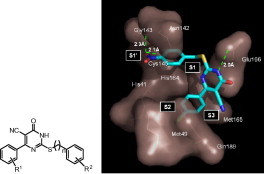
Graphical abstract
2-(Benzylthio)-6-oxo-4-phenyl-1,6-dihydropyrimidine derivatives have been prepared and their inhibition activities against SARS-CoV 3CL protease were evaluated.
Keywords: SARS-CoV, Pyrimidines
Abstract
A series of 2-(benzylthio)-6-oxo-4-phenyl-1,6-dihydropyrimidine as SARS-CoV 3CL protease inhibitors were developed and their potency was evaluated by in vitro protease inhibitory assays. Two candidates had encouraging results for the development of new anti-SARS compounds.
Severe acute respiratory syndrome (SARS) has been recognized as a global threat. SARS is characterized by high fever, malaise rigor, headache, chills, cough, and progressive radiographic changes of the chest and lymphophenia.1, 2, 3 The initial outbreak of SARS was first identified in Guangdong Province, China in November 2002. This outbreak spread to several countries and has had significant health and economic impact. The mortality rate is nearly 10%.4 With rigorous effort by the world health organization (WHO), researchers found that SARS is caused by a novel coronavirus, SARS-CoV.1, 2, 5 The SARS-CoV is a positive-strand RNA virus and the genome is ∼30 kb (Tor2 strain). The genome is constituted of five major open reading frames namely replicase polyproteins, nucleocapsid proteins, spike (S), envelope (E), and membrane (M) glycoproteins.
Resulting of structural and functional studies of coronaviral lifecycle has provided a number of significant targets for ceasing the viral replication. During the viral replication, the replicase polyprotein undergoes extensive processing by two viral proteases namely, chymotrypsin-like protease (3CLpro) and papain-like protease (PLpro), reside within the polyprotein. They catalyze their own release from the polyprotein and other non-structural proteins (nsps) from the polyproteins and initiate virus mediated RNA replication.6 Because of their essential roles in viral replication, both proteases are recognized as attractive targets for development of anti-SARS agents.
To date various SARS-CoV protease inhibitors have been reported from both screened compound libraries and designed compounds based on the substrate structure or active site properties. Their scaffolds are diverse, including C 2-symmetric diols,7 3-quinolinecarboxylic acid derivatives,8 thiophene-2-carboxylate derivatives,9 cinanserin,10 calmodulin,11 keto-glutamine analogues,12 anilide,13 bifunctional boronic acid compounds,14 isatin derivatives,15 benzotriazole16 as well as glutamic acid and glutamine peptides possessing a trifluoromethyl ketone group,17 α,β-unsaturated esters,18 and etacrynic acid derivatives.19 With metal-conjugated structures, some molecules make a coordinate bond with Cys-145 at the active site of SARS-CoV 3CLpro.20 However, no effective therapy has been developed so far and recent isolation of strains of SARS-CoV emphasizes the possibility of a reemergence. Therefore, it is still a great challenge to explore new chemical classes of SARS-CoV 3CLpro inhibitors that can be used in anti-SARS therapy in case the disease re-emerges.
In our previous study, from high throughput screening we have identified various heterocycles as novel anti-SARS agents with selective inhibition ranging from IC50 2–10 μM against SARS–CoV 3CL protease (compounds 1–4, Fig. 1 ).21, 22 In this paper, as a part of our ongoing efforts to delineate a complete pharmacophore model, we designed several 2-(benzylthio)-6-oxo-4-phenyl-1,6-dihydropyrimidine derivatives as anti-SARS agents (compounds 6a–n, Fig. 1).
Figure 1.
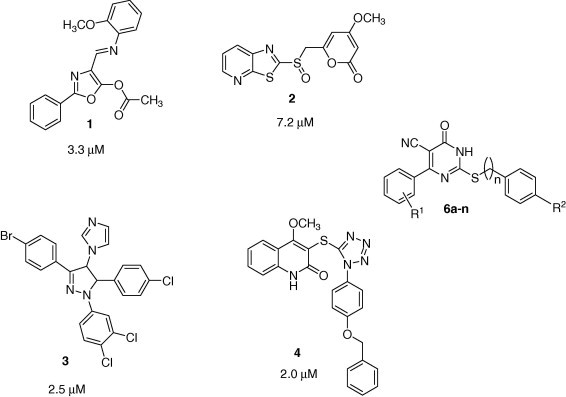
Hits from this study (6) and previous studies21, 22 in our laboratory.
From a synthetic point view, the preparation of the target compounds was envisioned following the synthetic routes illustrated in Scheme 1 . The synthesis of 6-aryl-5-cyano-2-thiouracils 5a–n was prepared by reaction between substituted benzaldehyde, ethyl cyanoacetate, and thiourea using the literature procedure.23 The regioselective S-alkylation of 2-thiouracil 5a–n achieved by slow addition of the respective halides to a solution of 5a–n in DMF using K2CO3 as a base at 0–5 °C to yield the corresponding compounds 6a–n.24 Both analytical and spectral data of all target compounds are accordant with the structures.
Scheme 1.
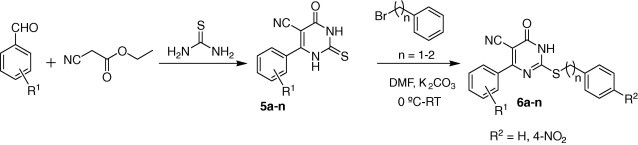
Synthesis of Inhibitors 6a–n.
The target compounds were tested for anti-SARS activity against SARS-CoV 3CLpro, using previously developed assay method25 containing 0.05 μM SARS 3CLpro, 6 μM fluorogenic substrate Dabcyl-KTSAVLQSGFRKME-Edans, and 50 μM of test compounds. Enhanced fluorescence of the reactions in the buffer of 20 mM Bis–Tris at pH 7.0 was monitored at 538 nm with excitation at 355 nm using a fluorescence plate reader. The compounds which inhibited more than 50% of the protease activity at 50 μM were selected for the next assay run at 10 μM for IC50 calculation. Compound 6m with R2 group of nitro functionality at C-4 position is the most potent inhibitor with an enzyme inhibitory activity against SARS-CoV 3CLpro with an IC50 of 6.1 μM. The structure and IC50 values are given in Table 1 . The cytotoxicity of the test compounds was tested by performing the MTT assay and found that all compounds are devoid of cytotoxicity.24
Table 1.
Structure and activity of compounds 6a–n.
| Compound | R1 | R2 | n | IC50 (μM) |
|---|---|---|---|---|
| 6a | H | H | 1 | >50 |
| 6b | H | H | 2 | >50 |
| 6c | H | 4-NO2 | 1 | 35.2 |
| 6d | 4-OCH3 | H | 1 | >50 |
| 6e | 4-OCH3 | H | 2 | 20.3 |
| 6f | 4-OCH3 | 4-NO2 | 1 | 26.3 |
| 6g | 4-CH3 | H | 1 | >50 |
| 6h | 4-CH3 | H | 2 | >50 |
| 6i | 4-CH3 | 4-NO2 | 1 | >50 |
| 6j | 3-NO2 | H | 2 | >50 |
| 6k | 3-NO2 | 4-NO2 | 1 | 10.6±1.2 |
| 6l | 4-Cl | H | 2 | 16.9±1.3 |
| 6m | 4-Cl | 4-NO2 | 1 | 6.1±1.1 |
| 6n | 3-Cl | H | 1 | >50 |
To obtain molecular insight into the binding properties of these active compounds, we conducted docking studies in the 3CLpro active site. For modeling analysis, the crystal structure of SARS 3CLpro in complex with a peptide inhibitor (PDB code 1UK4) was used.26 Docking process was performed using an automated ligand-docking subprogram of the Discovery Studio Modeling 1.2 SBD (Accelrys Inc., San Diego, CA), with a set of parameters chosen to control the precise operation of the genetic algorithm. Docking runs were carried out using standard default settings ‘grid resolution’ of 5 Å, ‘site opening’ of 12 Å, and ‘binding site’ selected for defining the active site cavity.
In search of obtaining a model of the associated complex between the compound 6m and protein, the distance between the NH of the pyrimidine ring and oxygen atom of Glu-166 was constrained in the distance of 2.0 Å. The orientation of the ligand has the nitro phenyl group situated in the S1 pocket, with the nitro group pointing towards the surface of the protein. One of the oxygen of the nitro group is in close proximity 2.3 Å to the Gly-143 and the other oxygen atom is forming hydrogen bond with Cys-145 at the distance of 2.1 Å (Fig. 2 ). The chlorophenyl ring fits into the S2 pocket and having hydrophobic interactions with Met-49 and Gln-189. The results of the docking studies presented here suggest that the nitro phenyl group of 6m can potentially occupy the active site cysteine residue in the enzyme. The oxygen of the nitro group formed a hydrogen bond with the side chain of Gly-143 and Cys-145 that was important for inhibition activity. The compounds lacking nitro functionality in the aryl ring lost the activity. Moderate electron withdrawing substituent R1 like chloro in the compounds 6l and 6m favors the inhibitory activity when compared to the electron donating groups like methyl and methoxy in the compound (see Table 1). This result suggests that the substituent R1 can be electron withdrawing group to increase the inhibitory action.
Figure 2.
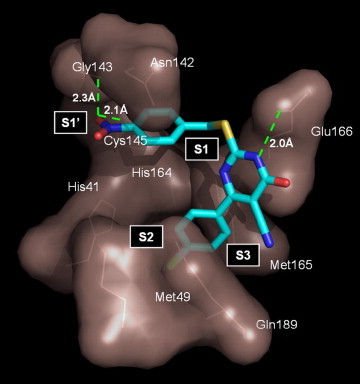
Computer modeling of 6m binding in the active site of SARS 3CLpro.
In conclusion, we disclosed the inhibitory potency of compound 6m, containing pyrimidine unit, as a SARS-CoV 3CLpro inhibitor. The measured inhibitory activity coupled with possible structure modifications revealed by 3D docking give us new directions for a fast development of much more potent inhibitors. Further investigations on this new family of compounds are currently in progress in our laboratory.
References and notes
- 1.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Shieh W.-J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.-Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Peiris J.S.M., Lai S.-T., Poon L.L.-M., Guan Y., Yam L.Y.-C., Lim W., Nicholls J., Yee W.K.-S., Yan W.W., Cheung M.-T., Cheng V.C.-C., Chan K.-H., Tsang D.N.-C., Yung R.W.-H., Ng T.K., Yuen K.-Y. Lancet. 2003;361:1319. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.-H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.He J.-F., Peng G.-W., Min J., Yu D.-W., Liang W.-L., Zhang S.-Y., Xu R.-H., Zheng H.-Y., Wu X.-W., Xu J., Wang Z.-H., Fang L., Zhang X., Li H., Yan X.-G., Lu J.-H., Hu Z.-H., Huang J.-C., Wan Z.-Y., Hou J.-L., Lin J.-Y., Song H.-D., Wang S.-Y., Zhou X.-J., Zhang G.-W., Gu B.-W., Zheng H.-J., Zhang X.-L., He M., Zheng K., Wang B.-F., Fu G., Wang X.-N., Chen S.-J., Chen Z., Hao P., Tang H., Ren S.-X., Zhong Y., Guo Z.-M., Liu Q., Miao Y.-G., Kong X.-Y., He W.-Z., Li Y.-X., Wu C.-I., Zhao G.-P., Chiu R.W.K., Chim S.S.C., Tong Y.-K., Chan P.K.S., Tam J.S., Lo Y.M.D. Science. 2004;303:1666. [Google Scholar]
- 5.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 6.Baker S.C. 3rd ed. Vol. 1. Academic Press; New York: 2008. Coronaviruses: Molecular Biology; pp. 554–562. (Encyclopedia of Virology). [Google Scholar]
- 7.(a) Wu C.-Y., Jan J.-T., Ma S.-H., Kuo C.-J., Juan H.-F., Cheng E.Y.-S., Hsu H.-H., Huang H.-C., Wu D., Brik A., Liang F.-S., Liu R.-S., Fang J.-M., Chen S.-T., Liang P.-H., Wong C.-H. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10012. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shao Y.-M., Yang W.-B., Peng H.-P., Hsu M.-F., Tsai K.-C., Kuo T.-H., Wang A.H.-J., Liang P.-H., Lin C.-H., Yang A.-S., Wong C.-H. ChemBioChem. 2007;8:1654. doi: 10.1002/cbic.200700254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kao R.Y., Tsui W.H.W., Lee T.S.W., Tanner J.A., Watt R.M., Huang J.D., Hu L.H., Chen G.H., Chen Z.W., Zhang L.Q., He T., Chan K.H., Tse H., To A.P.C., Ng L.W.Y., Wong B.C.W., Tsoi H.W., Yang D., Ho D.D., Yuen K.Y. Chem. Biol. 2004;11:1293. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. Chem. Biol. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Chen L.L., Gui C.S., Luo X.M., Yang Q.G., Gunther S., Scandella E., Drosten C., Bai D., He X.C., Ludewig B., Chen J., Luo H.B., Yang Y.M., Yang Y.F., Zou J.P., Thiel V., Chen K., Shen J.H., Xu S., Jiang H.L. J. Virol. 2005;79:7095. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yang Q., Chen L., He X., Gao Z., Shen X., Bai D. Chem. Pharm. Bull. 2008;56:1400. doi: 10.1248/cpb.56.1400. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z., Huang C., Fan K., Wei P., Chen H., Liu S., Pei J., Shi L., Li B., Yang K., Liu Y., Lai L. J. Chem. Inf. Model. 2005;45:10–17. doi: 10.1021/ci049809b. [DOI] [PubMed] [Google Scholar]
- 12.Jain R.P., Petterson H.I., Zhang J., Aull K.D., Fortin P.D., Huitema C., Eltis L.D., Parrish J.C., James M.N.G., Wishart D.S., Vederas J.C. J. Med. Chem. 2004;47:6113. doi: 10.1021/jm0494873. [DOI] [PubMed] [Google Scholar]
- 13.Shie J.-J., Fang J.-M., Kuo T.-H., Kuo C.-J., Liang P.-H., Huang H.-J., Yang W.-B., Lin C.-H., Chen J.-L., Wu Y.-T., Wong C.-H. J. Med. Chem. 2005;48:4469. doi: 10.1021/jm050184y. [DOI] [PubMed] [Google Scholar]
- 14.Bacha U., Barrila J., Velasquez-Campoy A., Leavitt S.A., Freire E. Biochemistry. 2004;43:4906. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 15.Chen L.-R., Wang Y.-C., Lin Y.-W., Chou S.-Y., Chen S.-F., Liu L.-T., Wu Y.-T., Kuo C.-J., Chen T. S.-S., Juang S.-H. Bioorg. Med. Chem. Lett. 2005;15:3058. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C.-Y., King K.-Y., Kuo C.-J., Fang J.-M., Wu Y.-T., Ho M.-Y., Liao C.-L., Shie J.-J., Liang P.-H., Wong C.-H. Chem. Biol. 2006;13:4469. doi: 10.1016/j.chembiol.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Bacha U., Barrila J., Gabelli B., Kiso Y., Amzel L.M., Freire E. Chem. Biol. Drug Des. 2008;72:34. doi: 10.1111/j.1747-0285.2008.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Regnier T., Sarma D., Hidaka K., Bacha U., Freire E., Hayashi Y., Kiso Y. Bioorg. Med. Chem. Lett. 2009;19:2722. doi: 10.1016/j.bmcl.2009.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Ghosh A.K., Xi K., Ratia K., Santarsiero B.D., Fu W., Harcourt B.H., Rota P.A., Baker S.C., Johnson M.E., Mesecar A.D. J. Med. Chem. 2005;48:6767. doi: 10.1021/jm050548m. [DOI] [PubMed] [Google Scholar]; (b) Shie J.-J., Fang J.-M., Kuo T.-H., Kuo C.-J., Liang P.-H., Huang H.-J., Wu Y.-T., Jan J.-T., Cheng E. Y.-S., Wong C.-H. Bioorg. Med. Chem. 2005;13:5240. doi: 10.1016/j.bmc.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ghosh A.K., Xi K., Grum-Tokars V., Xu X., Ratia K., Fu W., Houser K.V., Baker S.C., Johnson M.E., Mesecar A.D. Bioorg. Med. Chem. Lett. 2007;17:5876. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaeppler U., Stiefl N., Schiller M., Vicik R., Breuning A., Schmitz W., Rupprecht D., Schmuck C., Baumann K., Ziebuhr J., Schirmeister T.A. J. Med. Chem. 2005;48:6832. doi: 10.1021/jm0501782. [DOI] [PubMed] [Google Scholar]
- 20.Hsu J.T.A., Kuo C.J., Hsieh H.P., Wang Y.C., Huang K.K., Lin C.P.C., Huang P.F., Chen X., Liang P.H. FEBS Lett. 2004;574:116. doi: 10.1016/j.febslet.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo C.J., Liu H.G., Lo Y.K., Seong C.M., Lee K.I., Jung Y.S., Liang P.H. FEBS Lett. 2009;583:549. doi: 10.1016/j.febslet.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn T.Y., Kuo C.J., Liu H.G., Ha D.C., Liang P.H., Jung Y.S. Bull. Korean Chem. Soc. 2010;31:87. [Google Scholar]
- 23.Ram V.J., Vanden Berghe D.A., Vlietinck A.J. J. Heterocycl. Chem. 1984;21:307. [Google Scholar]
- 24.Ramajayam, R.; Mahera, N. B.; Neamati, N.; Yadav, M. R.; Giridhar, R.; Arch. Pharm.2009, 342, 710. General procedure for the synthesis of compound 6a–n: To a mixture of 6-aryl-5-cyano-2-thiouracil (1 mmol) and K2CO3 (1.5 mmol) in DMF (10 mL), alkyl iodide (1.2 mmol) was added dropwise with stirring while maintaining the temperature of the reaction mixture at 0–5 °C. Stirring was continued for 3 h at this temperature and continued for additional 2 h at room temperature. Water was added to the mixture and filtered. The aqueous filtrate was neutralized with acetic acid and the precipitate was filtered and purified.The selected data of representative compounds 6k and 6m were as follows:Compound 6k: Yield: 51%; mp 236–237 °C; IR (KBr): 3078, 2221, 1666, 1521, 1473, 1346, 1249, 1112, 1010, 916, 891, 785 cm−1; 1H NMR (400 MHz, DMSO-d6) d: 4.59 (s, 2H, CH2), 7.63 –7.65 (d, J = 8.72 Hz, 2H, ArH), 7.79 (t, J = 8.04 Hz, 1H, ArH), 8.17 –8.20 (d, J = 8.72 Hz, 2H, ArH), 8.31–8.33 (d, J = 8.0 Hz, 1H, ArH), 8.41–8.44 (m, 1H, ArH), 8.69 –8.70 (m, 1H, ArH), 12.9 (br, 1H, NH); MS (CI) m/z: 410 [M+H] +. Anal. Calcd for C18H11N5O5S: C, 52.81; H, 2.71; N, 17.11. Found: C, 52.68; H, 2.85; N, 17.04;Compound 6m: Yield: 66%; mp 254–257 °C; IR (KBr): 3000, 2218, 1651, 1517, 1467, 1346, 1244, 1009, 1004, 997, 856, 779 cm−1; 1H NMR (400 MHz, DMSO-d6) d: 4.60 (s, 2H, CH2), 7.51–7.53 (d, J = 8.64 Hz, 2H, ArH), 7.60–7.62 (d, J = 8.76 Hz, 2H, ArH), 7.89–7.91 (d, J = 8.64 Hz, 2H, ArH), 8.13–8.15 (d, J = 8.72 Hz, 2H, ArH), 12.88 (br, 1H, NH); MS (CI) m/z: 399 [M+H]+. Anal. Calcd for C18H11ClN4O3S: C, 54.21; H, 2.78; N, 14.05. Found: C, 54.18; H, 2.85; N, 14.01.
- 25.Kuo C.J., Chi Y.H., Hsu J.T.A., Liang P.H. Biochem. Biophys. Res. Commun. 2004;318:862. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anard K., Bartlam M., Hilgenfeld R., Rao Z. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13190. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]


