Abstract
RNA interference (RNAi) is a double-stranded RNA (dsRNA)-triggered mechanism for suppressing gene expression, which is conserved in evolution and has emerged as a powerful tool to study gene function. Rotaviruses, the leading cause of severe diarrhea in young children, are formed by three concentric layers of protein, and a genome composed of 11 segments of dsRNA. Here, we show that the RNAi machinery can be triggered to silence rotavirus gene expression by sequence-specific short interfering RNAs (siRNAs). RNAi is also useful for the study of the virus-cell interactions, through the silencing of cellular genes that are potentially important for the replication of the virus. Interestingly, while the translation of mRNAs is readily stopped by the RNAi machinery, the viral transcripts involved in virus genome replication do not seem to be susceptible to RNAi. Since gene silencing by RNAi is very efficient and specific, this system could become a novel therapeutic approach for rotavirus and other virus infections, once efficient methods for in vivo delivery of siRNAs are developed. Although the use of RNAi as an antiviral therapeutic tool remains to be demonstrated, there is no doubt that this technology will influence drastically the way postgenomic virus research is conducted.
Keywords: Rotavirus, RNA interference, siRNAs, Antiviral agents, Reoviridae
1. Introduction
Acute, infectious diarrhea is the most common cause of morbidity and mortality among young children living in developing countries, accounting for as many as one billion illnesses and between 2.5 and 3.2 million deaths annually (Parashar and Glass, 2003). Rotaviruses are the leading etiologic agent of severe diarrheal disease in infants and young children worldwide. In developed countries, rotaviruses have been detected in 30–50% of infants hospitalized with acute diarrhea. In less developed countries, rotaviruses are also the most frequently detected pathogen in children with severe gastroenteritis. While the mortality from rotavirus disease in developed countries is very low, rotavirus causes an estimated 500,000–600,000 deaths each year (Parashar and Glass, 2003). The frequency of rotavirus infection is remarkably similar in both settings. Since rotaviruses play such an important role in severe dehydrating gastroenteritis, and because even advanced levels of hygiene seem unable to control the spread of rotavirus infections, there is an urgent need to develop effective vaccination and therapeutic strategies. Fundamental to these developments is a basic understanding of the molecular mechanisms by which rotaviruses interact with their host cell.
Rotaviruses, a genus of the Reoviridae family, are formed by three concentric layers of protein that enclose a genome composed of 11 segments of double stranded RNA (dsRNA), ranging in size from approximately 660–3300 base pairs (bp). The total genomic size is about 18,500 bp. The innermost layer of the virion is formed by 120 molecules of protein VP2, which surrounds the viral genome. Twelve copies each of VP1, the RNA polymerase, and VP3, a guanylyltransferase and methylase, constitute the core of the virus. The addition of 260 trimers of VP6 on top of the VP2 layer produces double-layered particles (DLPs). The outermost layer, characteristic of triple-layered particles (TLPs), is composed of two proteins, VP4 and VP7. The smooth external surface of the virus is made up of 780 copies of glycoprotein VP7, organized as trimers, while 60 spike-like structures, formed by dimers of VP4, extend about 12 nm from the VP7 surface. The mature virus particle is approximately 100 nm in diameter and contains 132 porous channels, which allow the influx of compounds in aqueous solution to the inside of the capsid and the efflux of newly formed mRNAs (Pesavento et al., 2003).
VP4 has essential functions in the virus life cycle, including receptor binding and cell penetration. The role of VP7 during the initial interactions of the virus with the cell is less clear, although it has been recently shown that VP7 interacts with cell surface molecules at a step subsequent to the initial attachment of the virus through the spike protein (Graham et al., 2003, Zarate et al., 2002). After the virus attaches to the cell surface, it has to penetrate the plasma membrane to productively infect the cell. This penetration is increased by, and most probably dependent on, trypsin treatment of the virus that results in the specific cleavage of VP4 to polypeptides VP8 and VP5. The cleavage of VP4 does not affect cell binding and is rather associated with the entry of the virus into the cell (Arias et al., 2001, Estes, 2001).
During, or shortly after cell entry, the infecting TLP is uncoated, loosing the two proteins of the outer layer, and yielding a DLP, which is transcriptionally active. VP1 synthesizes the primary viral transcripts, which are extruded into the cell’s cytoplasm through the class I channels located at the icosahedral five-fold vertices of the particle (Pesavento et al., 2003).
The RNA transcripts direct the synthesis of six structural and six non-structural viral proteins (i.e., function as mRNAs) and also serve as RNA templates (RNA(+)) for the synthesis of the RNA negative strands (RNA(−)), to form the dsRNA genome segments. Once a critical mass of viral proteins is accumulated into structures known as viroplasms, core replication intermediate (RI) particles assemble. The synthesis of RNA(−) has been proposed to occur concurrently with the packaging of RNA(+) into the core RIs, and is highly coordinated in such a way that packaging and replication lead to the formation of cores containing one copy of each of the 11 genome segments of dsRNA (Patton and Spencer, 2000). In addition to the virus RNA polymerase, the non-structural proteins NSP2 and NSP5 are thought to be essential for the early steps of morphogenesis. All these processes lead to the production of transcriptionally active, dsRNA-containing double-layered RI particles. These particles are responsible for an enhanced second round of transcription, which results in a second wave of assembly of double-layered RI particles, which then bud through the membrane of the ER. During this budding, which is mediated by the interaction of the double-layered RIs with the ER membrane-associated rotavirus protein NSP4, the particles acquire a transient membrane envelope (Estes, 2001). The transiently enveloped particles contain, in addition to NSP4, the virus surface proteins VP4 and VP7, as well as minor amounts of other non-structural proteins (Poruchynsky and Atkinson, 1991). The lipid envelope is then removed by a largely unknown mechanism, to yield mature TLPs (Fig. 1 ).
Fig. 1.
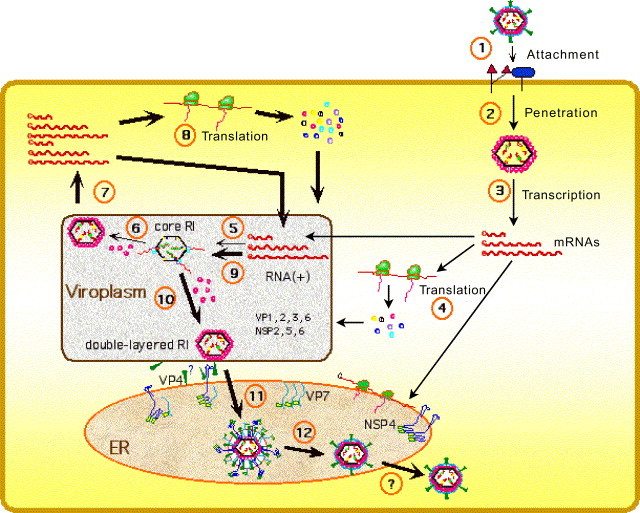
Replication cycle of rotaviruses. The different steps in the replication cycle of the virus are indicated by numbers—1: attachment of the virion to the cell surface; 2: penetration and uncoating of the virus particle to yield DLPs; 3: primary transcription of the genomic dsRNA; 4: synthesis of viral proteins; 5: assembly of core RIs and negative strand RNA synthesis; 6: assembly of double-layered RIs; 7: secondary transcription from double-layered RIs; 8: secondary, enhanced synthesis of viral proteins; 9: secondary, increased assembly of core RIs and negative strand RNA synthesis; 10: secondary, increased assembly of double-layered RIs; 11: budding of double-layered RIs through the membrane of the endoplasmic reticulum (ER), and acquisition of a membrane envelope; 12: loss of the membrane envelope and generation of mature triple-layered virions.
Apart from glycoprotein NSP4, the other five nonstructural proteins (NSP1 to NSP3, NSP5, and NSP6) have the ability to bind RNA (Patton, 1995) and are thought to be involved in the replication of the viral genome, although their precise function is not known. Despite the efforts of many laboratories, at present there is no reverse genetics system available for rotaviruses. Thus, the in vivo characterization of rotavirus gene function has been limited so far to the characterization of antibody escape variants, temperature-sensitive mutants, and reassortants.
2. RNA interference and rotaviruses
RNA interference (RNAi) has become a powerful and widely used tool for the analysis of gene functions. This evolutionarily conserved mechanism, triggered by double-stranded RNA (dsRNA), specifically suppresses gene expression by selectively degrading mRNAs matching the sequence of the dsRNA that triggered the response, without affecting the expression of other genes. During RNAi, long dsRNA molecules are processed into 21–25 bp RNAs known as short-interfering RNAs (siRNAs) that serve as guides for enzymatic cleavage of complementary RNAs, thus offering an exquisite mode to specifically knockdown the expression of a particular gene.
This phenomenon has been observed in invertebrates and plants (Hannon, 2002, Sharp, 2001, Zamore, 2001); however, the demonstration of an RNAi-like response in somatic mammalian cells had been hampered by dsRNA-induced mechanisms, which non-specifically inhibit gene expression. In mammalian cells, dsRNA fragments longer than 30 bp induce components of the interferon response, including the dsRNA-dependent protein kinase (PKR), the 2′,5′-oligoadenylate synthetase (OAS) and the non-specific RNAse L (Katze et al., 2002). These factors mediate a generalized inhibition of gene expression through the non-specific shutdown of protein synthesis and mRNA degradation, what eventually leads to cell apoptosis. The recent demonstration that synthetic 21-nt siRNA duplexes effectively inhibit mammalian endogenous gene expression in a sequence-specific manner without activating the nonspecific dsRNA responses (Caplen, 2002, Elbashir et al., 2001) offers great opportunities to use this pathway of gene silencing to study the function of mammalian virus genes.
Rotaviruses have a segmented dsRNA genome, which is replicated in the cytoplasm of cells. The dsRNA genome, however, is masked at all times during the virus replication cycle, being found always associated with subviral particles and not free in the cellular cytosol. This probably prevents the rotavirus genome to be detected and targeted by general defense mechanisms of the cell. In addition, the virus replication cycle is highly lytic and rapid, being completed in about 12 h. The features of rotavirus replication represented a challenge for the RNAi machinery to silence the virus gene expression and to block the replication of the virus.
We showed that a siRNA with a sequence homologous to the gene encoding the virus spike protein VP4 (siRNAVP4) efficiently inhibited the synthesis of VP4 to a barely detectable level in cells transfected with the siRNA, whereas all other structural proteins remained unaltered (Dector et al., 2002). Similarly, the yield of progeny virus that resulted from an infection carried out in the presence of siRNAVP4 was reduced to 15–25% that of a control infection performed in the presence of an unrelated siRNA (homologous to the nuclear proteins laminA/C). The residual infectious virus produced in the presence of siRNAVP4 was mostly the result of virus replication in those cells that were not transfected with the siRNA, the transfection efficiency achieved being about 75%. As result of the inhibition of VP4 synthesis, non-infectious TLPs, lacking the VP4 protein (appearing as “spike-less” TLPs), were obtained (Dector et al., 2002).
We have also addressed the question of the duration of an effective RNAi response in a rotavirus infection. For this, we transfected MA104 cells with either a siRNA to VP4 or to VP7, and then infected with rotavirus at different times post-transfection. The inhibition of the synthesis of both viral proteins could be achieved even when the cells were infected 7 days after transfection (shown up to day 5 in Fig. 2A ), with the consequence of a significant decrease on the yield of viral progeny (Fig. 2B). The inhibitory effect could probably last even longer, however, after the fifth day cells in culture were notoriously damaged, even in the untransfected controls, so that it was difficult to have reproducible results at later times. Of interest, we consistently found that the highest inhibition was achieved when the cells were infected 72 h after transfection with the siRNA, suggesting that some of the elements of the RNAi response could be induced or activated by the presence of the siRNA, increasing their effective concentration inside the cell. Our observations are in agreement with other studies carried out with different viruses and different cell lines, where the RNAi effect was reported to last also about 5 days (Flores-Jasso et al., 2004, Ge et al., 2004, Kapadia et al., 2003). It is not clear, however, whether this effect is due to the fact that the siRNAs are stable within the cells, or whether they are slowly released from the membrane-associated transfection mixture (Gitlin and Andino, 2003).
Fig. 2.
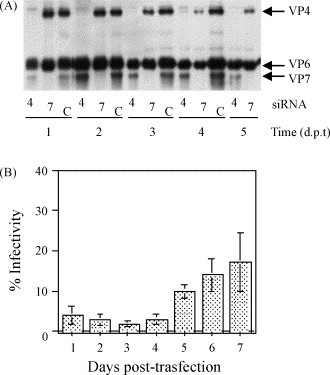
The RNAi response lasts up to 7 days. MA104 cells in 48-well plates were transfected with an siRNA directed to VP4 (4), VP7 (7), or lamin A/C (C), and after 8 h the lipofectamine–siRNA mixture was removed and replaced by medium. At the indicated times (days post-transfection, d.p.t.), the cells were infected with rotavirus RRV at an MOI of 1 and 12 h post-infection (p.i.), the cells were harvested and processed for electrophoresis or infectivity assays. (A) Western blot stained with a rabbit polyclonal antibody to rotavirus TLPs. Arrows indicate the positions of VP4, VP6, and VP7. (B) The yield of progeny virus obtained after transfection with the siRNA to VP4 was determined by an immunoperoxidase assay, as described (Lizano et al., 1991). Data are expressed as the percentage of the infectivity obtained when the cells were transfected with the control siRNA to lamin A/C.
The results obtained in our work indicate that rotaviruses are sensitive to the RNAi response of the cell, as has been described for viruses belonging to other families, such as poliovirus, hepatitis B and C viruses, human immunodeficiency virus, respiratory syncytial virus, vesicular stomatitis virus, papillomavirus, herpes virus, influenza virus, dengue virus, and baculovirus, some of which are discussed in accompanying articles of this Issue (Adelman et al., 2002, Bitko and Barik, 2001, Capodici et al., 2002, Coburn and Cullen, 2002, Ge et al., 2003, Gitlin et al., 2002, Hall and Alexander, 2003, Hamasaki et al., 2003, Hu et al., 2002, Jacque et al., 2002, Jia and Sun, 2003, Jiang and Milner, 2002, Kapadia et al., 2003, Klein et al., 2003, Martinez et al., 2002, McCaffrey et al., 2003, Novina et al., 2002, Randall et al., 2003, Shlomai and Shaul, 2003, Valdes et al., 2003, Wilson et al., 2003).
3. Is there a suppression of the RNAi response during rotavirus infection?
It has been found that many plant viruses, and at least one insect virus (flock house virus) are able to counteract the RNA interference of the cell by producing proteins that suppress this response at different levels (Ding et al., 2004, Li et al., 2002, Roth et al., 2004, Vance and Vaucheret, 2001). Although it is not yet known whether the RNAi has antiviral functions in mammalian cells, it is tempting to speculate that one of the rotavirus proteins could act as a suppressor of a cellular RNAi response triggered by the viral infection.
In a previous publication we described that the synthesis of a viral protein in MA104 cells lipofected with a siRNA specific for a viral protein 48–72 h prior to infection, was silenced in a sequence-specific manner (Dector et al., 2002, unpublished results), suggesting that rotavirus infection cannot suppress the interfering process. However, since in those experiments the siRNA was added to the cells before virus infection, we reasoned that for a virus-encoded suppressor of RNAi, there might have not been enough time for its synthesis before the RNAi response (triggered by the siRNA) inhibited the virus replication. To address this issue, we evaluated the capacity of the RNAi response to silence rotavirus gene expression when the siRNAs were transfected into cells after infection with the virus. We found that the synthesis of VP4 was silenced even when the siRNAVP4 was added at 4 h post-infection (p.i.), a time point at which viral protein synthesis is already predominant in the infected cells (Fig. 3 ). These results suggest that the RNAi machinery is active and effective even in the presence of previously synthesized rotaviral proteins, and thus that rotaviruses do not encode an RNAi suppressor. However, in order to formally demonstrate that rotaviruses do not code for suppressors of RNAi, further experiments need to be done. For example, it is necessary to determine if rotavirus infection could block an RNAi response triggered by short hairpin RNAs, which have been shown to specifically induce the RNAi system. These short hairpin RNAs are substrates for the cellular enzyme Dicer (Brummelkamp et al., 2002), which is responsible for processing long dsRNA molecules into siRNAs. Also, it could be helpful to assay candidate viral proteins as RNAi suppressors in a heterologous system, like plants. With such an approach, it was recently reported that the σ3 protein of reovirus, which is a dsRNA binding protein, has a supressor activity that blocks the synthesis of GFP in an assay for suppressor silencing in tobacco plants (Lichner et al., 2003, Roth et al., 2004).
Fig. 3.
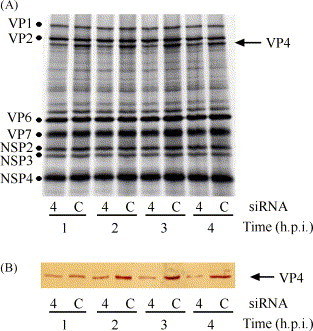
Rotavirus gene expression is efficiently inhibited even if RNAi is induced after viral infection. MA104 cells in 48-well plates were infected for 1 h with rotavirus RRV, at an MOI of 3. At the indicated times post-infection (hours post-infection, h.p.i.), the cells were transfected with a siRNA to VP4 (4), or lam A/C(C) as control, for 8 h. At 12 h.p.i. the cells were labeled with [35S]-methionine for 1 h, lysed, and the proteins separated by gel electrophoresis and analyzed by fluorography (A), or by Western blot using a monoclonal antibody to VP4 (B). The dots in (A) indicate the migration of viral proteins other than VP4.
4. RNA interference to study the function of rotavirus proteins
The segmented nature of the genome of the viruses in the Reoviridae family makes them particularly amenable to analysis by RNAi. In the case of rotaviruses, each of the 11 segments of dsRNA is transcribed into a single mRNA, each encoding a single protein (with the exception of one bicistronic segment). This makes it possible to silence the expression of individual genes without affecting the expression of the others. The analysis of the phenotypes generated should allow the characterization of the function of proteins encoded by the silenced genes.
This sort of individual gene function analysis is more difficult to carry out in viruses with a positive strand RNA genome, since, as it has been shown for poliovirus, hepatitis C (Randall and Rice, 2004, Saleh et al., 2004), and SARS-coronavirus (Zhang et al., 2003), their genome is targeted by the RNAi machinery, inhibiting the replication of the virus. Since the genome of these viruses functions as mRNA to direct the synthesis of precursor polyproteins which are post-translationally cleaved to yield the individual viral polypeptides, any siRNA directed against the mRNA (represented by either the genomic RNA, or the subgenomic mRNAs in some virus families) would result in a decreased synthesis of the entire polyprotein, preventing the study of the role of individual polypeptides. In this regard, it is of interest that in viruses with a non-segmented, negative strand RNA genome, like respiratory syncytial virus, vesicular stomatitis virus, and human parainfluenza virus, the genomic RNA as well as the replication intermediate RNA(+), are protected from the action of RNAi, while only the mRNAs are degraded (Barik, 2004, Bitko and Barik, 2001). This also seems to be the case for influenza A viruses which have a genome composed of eight single-stranded RNA segments of negative polarity (Ge et al., 2003). The functions of proteins of DNA viruses and retroviruses, whose genome is in general transcribed into monocistronic mRNAs, is also amenable to dissection by RNAi, as has been shown for baculovirus, human immunodeficiency virus, and retroviruses in this issue, and for herpes virus, papillomavirus and the hepatitis delta agent (Chang and Taylor, 2003, Hall and Alexander, 2003, Jia and Sun, 2003).
In the case of rotaviruses, one could explore the function of the viral proteins in three different conceptual areas: (i) proteins that do not participate in the replication of the virus genome, but are involved in the late steps of virus morphogenesis; (ii) proteins involved directly or indirectly in the replication of the genomic RNA; and (iii) proteins that, although not directly involved in virus replication, contribute to make the replication cycle more efficient. Some viral proteins could certainly have activities that could fit into more than one of the areas described.
Silencing the viral genes that encode proteins involved in the late steps of morphogenesis, like VP4 and VP7, should allow the replication cycle to reach the synthesis of the second wave of double-layered RIs. However, after this point the morphogenesis of the virus should be impaired. In agreement with this hypothesis, we have shown that the silencing of the VP4 gene allowed the synthesis of abundant double-layered RIs, that efficiently bud through the ER membrane, eventually loosing the transient lipid membrane and incorporating the outer protein layer formed by VP7, to form spike-less TLPs. Of interest, the VP7 outer layer assembled on the spike-less TLPs was found by cryoelectron microscopy to have an structure identical to that of the outer layer in wild-type TLPs (B.V.V. Prasad, unpublished results), clearly showing that the assembly of VP7 on DLPs is a process independent of the assembly of VP4. In addition, these findings indicated that VP4 is neither required for the budding process nor for the removal of the lipid membrane from the enveloped particles, as had been suggested (Estes, 2001). Similarly, silencing the VP7 gene prevented the formation of TLPs, and caused DLPs to accumulate (Patton, 2003; Camacho et al., manuscript in preparation). A priori, one would expect that inhibition of the synthesis of NSP4, the ER membrane receptor for DLPs, should also lead to an accumulation of DLPs. However, since it is known that this protein alters the calcium homeostasis of cells (Tian et al., 1994, Tian et al., 1995), its absence might have additional effects on the virus replication cycle. Silencing the NSP4 and VP7 genes should allow us to establish directly the role of these proteins in the translocation of DLPs into the lumen of the ER and in the removal of the intermediary lipid envelope.
RNAi should also be useful to study in vivo the role of proteins that have been suggested to be involved in the early stages of the virus replication cycle, like the viral polymerase VP1, the protein that forms the innermost shell of the virion VP2, and the nonstructural proteins NSP2 and NSP5, all of which are thought to be necessary to generate core RIs containing newly formed genomic dsRNA segments (Patton and Spencer, 2000). One would expect that silencing the expression of either of these genes would block the virus secondary transcription, with the concomitant inhibition of the synthesis of all viral proteins, as it has been found for VP1 and NSP5 (Campagna et al., 2003; T. López and M. Arias, manuscript in preparation). A more detailed analysis of these data should allow us to define the role of each protein in the formation of functional cores. Also, since most viral proteins do not have a random distribution within the cell, the inhibition of the synthesis of particular proteins should help to identify the role they might have in the overall intracellular organization of the viral polypeptides.
In addition, RNAi could serve to approach the characterization of the role of viral proteins that have been proposed to favor the replication cycle of the virus through, for instance, inhibiting the cellular protein synthesis (NSP3), increasing the translation rate of the viral proteins (Piron et al., 1998), counteracting cellular activities that might be noxious to virus replication, such as the interferon response (NSP1) (Graff et al., 2002), or promoting the cytopathic effect to facilitate the release of virus particles (NSP4) (Estes, 2003).
It is foreseen that in the near future many new studies using this methodology will help to unravel the role of all rotaviral proteins in vivo, both in cell culture and in animal models. The feasibility of inhibiting virus replication in complete animals has already been shown for hepatitis C using the mouse as a model (McCaffrey et al., 2002).
5. RNAi to study rotavirus–host cell interactions
Recently, a number of cellular proteins have been suggested to participate in the virus replication cycle. These proteins range from virus cellular receptors to chaperones and transcription factors. It is clear that RNAi will help to elucidate the role of these cell molecules during the infectious process.
The entry of rotaviruses into cells has been described as a multistep process where at least four interactions between the viral surface proteins and cellular surface molecules occur. The molecules implicated as virus receptors include gangliosides GM1 and GM3, integrins α2β1, αxβ2, and αvβ3, and the heat shock cognate protein hsc70 (Arias et al., 2001). Silencing of individual receptors or combinations of them should help to establish their role during virus entry. In fact, preliminary experiments have shown that inhibition of αvβ3 expression decreases the ability of rotaviruses to infect epithelial cells (P. Isa, unpublished results).
It has been shown that rotavirus infection induces the expression of a large set of genes, including those encoding heat shock proteins hsp90 and hsp70, as well as the glucose regulated proteins grp78 and grp94 (Cuadras et al., 2002, Xu et al., 1998; L. Maruri, unpublished). These findings, together with the fact that cells that are poorly susceptible to rotavirus infection increase their susceptibility to the virus up to 100-fold when subjected to a heat shock (T. Lopez, unpublished results), suggest that stress proteins might be important for the efficient replication of the virus. The role of these proteins in the virus replication cycle should be amenable to analysis by RNAi.
Knockdown of the caveolin-1 gene and other genes encoding proteins involved in different types of cell endocytosis, like those encoding clathrin and Eps15, should also help to study the role of these proteins in virus entry. Using this approach, together with pharmacological and other genetic approaches, we have recently described that rotavirus cell entry occurs through a caveolae- and clathrin-independent endocytosis, (Sánchez-SanMartín et al., submitted).
6. RNAi induces the degradation of only a subset of the viral transcripts
During rotavirus infection, the DLPs derived from the infecting viruses synthesize 11 viral transcripts. Since these transcripts function both as mRNA and as RNA(+), it was expected that siRNAs directed to a given transcript would impair the synthesis of both the corresponding protein and the corresponding dsRNA segment. However, we and others have found that when silencing rotavirus genes encoding proteins not involved in the replication of the viral genome, such as VP4 and VP7, the newly assembled particles, either spike-less TLPs (in the case of silencing VP4) or DLPs (in the case of silencing VP7), contain all 11 RNA segments in equimolar amounts (Dector et al., 2002, Patton, 2003; Camacho et al., manuscript in preparation), despite the fact that the synthesis of the target protein is almost completely ablated, indicating that the mRNA has been degraded.
The previous observations have been taken to suggest that viral transcripts establish two, functionally different and physically separated pools (Patton, 2003; Camacho et al., manuscript in preparation), one that directs the synthesis of proteins, and the other that is used as template for replication of the virus genome. The mRNAs are accessible in the cytoplasm to be translated (where they can be efficiently targeted by RNAi), whereas the RNA(+) molecules acting as template for dsRNA replication could be either kept within the so-called viroplasms (where the synthesis of the RNA(−) is thought to take place) or could be associated in the cytoplasm with RNA binding proteins, making them inaccessible to the RNAi machinery. Even though the existence of two separated viral transcripts pools would appear to be logical, the common belief is that viral transcripts represent a single pool of RNA that could be used at any time, and stochastically, for either of the two functions. The hypothesis that a mRNA pool could be physically separated from the RNA(+) (replication) pool might explain why the attempts to develop a reverse genetic system for rotaviruses have so far failed; the introduction of exogenous transcripts that represent rotavirus mRNAs (obtained by in vitro synthesis), or the production inside the cell of these transcripts from recombinant vectors, might not reach the compartment where such molecules are used as templates for replication.
7. Perspectives for RNAi in rotavirus research
The ability to knock down the expression of individual rotavirus genes should provide the basis to develop phenotypic complementation systems in the short term. In principle, given the high efficiency of RNAi to prevent the synthesis of a given viral protein, it is conceivable that an almost homologous polypeptide (encoded by a gene having a few nucleotide differences in the siRNA target site) could be synthesized from a recombinant expression vector, to phenotypically rescue the activity of the missing protein. Once established, such system should allow the expression of modified versions of the protein to dissect its function. In the absence of a reverse genetics system, this approach should be very useful to carry out functional genomics in rotaviruses, and in viruses of other genera of the Reoviridae. In addition, it can be forseen that the studies employing RNAi will soon extend to the analysis of a number of cellular genes whose protein products influence the replication of the virus in a positive or negative manner, and more in the mid term, to the analysis of viral gene function in complete animal models.
One of the limiting steps using RNAi to silence viral genes is the siRNA transfection efficiency. Thus, the RNAi studies, including the phenotypic rescue system described above, will be greatly improved and simplified if all cells in a culture could harbor the silencing siRNA. This could be achieved by constructing stable cell lines expressing the siRNA (either as two complementary strands that will hybridize inside the cell, or as precursor short hairpin RNAs; Miyagishi et al., 2004), or alternatively, by developing better systems to deliver the siRNAs into the cells, for instance, using lentiviruses or flavivirus replicons as vectors to direct the intracellular synthesis of the siRNAs. These approaches should be evaluated carefully to avoid the induction of the non-specific interferon response by the siRNAs, as has been recently reported (Bridge et al., 2003, Sledz et al., 2003).
The sequence specificity of RNAi is remarkable; siRNAs that differ from their target sequence in one or more bases, depending on the position of the mismatch, do not efficiently silence the expression of that gene. This fact has been regarded at as a drawback for the use of siRNAs as potential therapeutic antiviral compounds, since variant viruses could escape the RNAi response, as has been shown for poliovirus in cell culture (Gitlin et al., 2002). However, based on the high specificity of RNAi, an interesting potential application could be the use of siRNAs directed to a specific region of a gene, to select viral escape mutants that will contain nucleotide changes in the specific targeted region. SiRNAs could then be used as region-specific mutagens to generate pre-designed mutants.
On the other hand, the inhibition of viral gene expression by siRNAs offers the potential for a novel therapeutic approach for infections by rotaviruses and other viruses, once efficient methods for in vivo delivery of siRNAs have been developed. In addition to the standard management of rotavirus infection symptoms, directed towards restoration of fluid and electrolyte balance until the infection resolves (symtomatic treatment), specific interventions aimed at inhibiting the viral replication could be designed. The introduction of siRNAs preventing the replication of the virus (for instance silencing genes involved in the replication of the viral genome) could shorten the duration of the illness and possibly help to prevent dehydration, the principal cause of death of a rotavirus infection. Another attractive target for antiviral therapy could be NSP4, since this nonstructural protein, in addition to its important role during virus morphogenesis, has been shown to function as a viral enterotoxin that contributes to the diarrheal illness (Estes, 2003).
Although the use of RNAi as an antiviral therapeutic principle and method still remains to be demonstrated, there is no doubt that this technology will influence drastically the way postgenomic viral and cellular research is conducted.
Acknowledgements
We thank Ulrich Desselberger for critical reading the manuscript. The excellent technical assistance of Pedro Romero is acknowledged. This work was partially supported by grants 55003662 and 55000613 from the Howard Hughes Medical Institute, and by grant G37621N from the National Council for Science and Technology—Mexico.
References
- Adelman Z.N., Sanchez-Vargas I., Travanty E.A., Carlson J.O., Beaty B.J., Blair C.D., Olson K.E. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 2002;76:12925–12933. doi: 10.1128/JVI.76.24.12925-12933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C.F., Guerrero C.A., Mendez E., Zarate S., Isa P., Espinosa R., Romero P., Lopez S. Early events of rotavirus infection: the search for the receptor(s) Novartis Found. Symp. 2001;238:47–60. doi: 10.1002/0470846534.ch4. discussion 60–43. [DOI] [PubMed] [Google Scholar]
- Barik, S., 2004. Control of nonsegmented negative-strand RNA virus replication by siRNA. Virus Res., this issue. [DOI] [PubMed]
- Bitko V., Barik S. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 2001;1:34. doi: 10.1186/1471-2180-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge A.J., Pebernard S., Ducraux A., Nicoulaz A.L., Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Campagna, M., Vascotto, F., Eichwald, C., Burrone, O.R., 2003. Interfering with rotavirus NSP5. In: Proceedings of 8th International Symposium on Double-Stranded RNA Viruses, Lucca, Italy.
- Caplen N.J. A new approach to the inhibition of gene expression. Trends Biotechnol. 2002;20:49–51. doi: 10.1016/s0167-7799(01)01900-x. [DOI] [PubMed] [Google Scholar]
- Capodici J., Kariko K., Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- Chang J., Taylor J.M. Susceptibility of human hepatitis delta virus RNAs to small interfering RNA action. J. Virol. 2003;77:9728–9731. doi: 10.1128/JVI.77.17.9728-9731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn G.A., Cullen B.R. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadras M.A., Feigelstock D.A., An S., Greenberg H.B. Gene expression pattern in Caco-2 cells following rotavirus infection. J. Virol. 2002;76:4467–4482. doi: 10.1128/JVI.76.9.4467-4482.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dector M.A., Romero P., Lopez S., Arias C.F. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 2002;3:1175–1180. doi: 10.1093/embo-reports/kvf234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W., Li, H., Lu, R., Li, F., Li, W.X., 2004. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res., this issue. [DOI] [PubMed]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Estes, M.K., 2001. Rotaviruses and their replication. In: Knipe, D.N., Howley, P.M. (Eds.), Virology, fourth ed. Lippincott Williams and Wilkins, Philadelphia, PA, pp. 1747–1785.
- Estes, M.K., 2003. The rotavirus NSP4 enterotoxin: current status and challenges. In: Desselberger, U., Gray, J. (Eds.), Viral Gastroenteritis. Elsevier Science, Amsterdam, pp. 207–224.
- Flores-Jasso, C.F., Valdes, V.J., Sampieri, A., Valadez-Graham, V., Recillas-Targa, F., Vaca, L., 2004. Silencing structural and nonstructural genes in baculovirus by RNA interference. Virus Res., this issue. [DOI] [PubMed]
- Ge, Q., Eisen, H.N., Chen, J., 2004. Use of siRNAs to prevent and treat influenza virus infection. Virus Res., this issue. [DOI] [PubMed]
- Ge Q., McManus M.T., Nguyen T., Shen C.H., Sharp P.A., Eisen H.N., Chen J. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L., Andino R. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J. Virol. 2003;77:7159–7165. doi: 10.1128/JVI.77.13.7159-7165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L., Karelsky S., Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- Graff J.W., Mitzel D.N., Weisend C.M., Flenniken M.L., Hardy M.E. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 2002;76:9545–9550. doi: 10.1128/JVI.76.18.9545-9550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.L., Halasz P., Tan Y., Hewish M.J., Takada Y., Mackow E.R., Robinson M.K., Coulson B.S. Integrin-using rotaviruses bind a2b1 integrin a2 I domain via VP4 DGE sequence and recognize aXb2 and aVb3 by using VP7 during cell entry. J. Virol. 2003;77:9969–9978. doi: 10.1128/JVI.77.18.9969-9978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A.H., Alexander K.A. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J. Virol. 2003;77:6066–6069. doi: 10.1128/JVI.77.10.6066-6069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki K., Nakao K., Matsumoto K., Ichikawa T., Ishikawa H., Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543:51–54. doi: 10.1016/s0014-5793(03)00400-9. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hu W.Y., Myers C.P., Kilzer J.M., Pfaff S.L., Bushman F.D. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 2002;12:1301–1311. doi: 10.1016/s0960-9822(02)00975-2. [DOI] [PubMed] [Google Scholar]
- Jacque J.M., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q., Sun R. Inhibition of gammaherpesvirus replication by RNA interference. J. Virol. 2003;77:3301–3306. doi: 10.1128/JVI.77.5.3301-3306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- Kapadia S.B., Brideau-Andersen A., Chisari F.V. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze M.G., He Y., Gale M., Jr Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Klein C., Bock C.T., Wedemeyer H., Wustefeld T., Locarnini S., Dienes H.P., Kubicka S., Manns M.P., Trautwein C. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9–18. doi: 10.1016/s0016-5085(03)00720-0. [DOI] [PubMed] [Google Scholar]
- Li H., Li W.X., Ding S.W. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Lichner Z., Silhavy D., Burgyan J. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 2003;84:975–980. doi: 10.1099/vir.0.18987-0. [DOI] [PubMed] [Google Scholar]
- Lizano M., Lopez S., Arias C.F. The amino-terminal half of rotavirus SA114fM VP4 protein contains a hemagglutination domain and primes for neutralizing antibodies to the virus. J. Virol. 1991;65:1383–1391. doi: 10.1128/jvi.65.3.1383-1391.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M.A., Clotet B., Este J.A. RNA interference of HIV replication. Trends Immunol. 2002;23:559–561. doi: 10.1016/s1471-4906(02)02328-1. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Meuse L., Pham T.T., Conklin D.S., Hannon G.J., Kay M.A. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Nakai H., Pandey K., Huang Z., Salazar F.H., Xu H., Wieland S.F., Marion P.L., Kay M.A. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Miyagishi, M., Matsumoto, S., Taira, K., 2004. Generation of an RNA expression library against the human genome. Virus Res., this issue. [DOI] [PubMed]
- Novina C.D., Murray M.F., Dykxhoorn D.M., Beresford P.J., Riess J., Lee S.K., Collman R.G., Lieberman J., Shankar P., Sharp P.A. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Parashar, U.D., Glass, R.I., 2003. Viral causes of gastroenteritis. In: Desselberger, U., Gray, J. (Eds.), Viral Gastroenteritis. Elsevier Science, Amsterdam, pp. 9–22.
- Patton J.T. Structure and function of the rotavirus RNA-binding proteins. J. Gen. Virol. 1995;76(Pt 11):2633–2644. doi: 10.1099/0022-1317-76-11-2633. [DOI] [PubMed] [Google Scholar]
- Patton, J.T., 2003. Rotavirus genome replication: sites and signals. In: Proceedings of 8th International Symposium on Double-Stranded RNA Viruses, Lucca, Italy.
- Patton J.T., Spencer E. Genome replication and packaging of segmented double-stranded RNA viruses. Virology. 2000;277:217–225. doi: 10.1006/viro.2000.0645. [DOI] [PubMed] [Google Scholar]
- Pesavento, J.B., Estes, M.K., Prasad, B.V.V., 2003. Structural organization of the genome in rotavirus. In: Desselberger, U., Gray, J. (Eds.), Viral Gastroenteritis, first ed., vol. 9. Elsevier Science, Amsterdam, pp. 115–128.
- Piron M., Vende P., Cohen J., Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poruchynsky M.S., Atkinson P.H. Rotavirus protein rearrangements in purified membrane-enveloped intermediate particles. J. Virol. 1991;65:4720–4727. doi: 10.1128/jvi.65.9.4720-4727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G., Grakoui A., Rice C.M. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall, G., Rice, C.M., 2004. Interfering with hepatitis C virus RNA replication. Virus Res., this issue. [DOI] [PubMed]
- Roth, B.M., Pruss, G.J., Vance, V., 2004. Plant viral suppressors of RNA silencing. Virus Res., this issue. [DOI] [PubMed]
- Saleh, M., Van Rij, R.P., Andino, R., 2004. RNA silencing in viral infections: insights from poliovirus. Virus Res., this issue. [DOI] [PubMed]
- Sharp P.A. RNA interference—2001. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- Shlomai A., Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;9:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Tian P., Estes M.K., Hu Y., Ball J.M., Zeng C.Q., Schilling W.P. The rotavirus nonstructural glycoprotein NSP4 mobilizes Ca2+ from the endoplasmic reticulum. J. Virol. 1995;69:5763–5772. doi: 10.1128/jvi.69.9.5763-5772.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P., Hu Y., Schilling W.P., Lindsay D.A., Eiden J., Estes M.K. The nonstructural glycoprotein of rotavirus affects intracellular calcium levels. J. Virol. 1994;68:251–257. doi: 10.1128/jvi.68.1.251-257.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes V.J., Sampieri A., Sepulveda J., Vaca L. Using double-stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J. Biol. Chem. 2003;278:19317–19324. doi: 10.1074/jbc.M212039200. [DOI] [PubMed] [Google Scholar]
- Vance V., Vaucheret H. RNA silencing in plants—defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Wilson J.A., Jayasena S., Khvorova A., Sabatinos S., Rodrigue-Gervais I.G., Arya S., Sarangi F., Harris-Brandts M., Beaulieu S., Richardson C.D. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A., Bellamy A.R., Taylor J.A. BiP (GRP78) and endoplasmin (GRP94) are induced following rotavirus infection and bind transiently to an endoplasmic reticulum-localized virion component. J. Virol. 1998;72:9865–9872. doi: 10.1128/jvi.72.12.9865-9872.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. RNA interference: listening to the sound silence. Nat. Struct. Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- Zárate, S., Espinosa, R., Romero, P., Arias, C.F., López, S., 2002. Rotavirus interacts with avb3 through a novel binding site. In: Proceedings of Meeting of the Howard Hughes Medical Institute International Research Scholars, Palm Cove, Australia.
- Zhang R., Guo Z., Lu J., Meng J., Zhou C., Zhan X., Huang B., Yu X., Huang M., Pan X., Ling W., Chen X., Wan Z., Zheng H., Yan X., Wang Y., Ran Y., Liu X., Ma J., Wang C., Zhang B. Inhibiting severe acute respiratory syndrome-associated coronavirus by small interfering RNA. Chin. Med. J. 2003;116:1262–1264. [PubMed] [Google Scholar]


