultimately reveal the direct base pairing between fragments. Each PARIS read gives individual-molecule evidence of a duplex between two RNA fragments. The multiplicity of PARIS reads can thus reveal a single common structure, multiple alternative structures, or interactions between two RNAs in trans. As such, PARIS can provide information on RNA prevalent alternative conformation that, similarly to Shannon entropy, can be used to avoid structurally dynamic and poorly defined RNA regions for small molecule targeting.
The relative solvent accessible surface area is one of the parameters that has been established previously to predict the magnitude of binding-induced conformational changes from biomolecule structures.58, 59 The novel chemical probing methodology, namely Light Activated Structural Examination of RNA (LASER), informs on RNA solvent accessibility, and as such provides an additional layer of structural information, particularly in RNA-ligand complexes.60 LASER uses a light-generated nicotinoyl nitreniumion (NAz)47 to form covalent adducts with the C8 position of adenosine and guanosine, which can be quantified by either RT-stop or RT-mutate deep-sequencing. The authors verified the applicability of the method for ligand binding detection by probing the binding of an antimicrobial peptide on rRNA.
In addition to the above-mentioned methods, various bioinformatic tools, including Rsample61 and Swellix,62 can provide insights to RNA alternative conformation. Rsample considers that multiple copies of the same sequence can simultaneously fold into different conformations and focuses on the agreement between experimental mapping data and estimated mapping data by sampling RNA structure models, rather than interpreting data in their absence. This technique provides a principled approach for integrating thermodynamic prediction with mapping data. On the other hand, Swellix combines all possible helices with a combinatorial approach to the RNA folding problem in order to compute all possible non-pseudoknotted RNA structures. The software can include experimental constraints on global RNA structures, such as data from pairing constraints in phylogenetic analysis, SELEX experiments, or chemical and enzymatic probing experiments, as well as thermodynamic parameters. Both bioinformatic methods represent complementary tools to analyze RNA sequences with multiple folds. Overall, the abovementioned methodologies led to much-improved appreciation of the ubiquity and functional capabilities of RNA structures. As they provided not only the insight into RNA secondary, tertiary structure and conformational changes, they can inform on potential features for a given interaction between target RNA and small molecules and verify the specificity of such interaction.
4. Tackling RNA interactions
The diverse roles of RNAs, particularly ncRNAs, are often a consequence not only their complex structures, but most importantly their structure-mediated interactions with effector molecules, e.g. other RNAs, DNAs and proteins (Fig. 1B).51, 52, 53, 54, 55, 56, 57 These intermolecular contacts increase possible targetable structures – e.g. structure space for ligand binding, which might be missing with some less structured RNAs.
Recently, several biochemical methods have been developed to yield information on RNA interacting partners. RNA antisense purification (RAP-RNA)14 relies on the application of crosslinking reagents that fix endogenous RNA-RNA complexes in living cells, followed by RNA-specific affinity purification with biotinylated antisense oligonucleotides and high-throughput sequencing. RAP-RNA can be performed using various crosslinking reagents to differentiate between target RNA direct and indirect contacts with other transcripts. For instance, direct RNA-RNA interactions are specifically captured by UV and AMT crosslinking, while indirect interactions, involving protein intermediates, can be captured by crosslinking with formaldehyde. Compared to other similar techniques, the most distinctive feature of RAP is the use of long capture probes that tile across the target RNA, allowing for stringent hybridization and wash conditions that significantly reduce nonspecific background.
RAP method has been combined with quantitative mass spectrometry (RAP-MS) to identify proteins that directly interact with a specific RNA in vivo. 63 RAP-MS uses UV crosslinking to create covalent bonds between directly interacting RNA and protein, and purifies RNAs with target specific oligonucleotides in denaturing conditions to disrupt non-covalent interactions. This approach, which is used by other methods such as crosslinking and immunoprecipitation (CLIP),64 is known to identify only direct RNA–protein interactions and to separate interactions that are crosslinked in the cell from those that associate in solution. The application of RAP-RNA and RAP-MS revealed that Functional Intergenic Repeating RNA Element (FIRRE) interacts with a ∼5 Mb region that flanks their transcription site and with the nuclear matrix factor hnRNPU during the process of anchoring the inactive X chromosome to the nucleolus.65 Also, RAP-MS provided insight into novel protein factors that facilitate the activity of XIST the transcript that orchestrates XCI by coating and silencing one X chromosome in females.63 RAP-MS not only allowed to recognize proteins that activate histone deacetylase, but also components that recruit the polycomb repressive complex 2 (PCR2) across the X and exclude RNA polymerase II from the inactive X.66 This data, together with recently resolved secondary structure of the entire 18 kb XIST RNA,51 provides valuable findings that can accelerate small molecule targeting of XIST in various diseases.67, 68
Chromatin isolation by RNA purification (ChIRP),17 domain-specific chromatin isolation by RNA purification (dChIRP),69 and capture hybridization analysis of RNA targets (CHART)70 are techniques used to identify genomic binding sites of RNAs by purifying formaldehyde–crosslinked complexes through the use of short oligonucleotide probes directed against specific RNAs. The main difference between these techniques is that probes for ChIRP tile the entire target RNA, while probes for dChIRP are used as specific pools to characterize RNA at the domain level, and probes for CHART are experimentally determined after an RNase H assay, which defines the accessible hybridization regions. ChiRP has been applied to reveal the genomic occupancy of two therapeutically valuable lncRNAs, namely telomerase RNA (TERC) and HOX-transcript antisense RNA (HOTAIR). HOTAIR has been shown to function as a key regulator of chromatin states and dynamics by recruiting and affecting PRC2 occupancy on genes genome-wide. It has been associated with tumorigenesis, growth, invasion, cancer, stem cell differentiation, metastasis, and drug resistance, and as such it is considered as a biomarker for diagnostic and therapeutic purposes in several cancers.71, 72 TERC is a critical RNA component of telomerase polymerase that serves as a template for the enzyme telomerase reverse transcriptase (TERT) to elongate telomeres. Variants and copy-number changes at the TERC locus have been associated with cancer risk and progression73 and neurodegenerative diseases.74 Given the ability of small molecules to detect RNA structural elements and target specific biomolecular interactions, these tools are perfectly poised to expand the palette of achievable drug targets.
5. Epitranscriptomics, multi-layered world of RNA modifications
Recent studies have identified over 100 distinct epitranscriptomic RNA modifications, with the most prevalent involving the addition of a methyl group at certain positions on the nucleobase, as in N6-methyladenosine (m6A),75 5-methylcytosine (m5C)76 and N1-methyladenosine (m1A).77 Other predominant epitranscriptomic modifications include isomerization of uridine to pseudouridine (Ψ), ribose modifications, and adenosine-to-inosine editing.78 These post-transcriptional modifications expand RNA functional capacity beyond what can been recognized from its sequence and structure alone79 and, at the same time, offer an avenue of increasing specificity for small molecule targeting. Known to affect RNA conformation,79 function,80, 81 and metabolism,82 epitranscriptomics should be considered while developing adequate tools to study RNA druggability.
The different positions of methylation carry distinct structural consequences for RNA. For example, m6A destabilizes pairing with uracil by altering the energetics of the AU pair through steric hindrance, but without affecting the pattern of hydrogen-bonding donors and acceptors.83 The presence of m6A reduces RNA base pair stability both in vitro and in vivo,84 creating binding sites for proteins that preferentially recognize modified bases (Fig. 1C).83 Distinct human diseases, including developmental disorders,85 cancer and viral infections86 have been associated with dysregulation of m6A patterns in various cellular transcripts. Also, two m6A sites have been detected in the stem loop II of HIV-1 RRE,87 a viral RNA motif that has been the focus of many current and past targeting efforts,34 forcing scientists to rethink their strategies for effective drug-design. The regulatory functions of m5C are not yet fully understood.88 It has been shown, however, that in vitro cytosine-5 methylation can affect Mg2+ binding to tRNA molecules, which influences the anticodon stem loop conformation and stability of the secondary structure.89 Also, synthetic cytosine-5 methylated mRNAs exhibit increased stability, and the loss of methylation in the 3′ UTR of p16, an mRNA encoding tumor suppressor, has been reported to reduce its stability.88 Thus, it is likely that m5C affects RNA stability and function in living cells.76 The m1A modification carries a positive charge that protrudes from the Watson–Crick hydrogen-bonding face of adenine, resulting in the nucleotide remaining unpaired, and thus can dramatically alter protein-RNA interactions and RNA secondary structures through electrostatic effects. The m1A maps uniquely to positions near the translation start site and first splice site in coding transcripts, and it correlates with upregulation of translation.77 Isomerization of uridine, known as pseudouridine (Ψ),90 affects RNA secondary structure by increasing backbone rigidity and base stacking, due to a preference of the 3′-end conformation.91 The precise pseudouridylation pattern and their role in human RNAs remains only partially uncovered. However, based on the heat shock induced Ψ deposition in yeast mRNAs, it is assumed that Ψ increases mRNA stability by preventing the melting of RNA structures that protect the 3-end at increased temperatures.92
High-throughput mapping of modified nucleosides is now possible due to advances in deep-sequencing technologies.93 In general, these techniques fall into three categories: (1) immunoprecipitation of fragmented RNAs using modification-specific antibodies followed by sequencing, which has been used for mapping m6A (meRIPseq)93 and m1A (RIPSeq)77; (2) chemical treatment of RNA prior to sequencing, which exploits the differential reactivity of modified bases such as using sodium bisulfite for detection of m5C (Bisulfate-seq)88 or CMC (N-cyclohexyl-N9-(2-morpholinoethyl)-carbodiimidemetho-p-toluenesulphonate) for detection of pseudouridine (Ψ-seq)92; and (3) non-random mismatch signatures in RNA sequencing data produced during reverse transcription, which has been applied to m1A and m6A (miCLIP) modified DNA. To date, these technologies have produced transcriptome-wide maps for the above-mentioned epitranscriptomic modifications, while concurrently supporting the epitranscriptomic characterization of specific coding and nc transcripts with known therapeutic value.94
Furthering our understanding of the discriminatory potential of specific epitranscriptomic modifications holds immense promise for the informed design of RNA-based small molecule therapeutics. Also, since RNA modifications are known to contribute to immune system function by suppressing the signaling of innate RNA sensors,95 these findings have contributed to the ‘second coming’ of RNA therapeutics.96 Developing small-interfering RNAs, RNA-based vaccines, or mRNA therapeutics regularly includes modifying component RNA strands, which decreases nuclease sensitivity and reduces the activation of the innate immune response.
6. Druggable RNA targets
Until the last decade, the majority of small molecule based therapeutic strategies have been focusing on targeting proteins, mainly because the study of protein structure and function significantly predates that of RNA. Soon enough, the RNA revolution revealed that RNA function is complex, directly related to RNA folding, and often critical for the therapeutic interventions we pursue. To date, small molecules have been found to regulate approximately 30 unique disease-associated mammalian RNAs apart from the ribosome.97 These RNAs are known to be involved in fine-tuning of many cellular processes, including cellular differentiation, homeostasis, immune response and proliferation.98, 99, 100 Many of them contain conserved cis- and trans- acting motifs that act as scaffolding sites for other RNAs and proteins, influencing their subcellular localization, stability and functionality.101 Frequently these interactions are severely dysregulated in a variety of human disorders.102 In addition, many human diseases are caused by RNA viruses or DNA viruses whose transcripts dysregulate cellular functions, further expanding the palette of potential RNA targets. In this section, we highlight important RNA structural elements that have been at the forefront of RNA-targeted small molecule therapeutics and report recently discovered small molecules that modulate these disease-associated RNA elements.
6.1. miRNAs
miRNAs are post-transcriptional gene regulators that bind to the 3′ UTR of their target mRNA to inhibit translation or cause miRNA-mediated degradation. The biogenesis of these transcripts begins with nucleus localized primary miRNA (pri-miRNA) hairpins that are digested by Drosha to form precursor miRNA (pre-miRNA). The pre-miRNA is subsequently transported to the cytoplasm for further processing by Dicer, followed by loading of the duplex miRNA into the RNA-induced silencing complex (RISC).103 Dysregulated miRNAs contribute to a variety of human diseases,104 including cancer,105 and therefore they serve as potential small molecule target. For cancer research purposes, miRNAs can be divided into those that are over-expressed, which target tumor suppressor proteins, and those with decreased expression in cells, which target oncogenes. The principal goal of miRNA-targeting is to identify compounds that potently and specifically bind to miRNAs and/or its upstream precursors, thereby decreasing their levels.
In 2018, Li et al. described a novel small molecule, bleomycin A5 conjugate, which selectively binds to the precursor of miR-96106 (Fig. 2 A). OncomiR-96 is found to be upregulated in many breast cancers and has been associated with breast cancer proliferation, migration, and invasion in vitro, as well as tumor growth in vivo.107 The novel chemotype specifically inhibits Drosha processing of pri-miR-96, leading to the upregulation of its target FOXO1 and the induction of apoptosis in Michigan Cancer Foundation-7 (MCF-7) breast cancer cells. OncomiR-21 is another crucial cancer-associated miRNA that plays a pivotal role for the initiation, progression, and metastasizing of various cancers.108 In 2012, Gumireddy et al. discovered diazobenzene derivative, a small molecule that inhibits the transcription of pri-miR-21.109 Also, in 2017, Liu et al. described the novel small molecule sophocarpine (SC), which downregulates miR-21 expression in head and neck squamous cell carcinoma (HNSCC) by blocking Dicer-mediated miR-21 maturation in a dose-dependent manner (Fig. 2A).110 SC inhibits HNSCC cell proliferation, invasion, and metastasis through increased expression of tumor suppressors without tissue toxicity or damage, showing its promise as a small molecule anti-tumor therapeutic.
Fig. 2.
Small molecule binders of various RNA targets. [A] Ligands targeting miRNAs. Compound 1 and Compound 2 target oncomiR-21,113 Sophocarpine,111 Bleomycin A5 conjugate,113 and Targaprimir-96115 target oncomiR-96. Targaprimir-210 targets oncomiR-210.115 [B] Chemotypes, RG7800116 and LM1070116, 117 target alternative RNA splice site of SMN2 gene. [C] BRACO-19 and RHPS4 are G4-RNA-specific ligands used in G4RP-seq methodology.117 Compound 4 disrupts G4-RNA in the expanded repeat in ALS. [D] Chemotypes targeting viral RNA motifs, i.e. HIV-1 TAR,54, 119 SARS-CoV pseudoknot121, 122 and HCV IRES.10, 123
Despite the reported successes in miRNA-targeting, no small molecules are undergoing preclinical testing.113 This disparity can be attributed to similar secondary structures among different miRNAs and other cellular RNAs, that limits selectivity and specificity for RNA targets. Yet, we might be just at the verge of resolving that issue. For example, Costales et al.111 have recently identified a small molecule that binds two targets, the precursor hairpins of miR-515 and miR-885, which share a common target motif. In this new study, the group optimized the dual-selective molecule to bind only one of the targets. By using secondary structures of the two binding sites, the model identified an additional, adjacent binding site only present on miR-515, and used fragment-based assembly to create a molecule with over 3200-fold selectivity over the parent compound for the new site. The new molecule, Targaprimir-515, only bound miR-515 containing the original target site and the adjacent site. MiR-515 represses sphingosine kinase 1 (SK1), a key enzyme that phosphorylates sphingosine – and its downstream target S1P, thereby blocking a pathway that promotes cell proliferation and migration in human breast cancer.
6.2. RNA splice sites
The mechanism of pre-mRNA splicing is complex and requires interaction of the pre-mRNA molecule, small nuclear ribonucleoproteins, and splicing factor proteins through cis-acting regulatory sequence elements, trans-acting protein factors, and various cellular responses.123 To add another layer of complexity, alternative-splicing of pre-mRNAs will lead to the assemblage of various protein isotypes. Thus, mutations within splice sites and disrupted regulatory elements can result in loss of function, reduced specificity, and/or truncation of a protein, which are frequently associated with human diseases.124
Spinal Muscular Atrophy (SMA) is a severe neurodegenerative disease caused by a loss or dysfunction of survival motor neuron (SMN1) genes and is the leading cause of genetic-related infant death (Fig. 3 A).125 The loss of SMN1 results in the degradation of lower motor neurons in the spinal cord and causes symptoms such as hypotonia and severe weakness due to neuromuscular degradation. Interestingly, the sequence of the survival motor neuron 2 (SMN2) gene differs from SMN1 by a single nucleotide. This single nucleotide change increases the rate of exon 7 exclusion during splicing events, thus SMN2 can produce little to no functional SMN protein. Increasing exon 7 inclusion in SMN2 mRNA with a small molecule could be therapeutically beneficial. PTC Therapeutics discovered a molecule, RG7800, which increased the production of the inefficiently spliced SMN2 in a FRET-based assay (Fig. 2B).126 RG7800 was the first small molecule splicing modifier to enter clinical trials. The drug is being tested in Phase 1b/2a trials in SMA patients, although it is currently on hold due to safety findings.115 Novartis (Basel, Switzerland) also identified a small molecule, LMI070,122 which works via stabilization of the transient double-strand RNA structure formed by the SMN2 pre-mRNA and a small nuclear ribonucleic protein U1.125 LMI070 binding causes more efficient splicing of the SMN2 gene and therefore increases levels of functional SMN proteins. The ongoing first-in-human study (NCT02268552) is evaluating the safety, tolerability and potential benefit of LMI070 in type 1 SMA patients between 1 and 7 months of age. These discoveries demonstrate the feasibility of small molecule-mediated RNA-selective splice modulation, as well as the potential for leveraging this strategy in other splicing diseases such as cancer.127
Fig. 3.
Druggable RNA targets. [A] Mutated RNA splice sites can result in loss of function of a protein, e.g. survival motor neuron (SMN1) in Spinal Muscular Atrophy (SMA). RNA-specific ligand, e.g. LMI070 causes more efficient splicing of the SMN2 gene and therefore increases levels of functional SMN proteins. [B] Examples of RNA repeat expansion diseases (EPM – Unverricht–Lundborg disease, SCA – Spinocerebellar ataxia, TBP – TATA binding protein associated diseases, DRPLA – Dentatorubral-pallidoluysian atrophy) and the a ‘trans-dominant’ model of RNA toxicity. The RNAs that contain expanded repeats interact with different RNA-binding proteins (colored shapes) which interfere with their functions leading to abnormalities. [C] RNA G-quarduplexes are predicted to form in mRNA encoded by several oncogenes, current RNA-targeting efforts focus on identifying small molecules with high selectivity for the motif. [D] Examples of viral RNA motifs, e.g. HIV RRE-Rev, TAR-Tat, HCV IRES/Ribosome.
Another promising example of splicing-targeted small molecules is GQC-05, an ellipticine G4 ligand, which targets the two alternative 5′ splice sites of apoptosis regulator Bcl-X pre-mRNA.128 GQC-05 binding to Bcl-X pre-mRNA causes the direct alteration of the splicing sites Q2 and Q5 signified by a 15-fold fluorescent intensity increase and a shift in emission peak upon interaction of GQC-05 and the pre-mRNA, consistent with direct binding. The interaction of this small molecule with Bcl-X pre-mRNA affects the overall secondary structure close to the XS and XL sites at the Q2 and Q5 regions, respectively, to favor the formation of stable G4 conformations over the stem loop structure. Additionally, GQC-05 antagonizes the anti-apoptotic isoform of Bcl-X in favor of the pro-apoptotic isoform, showing the small molecule’s ability to induce apoptosis.
6.3. RNA expansion elements
Microsatellite diseases are a class of neurological and neuromuscular disorders caused by repeat expansions.129 These expansions can be located in (1) open reading frames (Huntington’s disease, HD), (2) untranslated regions (myotonic muscular dystrophy type 1, DM1; fragile X–associated tremor ataxia syndrome, FXTAS; and fragile X syndrome, FXS), and (3) introns (amyotrophic lateral sclerosis, ALS; and myotonic dystrophy type 2, DM2) (Fig. 3B).
Various groups have tackled targeting RNA repeat expansions with small molecules. The well-studied RNA repeat expansion is the trinucleotide repeat that causes DM1, r(CUG)exp, which binds and sequesters the RBP splicing regulator muscleblind-like 1 (MBNL1), among others.130 In 2009, Warf et al. identified a small molecule, pentamidine, which disrupted the MBNL1-CUG repeat complex in DM1.131 When pentamidine disrupts the CUG RNA foci, it releases the MBNL1 protein from sequestration and reverses aberrant alternative splicing that is commonly seen in DM1 mis-spliced pre-mRNAs. Originally, pentamidine was believed to bind to the r(CUG)exp. However, when Coonrod et al. attempted to identify the exact mechanism, biophysical experiments failed to demonstrate significant affinity between pentamidine and CUG RNA repeat.132 Through a series of experiments, scientists suggested that pentamidine may interact with the CTG*CAG repeat DNA to inhibit transcription; however, the mechanism of action for this small molecule is still unknown. Using a structure of r(CUG) repeats, the Zimmerman laboratory designed a small molecule comprised of an acridine and a triaminotriazine that interacts with the RNA via Janus-wedge hydrogen bonding.133 Optimization of the first-generation compound resulted in three bioactive compounds that improved mis-splicing and formation of nuclear foci in both DM1 cellular and Drosophila models.134 Furthermore, the Disney group has also been very active in identifying inhibitors of different RNA repeats including transcripts involved in both DM1 and DM2, FXTAS, and, most recently, amyotrophic lateral sclerosis (ALS).135 They reported a substituted naphthyridine that was identified by high-throughput screening and shown to inhibit the r(CUG)exp-MBNL1 interaction using a FRET-based assay.136 In cell models, the chemotype improved DM1-associated pre-mRNA splicing defects and caused reductions in nuclear foci formation. Subsequently, the compound has also been identified as an inhibitor of miR-544.137 Finally, the Disney laboratory also identified benzimidazoles by screening of an RNA-focused small-molecule library, as well as oligomeric Hoechst dye-like compounds that bind r(CUG)exp in cells and have promising effects on DM1-associated defects in cell models.138
The major advantage of targeting RNA expansion elements is that their structures are well documented leading a path forward for the effective design of multivalent molecules with high specificity for their target.139 Additionally, many repeat expansion disorders affect the central nervous system, where small molecules can more easily penetrate than competing antisense oligonucleotides. Currently, two therapeutic companies, namely Ribometrix and Arrakis Therapeutics Inc. lead the way in targeting RNA expansion elements.140 Ribometrix is pursuing Huntington’s disease (HD) and is targeting MYC mRNA for cancer, while Arrakis Therapeutics Inc. has programs on undisclosed repeat expansion disorders in neurology, as well as programs in oncology and rare genetic diseases.
6.4. RNA G-Quadruplexes
RNA can fold into alternate four stranded secondary structures in regions rich in guanine, called G-quadruplexes (G4-RNA, Fig. 3C).141 Each G-tetrad has four guanines arranged in a square planar arrangement and held together by Hoogsteen hydrogen bonding. Further stabilization of each G4-RNA is then achieved through the presence of a monovalent cation, most often potassium, which is localized in the center between each pair of tetrads.141 G-quadruplexes has been first described in DNA,142 yet there are key structural differences between DNA and RNA G4s that may further the therapeutic targeting of G4-RNA. In general, RNA G4s have higher stability attributed to differences in hydration and increased intramolecular hydrogen bonding due to 2′ of ribose sugar.143 Another key difference between DNA and RNA is the presence of a methyl group in uracil which is absent in its counterpart thymine present in DNA. Substitution of 2′ —OH by chemical analogues also has shown to destabilize G-quadruplexes further highlighting its importance in G-quadruplex structure.144 Utilizing these differences between RNA and DNA can increase the selectivity and specificity of available G-quadruplex ligands towards RNA.145 Recent publication by Kwok et al. provided a comprehensive overview of predictive algorithms and structure-based sequencing methods that can be utilized for transcriptome-wide detection of G4-RNA.146
Since G4-RNA are predicted to form in mRNA encoded by several oncogenes,141 current RNA-targeting efforts focus on identifying small molecules with high selectivity for the motif. A proof-of-concept was established using a reporter construct comprising the 5′ UTR of neuroblastoma RAS oncogene, which included the G-quadruplex element upstream of the firefly luciferase gene.147 The authors showed that the RR110 small molecule inhibits translation of the G-quadruplex-containing mRNA with high affinity and selectivity for the motif. Using a biotin-streptavidin pull-down assay, Xodo and colleagues identified small molecules that bind to G4-RNA in the 5′ UTR of Ki-ras2 Kirsten rat sarcoma (KRAS) oncogene.148 The human KRAS is mutated in ∼95% of patients with pancreatic ductal adenocarcinoma (PDAC),149 leading it to be considered a crucial target for anticancer drugs. The authors identified two compounds, anthrafurandiones and anthrathiophenediones, that suppressed luciferase expression from expression constructs, strongly induced apoptosis, and reduced both the metabolic activity and colony formation of Panc-1 cells carrying mutant KRAS.
Recently, a new small molecule chemotype has been described to bind to and inhibit G4 RNA formation in the expanded G4C2[(G4C2)exp] repeat of C9ORF72 in ALS.150 The RNA transcribed from this repeat expansion sequesters RNA-binding proteins that produce toxic dipeptide. The identified compound has been shown to bind to the GG internal loop in the hairpin structure of r(G4C2)exp to prevent the formation of G4 RNA. These results highlight the power of rationally designed small molecules to target therapeutically relevant RNA structures.
Due to their elusive nature, verifying the existence and function of RNA G-quadruplexes in biological systems has been a challenging task. However, a newly developed method for transcriptome-wide identification of RNA G-quadruplexes utilizing motif-specific small molecules has been recently reported.117 G4-RNA-specific precipitation with sequencing (G4RP-seq) uses a chemical crosslinking step, followed by affinity capture with BRACO-19 and RHPS4 chemotypes (Fig. 2C), and target identification by sequencing to capture global snapshots of transiently folded G4-RNA. This technique provides insight into the G4-RNA landscape in the human transcriptome, as well as a means for studying the dynamics of in vivo dynamic formation of G4-RNA under various biological conditions, in addition to the activity of G4-RNA-specific ligands.
Another challenge related to G-quadruplexes targeting is the limited selectivity of their typically large and flat aromatic ligands.151 These ligands are often decorated with protonated side chains which are necessary for loop and groove interactions but lower the overall selectivity of the molecule. Recent developments in crystallography and bioinformatic analyses may be the key into unlocking unique features of G4 structures, leading to the development of specific and potent ligands.152, 153
6.5. Viral RNA motifs
Viral genomes are highly economical when it comes to the genetic information they encode and, as such, provide only a narrow number of protein targets for therapeutic intervention. Recent studies aiming at elucidation of the structural conformation of viral RNA genomes52 and virus-encoded coding and nc transcripts49 greatly expanded the repertoire of drug targets.
HIV remains the most thoroughly studied virus, for which multiple RNA motifs have been investigated as drug targets (Fig. 3D). These include TAR154 and RRE,155 but also the dimer initiation sequence (DIS),156 the packaging signal (Ψ),157 and the Gag/Pol frameshifting signal.34 The TAR element resides within the 5′ UTR of the viral genome and serves as the binding site for the Tat protein. Formation of the TAR/Tat complex stimulates transcription elongation to yield full-length viral transcripts.33 A fragment-based screening approach to identify ligands of the HIV TAR has been described by Göbel and colleagues, who interrogated a set of 29 small molecules that were selected to represent molecular motifs beneficial for RNA recognition.118 Also, Benhida and coworkers pursued a design approach for TAR RNA‐binding ligands based on amino-phenylthiazole derivatives.54, 158 More recently, Schneekloth and colleagues have applied small molecule microarray (SMM) screening of a fluorescently labeled TAR hairpin to identify selective chemotypes (Fig. 2D).54 Other frequently targeted HIV motif include RRE, which is located in the second intron of the viral genomic RNA and serves as a high‐affinity binding site for the Rev protein. Rev binding to RRE is required for the nucleocytoplasmic export of full‐length and singly spliced viral transcripts.87 Only two studies report small molecules targeting the Rev-RRE complex.35 None of these approaches have produced inhibitors that show antiviral activity in cells. A recent study has shown that RRE carries m6A modifications that play a critical role in the activity of the RRE/Rev complex,34 which serves as a poignant reminder that authentic model systems are necessary for the study of RNA targets.
The internal ribosome entry site (IRES), which enables cap-independent translation, has been studied as therapeutically relevant target in both poliovirus and hepatitis C virus (HCV) (Fig. 3D).122 Seth and colleagues used mass spectrometry-based screening to identify 2‐aminobenzimidazole derivatives as ligands binding the internal loop RNA of subdomain IIa in the HCV IRES.126 Using an RNA-targeted small molecule library, the Hermann lab discovered a chemotype which binds to the IRES region and reduces viral replication.122 Scientists at Ibis Therapeutics also discovered functional inhibitors of HCV IRES RNA using a small molecule library and mass spectrometry screening methods (Fig. 2D).126
The severe acute respiratory syndrome coronavirus (SARS-CoV) frameshifting motif, which includes a slippery sequence followed by a pseudoknot that stalls the ribosome during translation, has been also explored as a valuable RNA target.121 Using an in silico screening approach, Park and colleagues identified 1,4-diazepane derivative that acted as an inhibitor of translational frameshifting both in vitro and in a cell-based assay (Fig. 2D).120
Recently, another therapeutic approach aiming at disruption of viral latency has emerged as a weapon against HIV-1 and herpesvirus-associated infections.159, 160, 161 Referred to as shock-and-kill therapy, this strategy aims to disrupt the viral reservoir to reactivate viral production, followed by antiviral treatment.162 Currently available latency-reversing agents against herpesvirus infections163, 164, 165 manipulate an epigenetic pathway, using histone epigenetic modifications to achieve viral reactivation. It would be interesting to see if specific small molecule could potentially bind to viral RNA and cause a latent-to-lytic switch in viral infection.13
7. Identification of RNA-specific small molecule therapeutics
RNA-specific small molecules represent a class of organic compounds with low molecular weight that can specifically bind to RNA secondary or tertiary structures and affect RNA-associated molecular processes, e.g. translation patterns, localization and stability.166, 167, 168 A desirable feature of such compounds is that they interact with RNA not through intercalation, sequence, or electrostatic complementarity, but by means of specific molecular recognition events unique to the particular RNA target. The use of small molecules as RNA-specific drug-like compounds has increased due to their frequently desirable physicochemical characteristics, including their good hydrophobicity and selectivity, as well as cell and tissue permeability. However, caution must be taken when examining their properties as they might follow a different set of principles compared to the ones that have been coined for protein-specific small molecules. Recently, Morgan et al. delved into understanding the physicochemical, structural, and spatial properties of RNA ligands and compiled the RNA-targeted BIoactive ligaNd Database (R-BIND), which contains a list of bioactive monovalent small molecules and multivalent ligands that target non-ribosomal RNAs.169, 170 The overall aim of this database is to use 20 cheminformatic parameters, e.g. oral availability, Lipinski’s rules, Veber’s rules, structural components, molecular complexity, and molecular recognition, to determine trends in bioactive RNA ligands.171 In this section, we will highlight novel technologies that pave the way to the discovery of new types of RNA-targeted therapeutics.
7.1. Fluorescent based assays
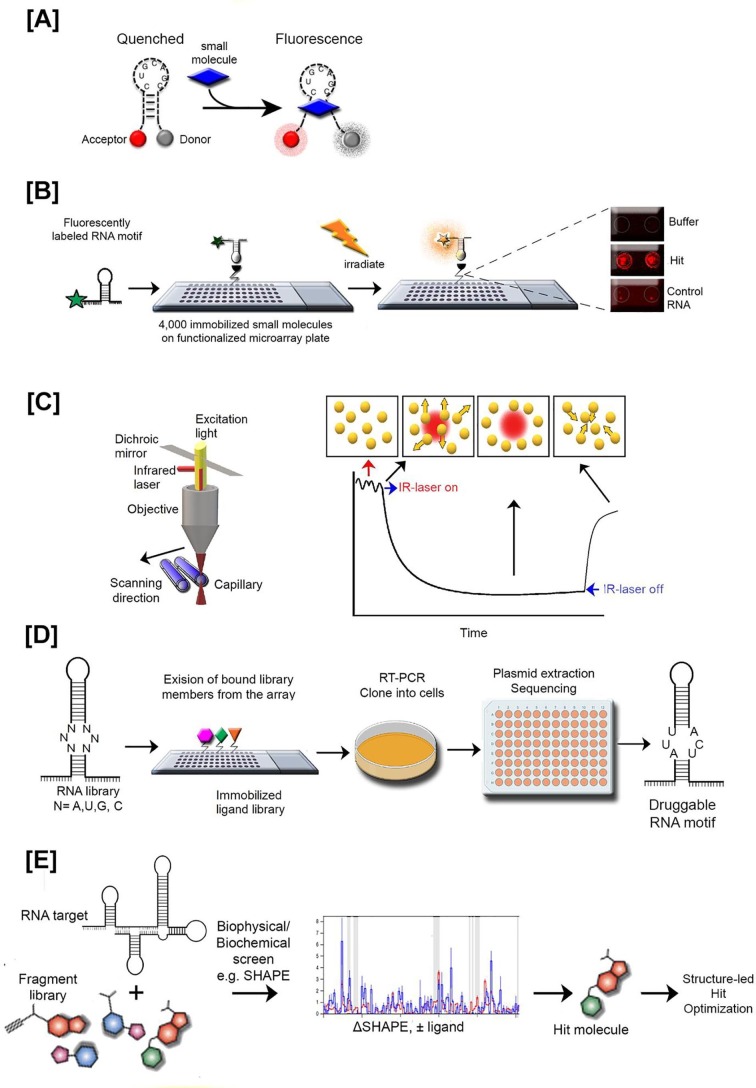
Fluorescence based assays are convenient and practical tools for identifying novel nucleic acid-binding ligands, mainly due to their sensitivity, speed, and versatility. Most conventional fluorescence assays rely on incorporation of fluorescent probes in the RNA sequence, which might impact its native conformation as a side effect.172 An alternative to that approach is the fluorescent indicator displacement assay (FID), where the indicator displays different fluorescence properties in the presence and absence of the oligonucleotide, and can therefore be utilized to measure the binding properties of various chemotypes.173, 174 FID is a tagless approach, meaning that the target RNA and the ligands under investigation do not have to be modified. Instead, the FID assay utilizes intercalating dyes, e.g. 3-methyl-2-((1-(3-(trimethylammonio)propyl)-4-quinolinylidene)methyl)benzothiazolium (TO-PRO)175 thiazole orange,176 which show high binding affinities to RNA but frequently require large quantities of RNA. Recent alternation to the method relies on the use of high affinity RNA-specific peptides, e.g., HIV Tat labeled with a Forster Resonance Enhancement Transfer (FRET) pair, against specific RNA-motif, e.g. TAR (Fig. 4 A).33, 119 When the peptide is bound to RNA, the fluorophores are distant in space, which facilitates the excitation of the donor and emission detection from the acceptor. When the small molecule probe displaces the peptide from the RNA, the fluorophores are proximal, and the emission of the acceptor is quenched. This quenching allows for quantification of the binding affinity of the small molecule towards TAR RNA. Recently, the Hargrove laboratory extended the development of fluorescent peptide displacement assay for screening small molecule ligands against four different RNA targets, namely Tat to HIV-1-TAR and HIV-2-TAR, bacterial ribosomal A-site RNA and the IIB domain HIV-1-Rev response element RNA (RRE-IIB).177 The authors validated the method by quantifying the binding of aminoglycosides and other known RNA binding small molecules, against all four RNA targets.
Fig. 4.
Examples of RNA-specific small molecule screening techniques. [A] Fluorescent peptide displacement assay. The fluorescently-labeled peptide bound to RNA has fluorophores distant in space facilitating excitation of donor and emission detection from acceptor. When the small molecule displaces the peptide from the RNA, the fluorophores are proximal, and the emission of acceptor is quenched allowing for quantification of ligand binding affinity. [B] Small molecule microarray screening approach (SMM). Fluorescently labeled target RNA motif is incubated with the array followed by the detection of fluorescence increase during binding event. Right side represents raw SMM images for hit structure (compounds are printed in duplicate), control and buffer. [C] Microscale thermophoresis (MST) is performed in thin capillaries (blue) in free solution. A microscopic temperature gradient is induced by an infrared laser (IR), and temperature related intensity change as well as thermophoresis are detected. Initially, the molecules are homogeneously distributed and a constant “initial fluorescence” is detected. After activation of the IR laser, a rapid change in fluorophore properties due to the fast temperature change is noted. Subsequently, thermophoretic movement of the fluorescencently labeled molecules out of the heated sample volume can be detected. The overall MST signal is plotted against the ligand concentration to obtain a dose-response curve (on the right). [D] Two-dimensional combinatorial screen (2DCS) selects privileged RNA motifs for a specific ligand by simultaneously screening a nucleic acid library and a small molecule library. The chemicals are immobilized in a microarray format in a gradient of concentrations. Each row represents decreasing concentrations of a unique chemical/small molecule. The RNA library is then allowed to react and hybridize on the microarray, and the unbound RNA is washed away. The bound fractions are then excised, RNA is extracted and sequenced to identify features which allow binding to each small molecule. [E] Fragment-based drug discovery (FBDD) includes unbiased diverse fragment set tested against target RNA using different biophysical, e.g. NMR and/or biochemical, e.g. SHAPE screen approaches leading to hit identification that is further structure-led optimized.
7.2. Mass spectrometry
Advancement of electrospray ionization (ESI) and matrix-assisted laser desorption-ionization (MALDI), which has decreased the sample requirement and increased the mass range, have resulted in mass spectrometry (MS) being increasingly utilized for screening of targets for small-molecule inhibitors. MS as a screening tool has the advantage that neither the target nor the potential ligands require labeling or deconvolution. Information such as stoichiometry, cooperativity, and relative binding affinities are determined quickly within a single sample solution at low concentrations.178
MALDI uses a pulsed laser for desorption of the ions and a time-of-flight analyzer, and has been successfully used in combination with ESI for the metabolite profiling of a model small interfering RNA (siRNA) duplex TSR#34.179 ESI mass spectrometry (ESI-MS) has been of greater utility for studying non-covalent molecular interactions because it generates molecular ions with little to no fragmentation. One of the first applications of ESI-MS resulted in identification of 2-aminobenzimidazole, that specifically binds to the internal ribosome entry site IIA subdomain of Hepatitis C Virus and results in the reduction of HCV RNA replication.126, 180 Also, ESI-MS was used by Dremann et al. to identify RNA-peptide complexes specific for bacterial ribosome helix 69 (H69).181 Fourier-transform ion cyclotron resonance (FT-ICR) MS provides high-resolution spectra, isotope-resolved precursor ion selection and accurate mass assignments. FT-ICR MS has been used to study the interaction between two closely related model RNA constructs corresponding to the decoding sites of the prokaryotic and eukaryotic rRNA and a collection of aminoglycoside antibiotics.182
The Automated Ligand Identification System (ALIS), a label-free AS-MS platform, is a novel MS-based approach that allows for high-throughput screening of small molecules as large combinatorial mixtures tested for binding to target macromolecules.183 By coupling fast size exclusion chromatography (SEC) to separate free ligands from target–ligand complexes to integrated LC-MS ligand identification, a single ALIS instrument can screen hundred thousand compounds a day with minimal, label-free target consumption. Unlike direct AS-MS methods that rely on MS to detect the intact target–ligand complex, ALIS is an “indirect” AS-MS technique that uses size-exclusion chromatography to resolve the target–ligand complex from the unbound species, then dissociates the ligand from the complex using denaturing conditions and employs MS to identify the previously bound ligand. ALIS has been recently used to identify and characterize two RNA ligands that competitively bind to the flavin mononucleotide (FMN) riboswitch and exhibit drug-like functional capabilities, potent analog of ellipticine (WG-1) and quinolone (WG-3) class of antibacterial, respectively.181 These MS-based screening aids in the discovery of novel chemical classes that bind RNAs that may be missed by nonbiophysical screening approaches.
7.3. Small molecule microarray (SMM)
SMM is a high-throughput technology for screening a large unbiased library of drug-like small molecules in a microarray format against a target RNA (Fig. 4B).54, 184 In this technique a collection of small molecules, i.e. primary and secondary alcohols and amines, are spatially arrayed with a robotic microarrayer and covalently linked to a glass surface at high density.112 The microarray is then incubated with fluorescently-labeled target RNA, followed with washing to remove any unspecific binding. The slides are imaged using a fluorescence scanner, and fluorescence intensity is quantified for each spot on the array. A statistical analysis then reveals spots with increased fluorescence upon incubation, corresponding with discrete molecular interactions between the target RNA and associated small molecule.
SMMs have become established as the choice platform for screening, lead discovery, and molecular characterization of RNA-specific binders. For example, SMM has been applied to discover a chemotype that specifically recognizes and binds the HIV TAR hairpin (Fig. 2D).54 The binding specificity of an identified small molecule has been validated by SHAPE probing performed on the HIV 5′ UTR in the presence and absence of the chemotype, confirming that the ligand binding was specific to the TAR hairpin. Also, recently, Connelly et al.112 used the SMM approach to identify novel small molecules that selectively bind in proximity to the apical loop of pre-miR-21 hairpin, preventing biogenesis of miR-21 through Dicer inhibition (Fig. 2A). The binding specificity and overall affinity of identified drug-like chemotypes has been validated through differential scanning fluorimetry (DSF), 2-aminopurine (2-AP) fluorescence titration, and fluorescence intensity assays.
Absorb Array is a recent advancement in SMM technology developed by the Disney laboratory.185 Here, small molecule compounds are non-covalently absorbed onto a hydrate agarose-coated microarray surface, creating a chemically unmodified library for binding to radiolabeled RNA motif libraries. This approach is meant to avoid the addition of functional groups for covalent immobilization of the chemotypes to the array, which could influence the molecular recognition of the parent compound’s natural targets. Absorb Array was applied to the NIH Clinical Collection, a library of RNA-focused small molecules, RNA splicing modulators, and a library of kinase inhibitors confirming that these drugs, in particular topoisomerase inhibitors, kinase inhibitors, and RNA splicing modulators bind to RNAs.114
7.4. Microscale thermophoresis (MST)
Understanding the biophysical characteristics of RNA-based small molecule therapeutics is equally important to the discovery of the lead compound itself. Microscale thermophoresis (MST) is a biophysical technique used to identify and quantify interactions between biomolecules including proteins, DNA, RNA, peptides, and small molecules, in close-to-native conditions (Fig. 4C).186, 187, 188, 189, 190, 191, 192 To quantify the intermolecular interactions, infrared-lasers are used to achieve precise microscale temperature gradients within the glass capillaries filled with the targeted solution, e.g. cell lysate, serum or bioliquids. The directed movement of biomolecules through the microscale temperature gradient allows for the quantification of changes within the chemical microenvironment of the fluorescently-labeled target biomolecule when it interacts with the non-fluorescently-labeled ligand, which, in turn, identifies binding affinities and discriminates between different binding sites of a targeted biomolecule.
The Schneekloth laboratory has recently explored the utility of MST for probing HIV RRE–neomycin and HIV RRE-Rev peptide interactions.189 They also examined competition in RRE RNA binding between neomycin and the Rev peptide via two different approaches. In all of these applications, MST effectively measured binding affinities and the results agreed with previously reported values determined via different biophysical techniques. Also, the Disney laboratory has utilized MST to characterize the binding affinity of a small molecule to the A and U bulge in oncomiR-21.193 This particular chemotype was found to inhibit levels of mature miR-21 and concomitantly increase levels of pre-miR-21, while reversing the invasive phenotype caused by elevated expression of miR-21 in triple-negative breast cancer cells.114
The capacity of MST to monitor therapeutically relevant RNA binding events and competitive displacement in connection with its high sensitivity and the small volume of sample required for analysis, makes it an attractive screening platform for discovering RNA-specific small molecules.
7.5. Two-Dimensional combinatorial screen (2DCS)
2DCS is a library-versus-library selection-based screening approach that identifies small molecule chemotypes with high-affinity and selection for small RNA secondary structures, i.e. hairpins, internal loops and bulges (Fig. 4D).194 First, a small molecule library is conjugated site-specifically onto an agarose microarray. This chemical array is probed for binding against a library of labeled RNA motifs, along with competitor oligonucleotides. After incubation, it is analyzed for RNA motif-ligand interactions and the hit RNA motifs are excised from the array, quantified, and sequenced. Following sequencing, the RNA binding-small molecule interaction is analyzed by structure-activity relationships through sequencing (StARTS) or high-throughput structure-activity relationships through sequencing (HiT-StARTS), which use statistical approaches to determine RNA motif fitness, affinity and selectivity for a specific small molecule.195, 196 StARTS can be utilized to identify positive and negative features of RNA motif targets that may impact small molecule affinity, and assign a score based upon the RNA motif-ligand interaction.195 HiT-StARTS defines RNA-small molecule affinity landscapes, scores the RNA-motif small molecule binding partners, and generates structure-activity relationships based on data collected from next-generation sequencing (RNA-seq), 2DCS, and experimentally determined affinities.196 StARTS and HiT-StARTS can be used in tandem with Inforna, a bioinformatics-based approach that integrates RNA motif–small molecule interactions identified via 2DCS and structural data of target RNAs.186, 196, 197 Inforna generates lead compounds for an RNA of interest by comparing the motifs found in the RNA target’s structure with the RNA motif–small-molecule interactions in its database.
The integration of 2DCS, HiT-StARTS, and Inforna led to the discovery of a small molecule chemotype that binds to the miR-96 and inhibits its biogenesis.197 Inforna was also applied to analyze secondary structures in miRNAs that can be matched with RNA motif–small molecule pairs.197 Precise linking of modules that bind near and in the Drosha processing site yielded Targaprimir-96 (Fig. 2A).114 This chemotype enables selective targeting of pri-miRNA-96 and subsequent inhibition of Drosha processing in breast cancer. A similar approach was applied to identify Targapremir-210 (Fig. 2A), a small molecule that modulates the production of miR-210, leading to increased apoptosis in triple-negative breast cancer cells via the hypoxia inducible factor (HIF) pathway.198 The results of these studies led the authors to propose broad guidelines for miRNA targeting to alleviate oncogenic phenotypes. They suggest that: (1) binding must occur in a functionally active region, e.g. Dicer or Drosha processing region, (2) the ligand must bind to that site with high affinity, and (3) both RNA abundance and molecule affinity can affect the target occupancy necessary to elicit a biological response.
7.6. Fragment-Based drug discovery (FBDD)
Fragment-based drug discovery (FBDD) is a biochemical and biophysical approach used to identify lead drug-like compounds though the identification and characterization of multiple small chemical fragments that bind to RNA (Fig. 4E). The small size of fragments enhances the hit rate against their target since they form less destructive steric interactions with their target compared to larger compounds. In addition, it has been argued that the size of fragment chemical space is substantially smaller than the chemical space of larger drug–like space such that it can be sampled more efficiently.199 Once identified, compounds may be elaborated via multiple strategies into potent, selective inhibitors. Overall, the FBDD screening approach includes: (1) fragment library, (2) target enablement, (3) screening chemical fragments with biophysical or biochemical assays to identify fragment hits, (4) generation of fragment-binding model by biophysical characterization and structure determination, and (5) fragment to lead drug-like compound optimization through medicinal chemistry and further characterization of the fragment.200
Early on, fragment linking studies identified novel small-molecule ligands for ribosomal antibiotic binding sites.201 A benzimidazole hit fragment for an internal loop within domain II of the HCV IRES was used to grow a set of ligands with binding affinity in the micromolar range. The series reduced HCV RNA levels in an HCV replicon assay, with minimal toxicity.126 Initial ligands for these efforts originated from large, generic high-throughput screen libraries. Recently, a fragment library has been designed specifically for fragment screening against RNA targets.202 An advantage of the approach used for the RNA-directed fragment library is that physicochemical properties of the fragments are much more favorable for classical medicinal chemistry than the original compounds.
More recently, the FBDD screening approach has been applied for targeting telomeric repeat-containing (TERRA) RNA, resulting in identification of 20 hits from a library of 355 fluorinated fragment compounds, 7 of which were able to specifically recognize the parallel propeller structure of G4-RNA.203 While all of the compounds interacted with the DNA analog of TERRA, the compounds were shown to favor the parallel conformation, which is the predominant conformation in the RNA G-quadruplexes present in TERRA. This result suggests that the ligands can select and have a strong preference for the parallel propeller-like conformation found in telomeric sequences.203 In the context of viral RNA, a FBDD approach has been used to identify ligands of the HIV TAR element.118 Göbel and colleagues interrogated a set of 29 small molecules that were selected to represent molecular motifs beneficial for RNA recognition. The fragments were rich in chemotypes that provide hydrogen bond donors and included amines, amidines, and guanidines, as well as benzene rings for stacking interactions. A fluorometric competition assay determined that seven small molecule compounds were able to displace a dye labeled Tat peptide from a TAR RNA. Also, Lee et al. reported a fragment-based screen for small molecules that bind to the influenza A virus promoter.204 Overall, the value of fragment-based approaches may not only be in developing leads for specific RNA structures, but also in helping to assess the druggability of newly discovered RNA structures. Compared to other chemotype screening methods, FBDD allows for the selection of a weak fragment to be highly specialized for the targeted RNA molecule based off not only the initial hits but also the targeted RNA’s NMR spectroscopy or crystal structure, thus creating a highly potent, RNA-specific therapeutic. Furthermore, FBDD can assess whether RNA target is druggable through the amount, strength, and specificity of weak binder hits, e.g. if there are little to no hits for a specific RNA then it will most likely have structural elements that inhibit the binding of a small molecule.
8. Conclusions
The variety of RNA targets for therapeutic intervention is staggering, spanning from viral regulatory RNA motifs to coding and ncRNAs involved in infectious and non-infectious human diseases. There is also the mostly unexplored potential of targeting novel RNA species, such as circular RNAs,205 antisense RNAs206 and lncRNAs.207 In addition, RNA-mediated interactions often dysregulated in variety of human disorders,208 further widen the pallete of druggable targets. Aberrant interactions of RNA with proteins is a well-recognized factor that leads to the RNA gain-of-function mechanism that underlines repeat expansion disorders as discussed. Here, to achieve a therapeutic effect, an RNA-specific chemotype would need to specifically recognize and bind the disrupted RNA to prevent protein binding. Binding to the protein itself would inhibit its cellular function and likely cause adverse effects. Conversely, in the case of loss-of-function disorders that decrease protein binding, a therapeutic small molecule needs to enhance protein binding, as shown by small molecule ligands that bind to SMN2 pre-mRNA and alter its alternative splicing. The advances in methods that enable characterization of native RNAs structure, function, cellular localization and intermolecular interaction will further expand the already broad selection of potential therapeutic targets for small molecules. Utilizing the information gained by studying RNAs from various biological and in vitro systems will certainly accelerate the progress of RNA-specific drug design. However, RNAs derived from different biological systems may represent distinct challenges as drug targets, regardless of their biological or pharmacological significance. Here, the relative expression of target RNA in a cell, its localization and structural accessibility of functional sites for ligand binding, will certainly affect its druggability.
Another challenge will be overcoming the issues of specificity and selectivity, which are major barriers for RNA-binding molecules. For example, rRNA constitutes the vast majority of cellular RNA while ncRNA collectively constitutes less than 5% of total cellular RNA. Thus, targeting one ncRNA selectively might be challenging. However, the implementation of structure-bases and high-throughput screening methods that we have discussed are proven to be effective at identifying new small molecule chemotypes with both good specificity and high affinity. Also, the flexibility of RNA-specific small molecules should aid the construction of effective multifunctional drugs that have more than one mode of action and affect multiple targets that could replace drug cocktails. Lessons learned from efforts of ligand discovery for structured RNA elements, including regulatory motifs in the viruses, bacterial rRNA and other ncRNA may inspire the future search for inhibitors that target RNA motifs in a wide range of human disorders. In addition, novel tools emerge as we speak, ranging from nanopore sequencing for detecting RNA base modification (https://www.biorxiv.org/content/early/2018/11/09/459529) to artificial intelligence-driven drug design.209 These will soon offer new insights to RNA biology and provide a significant step forward in the development of novel and much needed therapeutics.
Author contributions
J.S.-S. G.C.C., S.E.C. contributed to the manuscript writing and figure preparation.
Acknowledgments
J.S.-S. G.C.C. and S.E.C. are funded by Alabama Agricultural Experiment Station, Hatch Funding Program and start-up funds from the Department of Biological Sciences, College of Science and Mathematics, and Office of the Vice President for Research, Auburn University. Authors would like to thank Dr. William Sharpee and Dr. William Summers for their constructive criticism of the manuscript.
Biographies

Joanna Sztuba-Solinska, Ph.D. Assistant Professor of Virology, Auburn University, Auburn, AL. I was born on May 23, 1979, in Bydgoszcz, Poland. I graduated from the University of Technology and Life Sciences in Bydgoszcz with master’s degree in Biotechnology in 2003. In 2005, I joined the Department of Biological Sciences at Northern Illinois University to pursue my PhD degree. I conducted my doctoral research studying mechanisms of viral RNA recombination under Prof. Jozef Bujarski mentorship. In 2011, I joined the laboratory of Dr. Stuart Le Grice, at the National Institutes of Health, National Cancer Institute, where I pursued the elucidation of structure and function relationship of viral coding and non-coding RNAs and their potential as drug targets. My current research focuses on defining how structural aspects of viral long non-coding (lnc) RNAs, i.e., secondary structure, tertiary interactions, post-transcriptional modifications, govern their function and intermolecular interactions in cellular processes and viral pathogenesis. During the long-term virus-host evolutionary relationship, herpesviruses have acquired the largest number of lncRNAs, and as such represent an established model to address the principles, mechanisms, and processes that underline lncRNAs multifunctionality. The overall goal of my research is to find “weak-spots” in viral lncRNA interactome network that can be targeted with novel therapeutic strategies to combat viral infections.

Gabriela Chavez-Calvillo, Ph.D. Postdoctoral Scientist, Auburn University, Auburn, AL. I was born on August 8th, 1984, in Mexico. In 2008, I received my doctoral degree in Veterinary Medicine from the Autonomous University of Mexico. In 2009, I completed a master’s degree in Biotechnology, studying protein-protein interactions in viral capsids at the National Laboratory of Genomics for Biodiversity (Langebio), Cinvestav Irapuato, Guanajuato in Mexico. In 2011, I started my doctoral research in RNA Virology field in collaboration with the University of Helsinki in Finland working with positive sense single-stranded RNA viruses in mixed infections of Alfaflexiviridae and Potyviridae. In June 2016, I joined an NSF-PIRE project to study virus evolution during insect vector transmission. Since 2017, I have been working in Dr. Solinska laboratory focusing on structural and functional characterization of Kaposi's sarcoma-associated herpesvirus (KSHV) polyadenylated nuclear (PAN) lncRNA. My project aims at elucidating the influence of post-transcriptional modification on PAN lncRNA structure and function during KSHV latent-to-lytic switch. I am also addressing the intermolecular contact of PAN lncRNA with other coding and non-coding RNAs and proteins.

Sabrina Elizabeth Cline, Undergraduate Research, Auburn University, Auburn, AL. I was born on November 26, 1996 in Richmond, Virginia, but I have lived in various regions of the country due to my father’s military service. Through my travels, I was exposed to various scientific concepts and found myself drawn to molecular biology. This love of biology was cemented after a middle school summer camp at HudsonAlpha Institute of Biotechnology where I had two weeks of hands-on experience related to the molecular biology of DNA. Currently, I am a senior at Auburn University and pursuing a Bachelor of Science in Microbial, Cellular, and Molecular Biology with a concentration in Cellular and Molecular Biology. During the summer of 2018, I was selected for the prestigious BioTrain Internship at HudsonAlpha Institute of Biotechnology where I worked with Dr. Hongna Liu at iCubate to optimize the iC-GI Assay. I have joined Dr. Solinska laboratory as an undergraduate researcher in the Fall of 2017. Since then, I have been focusing on the elucidation of secondary structure of Epstein-Barr virus-encoded long non-coding (lnc) RNA BHLF1 by using high-throughput deep-sequencing SHAPE-MaP probing technology. Additionally, I have been involved in developing the immunofluorescence assay for detecting subcellular localization of KSHV PAN lncRNA. The overall goal of my research is to understand the structure-function relationship of viral lncRNAs which could be targeted with novel antiviral therapeutics.
References
- 1.Ozata D.M., Gainetdinov I., Zoch A., O’Carroll D., Zamore P.D. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2018 doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- 2.Arango D., Sturgill D., Alhusaini N., et al. Acetylation of cytidine in mRNA promotes translation efficiency in brief post-transcriptional acetylation of cytidines in mammalian mRNAs enhances RNA stability and translation. Article acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1–15. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C., Jia G. Reversible RNA Modification N1-methyladenosine (m1A) in mRNA and tRNA. Genom, Proteom Bioinforma. 2018;16(3):155–161. doi: 10.1016/j.gpb.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eken S.M., Jin H., Chernogubova E., Maegdefessel L. Making sense in antisense: therapeutic potential of noncoding RNAs in diabetes-induced vascular dysfunction. J Diabetes Res. 2013 doi: 10.1155/2013/834727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Ying, Ke Ke, Spitale Robert C., et al. Biochemical methods to image and analyze RNA localization: from one to many. Biochemistry. 2019;58(5):379–386. doi: 10.1021/acs.biochem.8b01087. [DOI] [PubMed] [Google Scholar]
- 6.Morgan B.S., Forte J.E., Hargrove A.E. Insights into the development of chemical probes for RNA. Nucleic Acids Res. 2018;46(16):8025–8037. doi: 10.1093/nar/gky718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandhi M., Caudron-Herger M., Diederichs S. RNA motifs and combinatorial prediction of interactions, stability and localization of noncoding RNAs. Nat Struct Mol Biol. 2018;25(12):1070–1076. doi: 10.1038/s41594-018-0155-0. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W., Szostak J.W., Huang Z. Nucleic acid crystallization and X-ray crystallography facilitated by single selenium atom. Front Chem Sci Eng. 2016 doi: 10.1007/s11705-016-1565-3. [DOI] [Google Scholar]
- 9.Fürtig B., Richter C., Wöhnert J., Schwalbe H. NMR spectroscopy of RNA. Chem Bio Chem. 2003 doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 10.Lukavsky P.J. Structure and function of HCV IRES domains. Virus Res. 2009;139(2):166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spitale R.C., Crisalli P., Flynn R.A., Torre E.A., Kool E.T., Chang H.Y. RNA SHAPE analysis in living cells. Nat Chem Biol. 2013;9(1):18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee B., Flynn R.A., Kadina A., Guo J.K., Kool E.T., Chang H.Y. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA. 2016 doi: 10.1261/rna.058784.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavez-Calvillo G., Martin S., Hamm C., Sztuba-Solinska J. The structure-to-function relationships of gammaherpesvirus-encoded long non-coding RNAs and their contributions to viral pathogenesis. Non-Coding RNA. 2018;4(4):24. doi: 10.3390/ncrna4040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engreitz J.M., Sirokman K., McDonel P., et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014;159(1):188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engreitz J., Lander E.S., Guttman M. NY: Springer New York; New York, NY: 2015. RNA Antisense Purification (RAP) for Mapping RNA Interactions with Chromatin. 10.1007/978-1-4939-2253-6. [DOI] [PubMed] [Google Scholar]
- 16.Lu Z., Zhang Q.C., Lee B., et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016;165(5):1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu C., Quinn J., Chang H.Y. Chromatin isolation by RNA purification (ChIRP) J Vis Exp. 2012;61:1–6. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen K.B., Darnell R.B. RNA-Protein Interaction Protocols. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3. [DOI] [Google Scholar]
- 19.Brosnan C.A., Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Cech T.R., Steitz J.A. The noncoding RNA revolution – Trashing old rules to forge new ones. Cell. 2014 doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Kligun E., Mandel-Gutfreund Y. Conformational readout of RNA by small ligands. RNA Biol. 2013;10(6):974–982. doi: 10.4161/rna.24682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider B., Morávek Z., Berman H.M. RNA conformational classes. Nucleic Acids Res. 2004;32(5):1666–1677. doi: 10.1093/nar/gkh333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo J., Westhof E. Base pairs and pseudo pairs observed in RNA-ligand complexes. J Mol Recognit. 2010;23(2):241–252. doi: 10.1002/jmr.978. [DOI] [PubMed] [Google Scholar]
- 24.Chow C.S., Bogdan F.M. A Structural Basis for RNA − ligand. Interactions. 1997 doi: 10.1021/cr960415w. [DOI] [PubMed] [Google Scholar]
- 25.Warner K.D., Hajdin C.E., Weeks K.M. Principles for targeting RNA with drug-like small molecules. Nat Rev Drug Discov. 2018;17(8):547–558. doi: 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermann T. Rational ligand design for RNA: the role of static structure and conformational flexibility in target recognition. Biochimie. 2002;84(9):869–875. doi: 10.1016/S0300-9084(02)01460-8. [DOI] [PubMed] [Google Scholar]
- 27.Johnson L.F., Abelson H.T., Penman S., Green H. The relative amounts of the cytoplasmic RNA species in normal, transformed and senescent cultured cell lines. J Cell Physiol. 1977;90(3):465–470. doi: 10.1002/jcp.1040900310. [DOI] [PubMed] [Google Scholar]
- 28.Clark M.B., Johnston R.L., Inostroza-Ponta M., et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012 doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J., Wang G., Zhang L.H., Ye X.S. Modifications of aminoglycoside antibiotics targeting RNA. Med Res Rev. 2007;27(3):279–316. doi: 10.1002/med.20085. [DOI] [PubMed] [Google Scholar]
- 30.Vicens Q., Westhof E. RNA as a drug target: the case of aminoglycosides. Chem Bio Chem. 2003;4(10):1018–1023. doi: 10.1002/cbic.200300684. [DOI] [PubMed] [Google Scholar]
- 31.Deigan K.E., FerrÉ-D’AmarÉ A.R. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011;44(12):1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blount K.F., Breaker R.R. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24(12):1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 33.Mei H.Y., Cui M., Heldsinger A., et al. Inhibitors of protein-RNA complexation that target the RNA: specific recognition of human immunodeficiency virus type 1 TAR RNA by small organic molecules. Biochemistry. 1998;37(40):14204–14212. doi: 10.1021/bi981308u. [DOI] [PubMed] [Google Scholar]
- 34.Fernandes J., Jayaraman B., Frankel A. The HIV-1 Rev response element. RNA Biol. 2012;9(1):6–11. doi: 10.4161/rna.9.1.18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lusvarghi S., Sztuba-Solinska J., Purzycka K.J., Pauly G.T., Rausch J.W., Le Grice S.F.J. The HIV-2 Rev-response element: determining secondary structure and defining folding intermediates. Nucleic Acids Res. 2013;41(13):6637–6649. doi: 10.1093/nar/gkt353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClorey G., Wood M.J. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr Opin Pharmacol. 2015;24:52–58. doi: 10.1016/j.coph.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Angelbello A.J., Chen J.L., Childs-Disney J.L., Zhang P., Wang Z.F., Disney M.D. Using genome sequence to enable the design of medicines and chemical probes. Chem Rev. 2018;118(4):1599–1663. doi: 10.1021/acs.chemrev.7b00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warner K.D., Hajdin C.E., Weeks K.M. Principles for targeting RNA with drug-like small molecules. Nat Rev Drug Discov. 2018;17:547. doi: 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziehler W.A., Engelke D.R. Probing RNA structure with chemical reagents and enzymes. Curr Protoc Nucleic Acid Chem. 2004 doi: 10.1002/0471142700.nc0601s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014 doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talkish J., May G., Lin Y., Woolford J.L., McManus C.J. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA. 2014 doi: 10.1261/rna.042218.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zubradt M., Gupta P., Persad S., Lambowitz A.M., Weissman J.S., Rouskin S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods. 2016 doi: 10.1038/nmeth.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waduge P., Sakakibara Y., Chow C.S. Chemical probing for examining the structure of modified RNAs and ligand binding to RNA. Methods. 2018 doi: 10.1016/j.ymeth.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Mortimer S.A., Weeks K.M. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc. 2009;4(10):1413–1421. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortimer S.A., Weeks K.M. Time-resolved RNA SHAPE chemistry. J Am Chem Soc. 2008;130(48):16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 46.Gherghe C.M., Mortimer S.A., Krahn J.M., Thompson N.L., Weeks K.M. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008;130(28):8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng C., Chan D., Joseph J., et al. Light-activated chemical probing of nucleobase solvent accessibility inside cells. Nat Chem Biol. 2018;14(3):276–283. doi: 10.1038/nchembio.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B., Mao Y.S., Diermeier S.D., et al. Identification and characterization of a class of MALAT1-like genomic loci. Cell Rep. 2017 doi: 10.1016/j.celrep.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sztuba-Solinska J., Rausch J.W., Smith R., Miller J.T., Whitby D., Le Grice S.F.J. Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res. 2017;45(11):6805–6821. doi: 10.1093/nar/gkx241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sztuba-Solinska J., Le Grice S.F.J. The “Topological Train Ride” of a viral long non-coding RNA. RNA Biol. 2018 doi: 10.1080/15476286.2017.1391442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smola M.J., Christy T.W., Inoue K., et al. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegfried N.A., Busan S., Rice G.M., Nelson J.A.E., Weeks K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11(9):959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauger D.M., Golden M., Yamane D., et al. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci. 2015 doi: 10.1073/pnas.1416266112. 201416266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sztuba-Solinska J., Shenoy S.R., Gareiss P., et al. Identification of biologically active, HIV TAR RNA-binding small molecules using small molecule microarrays. J Am Chem Soc. 2014;136(23):8402–8410. doi: 10.1021/ja502754f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mutate-and-map RNA structure characterization. Nat Methods. 2011;8:997. 10.1038/nmeth.1794. [DOI]
- 56.Tian S., Das R. RNA structure through multidimensional chemical mapping. Q Rev Biophys. 2016 doi: 10.1017/s0033583516000020. [DOI] [PubMed] [Google Scholar]
- 57.Lipson S.E., Hearst J.E. Psoralen cross-linking of ribosomal RNA. Methods Enzymol. 1988;164(C):330–341. doi: 10.1016/S0076-6879(88)64053-5. [DOI] [PubMed] [Google Scholar]
- 58.Janin J., Miller S., Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988;204(1):155–164. doi: 10.1016/j.robot.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 59.Marsh J.A., Teichmann S.A. Relative solvent accessible surface area predicts protein conformational changes upon binding. Structure. 2011;19(6):859–867. doi: 10.1016/j.str.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zinshteyn B., Chan D., England W., Feng C., Green R., Spitale R.C. Assaying RNA structure with LASER-Seq. Nucleic Acids Res. 2018;20:1–13. doi: 10.1093/nar/gky1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spasic A., Assmann S.M., Bevilacqua P.C., Mathews D.H. Modeling RNA secondary structure folding ensembles using SHAPE mapping data. Nucleic Acids Res. 2018;46(1):314–323. doi: 10.1093/nar/gkx1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sloat N., Liu J.W., Schroeder S.J. Swellix: a computational tool to explore RNA conformational space. BMC Bioinf. 2017;18(1):1–12. doi: 10.1186/s12859-017-1910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McHugh C.A., Chen C.K., Chow A., et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jensen K.B., Darnell R.B. CLIP: Crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008 doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hacisuleyman E., Goff L.A., Trapnell C., et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21(2):198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McHugh C.A., Chen C.K., Chow A., et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015 doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vawter M.P., Harvey P.D., DeLisi L.E. Dysregulation of X-linked gene expression in Klinefelter’s syndrome and association with verbal cognition. Am J Med Genet Part B Neuropsychiatr Genet. 2007;144(6):728–734. doi: 10.1002/ajmg.b.30454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weakley S.M., Wang H., Yao Q., Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35(8):1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinn J.J., Ilik I.A., Qu K., et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32(9):933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sexton A.N., Machyna M., Simon M.D. Capture hybridization analysis of DNA targets. Methods Mol Biol. 2016;1480:87–97. doi: 10.1007/978-1-4939-6380-5_8. [DOI] [PubMed] [Google Scholar]
- 71.Gupta R.A., Shah N., Wang K.C., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hajjari M., Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015 doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guilleret I., Yan P., Guillou L., Braunschweig R., Coindre J.M., Benhattar J. The human telomerase RNA gene (hTERC) is regulated during carcinogenesis but is not dependent on DNA methylation. Carcinogenesis. 2002;23(12):2025–2030. doi: 10.1093/carcin/23.12.2025. [DOI] [PubMed] [Google Scholar]
- 74.Samper E., Flores J.M., Blasco M.A. Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc-/- mice with short telomeres. EMBO Rep. 2001;2(9):800–807. doi: 10.1093/embo-reports/kve174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu N., Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol. 2016 doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 76.Hussain S., Aleksic J., Blanco S., Dietmann S., Frye M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013 doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016 doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacob R., Zander S., Gutschner T. The dark side of the epitranscriptome: chemical modifications in long non-coding rnas. Int J Mol Sci. 2017;18(11) doi: 10.3390/ijms18112387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boccaletto P., MacHnicka M.A., Purta E., et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018 doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meyer K.D., Patil D.P., Zhou J., et al. 5′ UTR m6A promotes cap-independent translation. Cell. 2015 doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015 doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014 doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roost C., Lynch S.R., Batista P.J., Qu K., Chang H.Y., Kool E.T. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015 doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kierzek E., Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003 doi: 10.1093/nar/gkg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du K., Zhang L., Lee T., Sun T. m6A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2018 doi: 10.1007/s12035-018-1138-1. [DOI] [PubMed] [Google Scholar]
- 86.Lichinchi G., Zhao B.S., Wu Y., et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bai Y., Tambe A., Zhou K., Doudna J.A. RNA-guided assembly of Rev-RRE nuclear export complexes. Elife. 2014 doi: 10.7554/eLife.03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Squires J.E., Patel H.R., Nousch M., et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen Y., Sierzputowska-Gracz H., Guenther R., Everett K., Agris P.F. 5-Methylcytidine Is Required for Cooperative Binding of Mg2+ and a Conformational Transition at the Anticodon Stem-Loop of Yeast Phenylalanine tRNA. Biochemistry. 1993 doi: 10.1021/bi00089a047. [DOI] [PubMed] [Google Scholar]
- 90.Li X., Zhu P., Ma S., et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11(8):592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 91.Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwartz S., Bernstein D.A., Mumbach M.R., et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014 doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grozhik A.V., Jaffrey S.R. Distinguishing RNA modifications from noise in epitranscriptome maps. Nat Chem Biol. 2018 doi: 10.1038/nchembio.2546. [DOI] [PubMed] [Google Scholar]
- 94.Liu N., Parisien M., Dai Q., Zheng G., He C., Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013 doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005 doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 96.Conde J., Artzi N. Are RNAi and miRNA therapeutics truly dead? Trends Biotechnol. 2015 doi: 10.1016/j.tibtech.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 97.Donlic A., Hargrove A.E. Targeting RNA in mammalian systems with small molecules. Wiley Interdiscip Rev RNA. 2018;9(4):1–21. doi: 10.1002/wrna.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frye Michaela, Harada Bryan T., Behm Mikaela, He Chuan. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. doi: 10.1126/science:aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Purnell B.A. Small RNA regulates glucose homeostasis. Science (80) 2016;354(6314):844–845. doi: 10.1126/science.354.6314.844-c. [DOI] [PubMed] [Google Scholar]
- 100.Schaefer J., Schnupp T. Non-coding RNA regulation of immunity. J Immunol. 2018;200(1):167.9. [Google Scholar]
- 101.Hughes T.A., Jones P.F. Regulatory RNAs: have mRNA untranslated regions joined the party? PLoS Med. 2008 doi: 10.1371/journal.pmed.0050110. [DOI] [Google Scholar]
- 102.Lee T.I., Young R.A. Transcriptional regulation and its misregulation in disease. Cell. 2013 doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014 doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 104.Tüfekci K.U., Öner M.G., Meuwissen R.L.J., Genç Ş. The role of microRNAs in human diseases. Methods Mol Biol. 2014 doi: 10.1007/978-1-62703-748-8_3. [DOI] [PubMed] [Google Scholar]
- 105.Price C., Chen J. MicroRNAs in cancer biology and therapy: current status and perspectives. Genes Dis. 2014 doi: 10.1016/j.gendis.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Y., Disney M.D. Precise small molecule degradation of a noncoding RNA identifies cellular binding sites and modulates an oncogenic phenotype. ACS Chem Biol. 2018 doi: 10.1021/acschembio.8b00827. acschembio.8b00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hong Y., Liang H., Rehman U.U., et al. MiR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci Rep. 2016;6(November):1–16. doi: 10.1038/srep37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kumarswamy R., Volkmann I., Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8(5):706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gumireddy Kiranmai, Young Douglas D.X.X. Small molecule inhibitors of MicroRNA miR-21 function. Angew Chem Int Ed Engl. 2012;47(39):7482–7484. doi: 10.1002/anie.200801555.Small. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu W., Zhang B., Chen G., et al. Targeting miR-21 with sophocarpine inhibits tumor progression and reverses epithelial-mesenchymal transition in head and neck cancer. Mol Ther. 2017;25(9):2129–2139. doi: 10.1016/j.ymthe.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adibekian A., Abegg D., Disney M.D., et al. A designed small molecule inhibitor of a non-coding RNA sensitizes HER2 negative cancers to herceptin. J Am Chem Soc. 2019 doi: 10.1021/jacs.8b10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Connelly Colleen M., Abulwerdi Fardokht A., Schneekloth John S. Discovery of RNA binding small molecules using small molecule microarrays. Methods Mol Biol. 2017:157–175. doi: 10.1007/978-1-4939-6584-7_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Angelbello A.J., Disney M.D. Bleomycin can cleave an oncogenic noncoding RNA. Chem Bio Chem. 2018;19(1):43–47. doi: 10.1002/cbic.201700581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Velagapudi S.P., Cameron M.D., Haga C.L., et al. Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1523975113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ratni H., Karp G.M., Weetall M., et al. Specific correction of alternative survival motor neuron 2 Splicing by Small Molecules: discovery of a potential novel medicine to treat spinal muscular atrophy. J Med Chem. 2016 doi: 10.1021/acs.jmedchem.6b00459. [DOI] [PubMed] [Google Scholar]
- 116.Palacino J., Swalley S.E., Song C., et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat Chem Biol. 2015 doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 117.Yang S.Y., Lejault P., Chevrier S., et al. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat Commun. 2018;9(1):4730. doi: 10.1038/s41467-018-07224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zeiger M., Stark S., Kalden E., et al. Fragment based search for small molecule inhibitors of HIV-1 Tat-TAR. Bioorganic Med Chem Lett. 2014;24(24):5576–5580. doi: 10.1016/j.bmcl.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 119.Gelus N., Bailly C., Hamy F., Klimkait T., Wilson W.D., Boykin D.W. Inhibition of HIV-1 Tat-TAR interaction by diphenylfuran derivatives: effects of the terminal basic side chains. Bioorganic Med Chem. 1999;7(6):1089–1096. doi: 10.1016/S0968-0896(99)00041-3. [DOI] [PubMed] [Google Scholar]
- 120.Park S.J., Kim Y.G., Park H.J. Identification of rna pseudoknot-binding ligand that inhibits the – 1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J Am Chem Soc. 2011;133(26):10094–10100. doi: 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- 121.Plant E.P., Pérez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005 doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Parsons J., Castaldi M.P., Dutta S., Dibrov S.M., Wyles D.L., Hermann T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat Chem Biol. 2009;5(11):823–825. doi: 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Douglas A.G.L., Wood M.J.A. RNA splicing: disease and therapy. Brief Funct Genom. 2011;10:3. doi: 10.1093/bfgp/elr020. [DOI] [PubMed] [Google Scholar]
- 124.Tazi J., Bakkour N., Stamm S. Alternative splicing and disease. Biochim Biophys Acta – Mol Basis Dis. 2009;1792(1):14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Calder A.N., Androphy E.J., Hodgetts K.J. Small molecules in development for the treatment of spinal muscular atrophy. J Med Chem. 2016;59(22):10067–10083. doi: 10.1021/acs.jmedchem.6b00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Seth P.P., Miyaji A., Jefferson E.A., et al. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J Med Chem. 2005;48(23):7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 127.Urbanski L.M., Leclair N., Anczuków O. Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA. 2018 doi: 10.1002/wrna.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weldon C., Dacanay J.G., Gokhale V., et al. Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 2018 doi: 10.1093/nar/gkx1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.La Spada A.R., Taylor J.P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010 doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fardaei M., Rogers M.T., Thorpe H.M., et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002 doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 131.Warf M.B., Nakamori M., Matthys C.M., Thornton C.A., Berglund J.A. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci. 2009;106(44):18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Coonrod L.A., Nakamori M., Wang W., et al. Reducing levels of toxic RNA with small molecules. ACS Chem Biol. 2013;8(11):2528–2537. doi: 10.1021/cb400431f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Arambula J.F., Ramisetty S.R., Baranger A.M., Zimmerman S.C. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc Natl Acad Sci. 2009 doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jahromi A.H., Fu Y., Miller K.A., et al. Developing bivalent ligands to target CUG triplet repeats, the causative agent of myotonic dystrophy type 1. J Med Chem. 2013 doi: 10.1021/jm400794z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee M.M., Childs-Disney J.L., Pushechnikov A., et al. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J Am Chem Soc. 2009 doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen C.Z., Sobczak K., Hoskins J., et al. Two high-throughput screening assays for aberrant RNA-protein interactions in myotonic dystrophy type 1. Anal Bioanal Chem. 2012 doi: 10.1007/s00216-011-5604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haga C.L., Velagapudi S.P., Strivelli J.R., Yang W.-Y.Y., Disney M.D., Small Phinney DG. Molecule inhibition of miR-544 biogenesis disrupts adaptive responses to hypoxia by modulating ATM-mTOR Signaling. ACS Chem Biol. 2015;10(10):2267–2276. doi: 10.1021/acschembio.5b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Childs-Disney J.L., Hoskins J., Rzuczek S.G., Thornton C.A, Disney M.D. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem Biol. 2012 doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bernat V., Disney M.D. RNA structures as mediators of neurological diseases and as drug targets. Neuron. 2015 doi: 10.1016/j.neuron.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martz L. Expansion into TNBC. Biocentury Innov From Idea IND. 2019 [Google Scholar]
- 141.Fay M.M., Lyons S.M., Ivanov P. RNA G-Quadruplexes in biology: principles and molecular mechanisms. J Mol Biol. 2017;429(14):2127–2147. doi: 10.1016/j.jmb.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gellert M., Lipsett M.N., Davies D.R. Helix Formation by guanylic acid. Proc Natl Acad Sci. 2006 doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arora A., Maiti S. Differential biophysical behavior of human telomeric RNA and dna quadruplex. J Phys Chem B. 2009 doi: 10.1021/jp810638n. [DOI] [PubMed] [Google Scholar]
- 144.Saccà B., Lacroix L., Mergny J.L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pandey S., Agarwala P., Maiti S. In: RNA Therapeutics. Garner A.L., editor. Springer International Publishing; Cham: 2018. Targeting RNA G-Quadruplexes for Potential Therapeutic Applications; pp. 177–206. 10.1007/7355_2016_22. [Google Scholar]
- 146.Kwok C.K., Marsico G., Balasubramanian S. Detecting RNA G-quadruplexes (rG4s) in the transcriptome. Cold Spring Harb Perspect Biol. 2018;10(7):1–14. doi: 10.1101/cshperspect.a032284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bugaut A., Rodriguez R., Kumari S., Hsu S.T.D., Balasubramanian S. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org Biomol Chem. 2010 doi: 10.1039/c002418j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Miglietta G., Cogoi S., Marinello J., et al. RNA G-Quadruplexes in Kirsten Ras (KRAS) oncogene as targets for small molecules inhibiting translation. J Med Chem. 2017 doi: 10.1021/acs.jmedchem.7b00622. [DOI] [PubMed] [Google Scholar]
- 149.Raphael B.J., Hruban R.H., Aguirre A.J., et al. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017 doi: 10.1017/9781107446724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang Z.-F., Ursu A., Childs-Disney J.L., et al. The hairpin form of r(G4C2)exp in c9ALS/FTD is repeat-associated Non-ATG translated and a target for bioactive small molecules. Cell Chem Biol. 2018:1–12. doi: 10.1016/j.chembiol.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ruggiero E., Richter S.N. Survey and summary G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018;46(7):3270–3283. doi: 10.1093/nar/gky187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Collie G.W., Parkinson G.N., Neidle S., Rosu F., De Pauw E, Gabelica V. Electrospray mass spectrometry of telomeric RNA (TERRA) Reveals the formation of stable multimeric G-quadruplex structures. J Am Chem Soc. 2010 doi: 10.1021/ja100345z. [DOI] [PubMed] [Google Scholar]
- 153.Martadinata H., Phan A.T. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J Am Chem Soc. 2009 doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- 154.Mei H.Y., Mack D.P., Galan A.A., et al. Discovery of selective, small-molecule inhibitors of RNA complexes – I. The Tat protein/TAR RNA complexes required for HIV-1 transcription. Bioorg Med Chem. 1997 doi: 10.1016/S0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 155.Chapman R.L., Stanley T.B., Hazen R., Garvey E.P. Small molecule modulators of HIV Rev/Rev response element interaction identified by random screening. Antiviral Res. 2002 doi: 10.1016/S0166-3542(01)00222-4. [DOI] [PubMed] [Google Scholar]
- 156.Chung J., Mujeeb A., Jiang Y., et al. A small molecule, Lys-Ala-7-amido-4-methylcoumarin, facilitates RNA dimer maturation of a stem-loop 1 transcript in vitro: Structure-activity relationship of the activator. Biochemistry. 2008 doi: 10.1021/bi800230m. [DOI] [PubMed] [Google Scholar]
- 157.Bell N.M., L’Hernault A., Murat P., Richards J.E., Lever A.M.L., Balasubramanian S. Targeting RNA-protein interactions within the human immunodeficiency virus type 1 lifecycle. Biochemistry. 2013 doi: 10.1021/bi401270d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chittapragada M., Roberts S., Ham Y.W. Aminoglycosides: molecular insights on the recognition of RNA and aminoglycoside mimics. Perspect Medicin Chem. 2009;2009(3):21–37. doi: 10.4137/pmc.s2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang H.C., Xing S., Shan L., et al. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest. 2009 doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Elliott J.H., Wightman F., Solomon A., et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014 doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tikhmyanova N., Schultz D.C., Lee T., Salvino J.M., Lieberman P.M. Identification of a new class of small molecules that efficiently reactivate latent Epstein-Barr virus. ACS Chem Biol. 2014 doi: 10.1021/cb4006326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bashiri K., Rezaei N., Nasi M., Cossarizza A. The role of latency reversal agents in the cure of HIV: a review of current data. Immunol Lett. 2018;196(January):135–139. doi: 10.1016/j.imlet.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 163.Kim Y., Anderson J.L., Lewin S.R. Getting the "Kill" into "Shock and Kill": strategies to eliminate latent HIV. Cell Host Microbe. 2018;23(1):14–26. doi: 10.1016/j.chom.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ghosh S.K., Perrine S.P., Williams R.M., Faller D.V. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood. 2012;119(4):1008–1017. doi: 10.1182/blood-2011-06-362434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Feng W.H., Kenney S.C. Valproic acid enhances the efficacy of chemotherapy in EBV-positive tumors by increasing lytic viral gene expression. Cancer Res. 2006;66(17):8762–8769. doi: 10.1158/0008-5472.CAN-06-1006. [DOI] [PubMed] [Google Scholar]
- 166.Walder R., Van Patten W.J., Ritchie D.B., et al. High-precision single-molecule characterization of the folding of an HIV RNA hairpin by atomic force microscopy. Nano Lett. 2018 doi: 10.1021/acs.nanolett.8b02597. [DOI] [PubMed] [Google Scholar]
- 167.McKnight K.L., Heinz B.A. RNA as a target for developing antivirals. Antivir Chem Chemother. 2003;14(2):61–73. doi: 10.1177/095632020301400201. [DOI] [PubMed] [Google Scholar]
- 168.Connelly C.M., Moon M.H., Schneekloth J.S. The emerging role of RNA as a therapeutic target for small molecules. Cell Chem Biol. 2016;23(9):1077–1090. doi: 10.1016/j.chembiol.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Morgan B.S., Forte J.E., Culver R.N., Zhang Y., Hargrove A.E. Discovery of key physicochemical, structural, and spatial properties of RNA-targeted bioactive ligands. Angew Chemie – Int Ed. 2017 doi: 10.1002/anie.201707641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Morgan B.S., Forte J.E., Hargrove A.E. Insights into the development of chemical probes for RNA. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 172.Paredes E., Evans M., Das S.R. RNA labeling, conjugation and ligation. Methods. 2011;54(2):251–259. doi: 10.1016/j.ymeth.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 173.Largy E., Hamon F., Teulade-Fichou M.P. Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures. Anal Bioanal Chem. 2011;400(10):3419–3427. doi: 10.1007/s00216-011-5018-z. [DOI] [PubMed] [Google Scholar]
- 174.Murata A., Harada Y., Fukuzumi T., Nakatani K. Fluorescent indicator displacement assay of ligands targeting 10 microRNA precursors. Bioorganic Med Chem. 2013;21(22):7101–7106. doi: 10.1016/j.bmc.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 175.Asare-Okai P.N., Chow C.S. A modified fluorescent intercalator displacement assay for RNA ligand discovery. Anal Biochem. 2011;408(2):269–276. doi: 10.1016/j.ab.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Boger D.L., Tse W.C. Thiazole orange as the fluorescent intercalator in a high resolution fid assay for determining DNA binding affinity and sequence selectivity of small molecules. Bioorganic Med Chem. 2001;9(9):2511–2518. doi: 10.1016/S0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]
- 177.Patwardhan N.N., Ganser L.R., Kapral G.J., et al. Amiloride as a new RNA-binding scaffold with activity against HIV-1 TAR. Med Chem Commun. 2017;8(5):1022–1036. doi: 10.1039/c6md00729e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Laughlin S., David Wilson W. May the best molecule win: competition ESI mass spectrometry. Int J Mol Sci. 2015 doi: 10.3390/ijms161024506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shimizu H., Jinno F., Morohashi A., et al. Application of high-resolution ESI and MALDI mass spectrometry to metabolite profiling of small interfering RNA duplex. J Mass Spectrom. 2012 doi: 10.1002/jms.3054. [DOI] [PubMed] [Google Scholar]
- 180.Kuersten S., Radek A., Vogel C., Penalva L.O.F. Translation regulation gets its “omics” moment. Wiley Interdiscip Rev RNA. 2013;4(6):617–630. doi: 10.1002/wrna.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rizvi N.F., Howe J.A., Nahvi A., et al. Discovery of selective RNA-binding small molecules by affinity-selection mass spectrometry. ACS Chem Biol. 2018;13(3):820–831. doi: 10.1021/acschembio.7b01013. [DOI] [PubMed] [Google Scholar]
- 182.Sannes-Lowery K.A., Drader J.J., Griffey R.H., Hofstadler S.A. Fourier transform ion cyclotron resonance mass spectrometry as a high throughput affinity screen to identify RNA binding ligands. TrAC – Trends Anal Chem. 2000 doi: 10.1016/S0165-9936(00)00029-7. [DOI] [Google Scholar]
- 183.Annis D.A., Athanasopoulos J., Curran P.J., et al. An affinity selection-mass spectrometry method for the identification of small molecule ligands from self-encoded combinatorial libraries: discovery of a novel antagonist of E. coli dihydrofolate reductase. Int J Mass Spectrom. 2004 doi: 10.1016/j.ijms.2003.11.022. [DOI] [Google Scholar]
- 184.Bradner J.E., McPherson O.M., Koehler A.N. A method for the covalent capture and screening of diverse small molecules in a microarray format. Nat Protoc. 2006;1(5):2344–2352. doi: 10.1038/nprot.2006.282. [DOI] [PubMed] [Google Scholar]
- 185.Velagapudi S.P., Costales M.G., Vummidi B.R., et al. Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chem Biol. 2018;25(9):1–9. doi: 10.1016/j.chembiol.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jerabek-willemsen M., André T., Wanner R., et al. MicroScale thermophoresis: interaction analysis and beyond. J Mole Struct. 2014;1077:101–113. [Google Scholar]
- 187.Gaffarogullari E.C., Krause A., Balbo J., Herten D.P., Jäschke A. Microscale thermophoresis provides insights into mechanism and thermodynamics of ribozyme catalysis. RNA Biol. 2013;10(12):1815–1821. doi: 10.4161/rna.27101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Seidel S.A.I., Dijkman P.M., Lea W.A., et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59(3):301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Moon M.H., Hilimire T.A., Sanders A.M., Schneekloth J.S. Measuring RNA-ligand interactions with microscale thermophoresis. Biochemistry. 2018;57(31):4638–4643. doi: 10.1021/acs.biochem.7b01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Baaske P., Wienken C.J., Reineck P., Duhr S., Braun D. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew Chemie – Int Ed. 2010;49(12):2238–2241. doi: 10.1002/anie.200903998. [DOI] [PubMed] [Google Scholar]
- 191.Seidel S.A.I., Wienken C.J., Geissler S., et al. Label-free microscale thermophoresis discriminates sites and affinity of protein-ligand binding. Angew Chemie – Int Ed. 2012;51(42):10656–10659. doi: 10.1002/anie.201204268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wienken C.J., Baaske P., Duhr S., Braun D. Thermophoretic melting curves quantify the conformation and stability of RNA and DNA. Nucleic Acids Res. 2011;39(8) doi: 10.1093/nar/gkr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Disney M.D., Labuda L.P., Paul D.J., et al. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008 doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 194.Disney M.D., Labuda L.P., Paul D.J., et al. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008;130(33):11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 195.Velagapudi S.P., Seedhouse S.J., Disney M.D. Structure-activity relationships through sequencing (StARTS) defines optimal and suboptimal RNA motif targets for small molecules. Angew Chemie Int Ed. 2010;49(22):3816–3818. doi: 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Velagapudi S.P., Luo Y., Tran T., et al. Defining RNA–small molecule affinity landscapes enables design of a small molecule inhibitor of an oncogenic noncoding RNA. ACS Cent Sci. 2017;3(3):205–216. doi: 10.1021/acscentsci.7b00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Disney M.D., Winkelsas A.M., Velagapudi S.P., Southern M., Fallahi M., Childs-Disney J.L. Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem Biol. 2016;11(6):1720–1728. doi: 10.1021/acschembio.6b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Costales M.G., Haga C.L., Velagapudi S.P., Childs-Disney J.L., Phinney D.G., Disney M.D. Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J Am Chem Soc. 2017;139(9):3446–3455. doi: 10.1021/jacs.6b11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Erlanson D.A., Fesik S.W., Hubbard R.E., Jahnke W., Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov. 2016;15(9):605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 200.Lamoree B., Hubbard R.E. Current perspectives in fragment-based lead discovery (FBLD) Essays Biochem. 2017;61(5):453–464. doi: 10.1042/EBC20170028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tor Y. Targeting RNA with small molecules. Chem Bio Chem. 2003;4(10):998–1007. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- 202.Bodoor K., Boyapati V., Gopu V., et al. Design and implementation of an ribonucleic acid (RNA) directed fragment library. J Med Chem. 2009;52(12):3753–3761. doi: 10.1021/jm9000659. [DOI] [PubMed] [Google Scholar]
- 203.Garavís M., López-Méndez B., Somoza A., et al. Discovery of selective ligands for telomeric RNA G-quadruplexes (TERRA) through19F-NMR based fragment screening. ACS Chem Biol. 2014;9(7):1559–1566. doi: 10.1021/cb500100z. [DOI] [PubMed] [Google Scholar]
- 204.Lee M.K., Bottini A., Kim M., et al. Correction: a novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication. Chem Commun. 2014;50(83):12578. doi: 10.1039/c4cc90361g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang M., Yu F., Wu W., et al. Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int J Biol Sci. 2017;13(12):1497–1506. doi: 10.7150/ijbs.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wenric S., Elguendi S., Caberg J.H., et al. Transcriptome-wide analysis of natural antisense transcripts shows their potential role in breast cancer. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-17811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lian Y., Yan C., Xu H., et al. A novel lncRNA, LINC00460, affects cell proliferation and apoptosis by regulating KLF2 and CUL4A expression in colorectal cancer. Mol Ther – Nucleic Acids. 2018;12(September):684–697. doi: 10.1016/j.omtn.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat Rev Genet. 2016;17(1):19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jordan A.M. Artificial intelligence in drug design – the storm before the calm? ACS Med Chem Lett. 2018 doi: 10.1021/acsmedchemlett.8b00500. acsmedchemlett.8b00500. [DOI] [PMC free article] [PubMed] [Google Scholar]