Abstract
Both helper dependent expression systems, based on two components, and single genomes constructed by targeted recombination, or by using infectious cDNA clones, have been developed. The sequences that regulate transcription have been characterized mainly using helper dependent expression systems and it will now be possible to validate them using single genomes. The genome of coronaviruses has been engineered by modification of the infectious cDNA leading to an efficient (>20 μg ml−1) and stable (>20 passages) expression of the foreign gene. The possibility of engineering the tissue and species tropism to target expression to different organs and animal species, including humans, increases the potential of coronaviruses as vectors. Thus, coronaviruses are promising virus vectors for vaccine development and, possibly, for gene therapy.
Keywords: Coronaviruses, Viral vectors, Transcription regulatory sequences, Virus tropism
1. Introduction
Coronaviruses have several advantages for use as vectors over other viral expression systems: (i) coronaviruses are single-stranded RNA viruses that replicate within the cytoplasm without a DNA intermediary, making integration of the virus genome into the host cell chromosome unlikely (Lai and Cavanagh, 1997); (ii) these viruses have the largest RNA virus genome and, in principle, have room for the insertion of large foreign genes (Masters, 1999, Enjuanes et al., 2000a); (iii) a pleiotropic secretory immune response is best induced by the stimulation of gut-associated lymphoid tissues. Since coronaviruses in general infect the mucosal surfaces, both respiratory and enteric, they may be used to target the antigen to the enteric and respiratory areas to induce a strong secretory immune response (Enjuanes and Van der Zeijst, 1995); (iv) the tropism of coronaviruses may be modified by the manipulation of the spike (S) protein allowing engineering of the tropism of the vector (Ballesteros et al., 1997, Leparc-Goffart et al., 1998, Sánchez et al., 1999, Kuo et al., 2000); (v) non-pathogenic coronavirus strains infecting most species of interest (human, porcine, bovine, canine and feline) are available to develop expression systems (Sánchez et al., 1992); and (vi) infectious coronavirus cDNA clones are available to design expression systems (Almazan et al., 2000, Yount et al., 2000, Thiel et al., 2001).
Vectors for the expression of heterologous genes have been developed from full-length cDNA clones of most of the positive-strand RNA viruses. These viruses can be classified according to the nature of their genome (one or more RNA fragments) and their expression strategy, for instance, a single mRNA encoding a polyprotein that is processed into functional proteins or a collection of mRNAs each encoding a protein. Expression systems based on positive-strand RNA viruses that are transcribed in a single mRNA molecule, such as picornaviruses (poliovirus) (Andino et al., 1994), and more recently flaviviruses (Khromykh and Westaway, 1994, Khromykh and Westaway, 1997, Chambers et al., 1999) have been developed. The alphaviruses (Togaviridae family), encoding a full-length mRNA and a subgenomic mRNA, are among the most advanced expression systems (Liljeström, 1994, Frolov et al., 1997, Pushko et al., 1997, Caley et al., 1999). The alphaviruses include the Sindbis virus, Semliki Forest virus (SFV) and Venezuelan equine encephalitis virus (VEEV), and are very efficient at eliciting humoral and cellular immune responses.
Two types of expression systems have been developed based on coronavirus genomes (Fig. 1 ), one requires two components (helper dependent expression system) and the other a single genome that is modified either by targeted recombination or by engineering a cDNA encoding an infectious RNA. Coronavirus derived expression systems are being developed for human, porcine, murine, bovine and avian coronaviruses. The first attempt to use coronavirus for heterologous gene expression was based on the mouse hepatitis virus (MHV) by using a helper dependent expression system (Lin and Lai, 1993). Group 1 coronaviruses, such as transmissible gastroenteritis virus (TGEV), and group 3 coronaviruses, such as infectious bronchitis virus (IBV), have also been used for foreign gene expression.
Fig. 1.

Coronavirus derived expression systems: A. Helper dependent expression system based on two components, the helper virus and a minigenome carrying the foreign gene (FG). An, poly A. B. Single genome engineered either by targeted recombination or by using an infectious coronavirus cDNA clone (pBAC-TGEVFL) derived from TGEV genome.
Among the positive-strand RNA viruses, coronaviruses have the largest genome size (around 30 kb) and, in principle, could have the largest cloning capacity (Enjuanes et al., 2000a). This review will focus on the description of the advantages and limitations of these novel coronavirus expression systems, the attempts to increase their expression levels by studying the transcription regulatory sequences (TRSs), and the proven possibility of modifying their tissue and species-specificity.
Limited progress on the understanding of replication and translation regulation in coronaviruses has been made. To obtain information on these aspects the reader is referred to recent reviews and selected papers where this issue has been addressed (Luytjes et al., 1988, Lai and Cavanagh, 1997, Tahara et al., 1998, O'Connor and Brian, 2000) since translation and replication will not be considered within this review.
2. Pathogenicity of coronaviruses
Coronaviruses comprise a large family of viruses infecting a broad range of vertebrates, from mammalian to avian species. Coronaviruses are associated mainly with respiratory, enteric, hepatic and central nervous system diseases. Nevertheless, organs such as kidney, heart, and eye can also be affected. In humans and fowl, coronaviruses primarily cause upper respiratory tract infections, while porcine and bovine coronaviruses (BCoVs) establish enteric infections that result in severe economical loss.
The human coronaviruses (HCoV) are responsible for 10–20% of all common colds (McIntosh et al., 1969), and have been implicated in gastroenteritis, high and low respiratory tract infections and rare cases of encephalitis. HCoV have also been associated with infant necrotizing enterocolitis (Resta et al., 1985, Luby et al., 1999) and are tentative candidates for multiple sclerosis (Talbot, 1997). However, HCoV have languished at the bottom of many lists of human pathogens because of the difficulty in isolating and characterizing the agents during outbreaks of illness (Denison, 1999). In addition, infections of man by coronaviruses seem to be ubiquitous, as coronaviruses have been identified wherever they have been looked for, including North and South America, Europe, and Asia and no other human disease has been clearly associated to them with the exception of the respiratory and enteric infections (Denison, 1999).
Epithelial cells are the main target of coronaviruses. Widely distributed cells such as macrophages are also infected by coronaviruses. These viruses have relatively restricted host ranges, infecting only their natural host and closely related animal species. Coronavirus biological vectors are not known.
3. Coronavirus members
The coronavirus and the torovirus genera form the Coronaviridae family, which is closely related to the Arteriviridae family. Both families are included in the Nidovirales order (Enjuanes et al., 2000a, Enjuanes et al., 2000b). Recently, a new group of invertebrate viruses, the okaviruses, with a genetic structure and replication strategy similar to those of coronaviruses, has been described (Cowley et al., 2000). The coronaviruses have been classified in three groups that comprise the members listed in Table 1 . The murine coronaviruses (MCoV) have been extremely useful to study gene expression using systems based in two components, a helper virus and a minigenome in which the heterologous gene was inserted. The advanced state of the research performed with coronaviruses and arteriviruses led to the development of single genome expression systems based in both virus families. Infectious cDNA clones are available for porcine (Almazan et al., 2000, Yount et al., 2000) and human (Thiel et al., 2001) coronaviruses, and for the arteriviruses equine infectious anemia virus (van Dinten et al., 1997, de Vries et al., 2000) and the porcine respiratory and reproductive syndrome virus (PRRSV) (Meulenberg et al., 1998). The availability of these cDNAs and the application of target recombination to coronaviruses (Masters, 1999) have been essential for the development of vectors based on the Nidovirales.
Table 1.
Coronaviridae family members
| Group 1 | |
| Human coronavirus 229E | HCoV-229E |
| Porcine enteric (transmisible gastroenteritis virus, TGEV; and porcine epidemic diarrhea virus, PEDV) and respiratory (PRCoV) coronavirus | PCoV |
| Canine coronavirus | CCoV |
| Feline coronavirus, including feline infectious peritonitis virus (FIPV) | FCoV |
| Group 2 | |
| Human coronavirus OC43 | HCoV-OC43 |
| Bovine coronavirus | BCoV |
| Turkey coronavirus BCoV related | TCoV-B |
| Murine coronaviruses including mouse hepatitis virus (MHV) | MCoV |
| Porcine hemagglutinating encephalomyelitis virus | HEV |
| Rat coronavirus including sialodacryoadenitis virus (SDAV) | RtCoV |
| Group 3 | |
| Avian coronavirus including infectious bronchitis virus (IBV) | ACoV |
| Turkey coronavirus IBV related | TCoV-I |
| Unclassified coronavirus | |
| Rabbit coronavirus | RbCoV |
4. Molecular biology of coronavirus
4.1. The coronavirus genome
Virions contain a single molecule of linear, positive-sense, single-stranded RNA (Fig. 2 B). The genomic RNA is the largest viral RNA genome known ranging from 27.6 to 31.3 kb in size. Coronavirus RNA has a 5′ terminal cap followed by a leader sequence of 65–98 nucleotides and an untranslated region of 200–400 nucleotides. At the 3′ end of the genome there is an untranslated region of 200–500 nucleotides followed by a poly(A) tail. The virion RNA, which functions as a mRNA and is infectious, contains ≈7–10 functional genes, four or five of which encode structural proteins. The genes are arranged in the order 5′-polymerase-(HE)-S-E-M-N-3′, with a variable number of other genes that are believed to be non-structural and largely non-essential, at least in tissue culture.
Fig. 2.
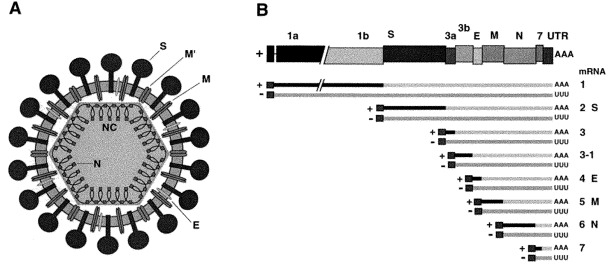
Structure and genome organization of coronaviruses: A. Schematic diagram of virus structure showing the envelope, the core and the nucleoprotein structure. S, spike protein; M and M′, M proteins with the amino-terminus facing the external surface of the virion and the carboxy-terminus towards the inside or the outside face of the virion, respectively; E, small envelope protein; N, nucleocapsid protein; NC, nucleocapsid. Some coronaviruses of group 2 have an additional protein, the haemagglutinin-esterase (HE) (not shown). B. Representation of a prototype TGEV coronavirus genome and subgenomic RNAs. Beneath the top bar a set of positive- and negative-sense mRNA species synthesized in infected cells is shown. The protein products obtained from each positive-sense RNA are indicated. Two products, polyproteins 1a and 1b, are translated from the genomic RNA by a ribosomal frameshifting mechanism. All other proteins are translated from the first open reading frame of each functionally monocistronic subgenomic RNA (dark lines). Poly(A) and Poly(U) tails are indicated by AAA or UUU. S, spike protein; E, envelope protein; M, membrane protein; N, nucleocapsid protein.
About two-thirds of the entire RNA comprises the ORF1a/b encoding the replicase gene. At the overlap between the ORF 1a and 1b regions, there is a specific seven-nucleotide ‘slippery’ sequence and a pseudoknot structure (ribosomal frameshifting signal), which are required for the translation of ORF 1b. In the 3′ end, one-third of the genome comprises the genes encoding the structural proteins and the other non-structural ones. Organization of the non-structural protein genes, which are interspersed between the known structural protein genes, varies significantly among different coronavirus strains (Enjuanes et al., 2000a). A pseudoknot structure is also predicted at the 3′ end of the coronaviral RNA (Williams et al., 1995, Hsue and Masters, 1997, Brian, 2001).
Coronavirus transcription occurs via an RNA-dependent RNA synthesis process in which mRNAs are transcribed from negative-stranded templates. Sequences at the 5′ end of each gene represent signals for the transcription of subgenomic mRNAs (Lai and Cavanagh, 1997, Sawicki and Sawicki, 1998). These sequences, known as TRSs include a stretch of a highly conserved sequence designed the core sequence (CS), located at sites immediately upstream of most of the genes. The CS presents some variation in sequence length among the coronaviruses, being 5′-CUAAAC-3′ for TGEV, or a related sequence, depending on the coronavirus (i.e. UCUAAAC for MHV). In previous reports the CS has been named intergenic sequence (IS). Since often genes overlap in the Nidovirales, the acronym IS does not seem appropriate in these cases and the acronym CS could reflect the nature of the highly conserved sequence contained within the TRS. Coronavirus mRNAs consist of six to eight types of varying sizes, depending on the coronavirus strain and the host species. The largest mRNA is the genomic RNA which also serves as the mRNA for ORF 1a and 1b and the remainder are subgenomic mRNAs. The mRNAs have a nested-set structure in relation to the genome structure (Fig. 2B). Except for the smallest mRNA, all of the mRNAs are structurally polycistronic. In general, only the 5′-most ORF of each mRNA is translated. However, there are exceptions: some mRNAs, e.g. mRNA 5 of MHV, mRNA 3 of IBV and BCoV nucleocapsid mRNA are translated by internal initiation into two or three proteins (Lapps et al., 1987, Krishnan et al., 1996).
4.2. Coronavirus proteins
Coronaviruses are enveloped viruses that contain a core that includes the ribonucleoprotein formed by the RNA and nucleoprotein N (Fig. 2A). The core is formed by the genomic RNA, the N protein and the carboxy-terminus of the membrane (M) protein. Most of the M protein is embedded within the membrane but its carboxy-terminus is integrated within the core and seems essential to maintain the core structure (Escors et al., 2001). The TGEV M protein presents two topologies. In one, both the amino- and the carboxy-terminus face the outside of the virion, while in the other the carboxy-terminus is inside (Risco et al., 1995). In addition, the virus envelope contains two or three other proteins, the S protein, the small membrane protein (E) and, in some strains, the hemagglutinin-esterase (HE) (Enjuanes et al., 2000a). The ratios of S:E:M:N proteins vary in different reports. For purified TGEV, these ratios have been estimated to be 20:1:300:140, respectively (Escors et al., 2001). The S protein is large, ranging from 1160 to 1452 amino acids, and in some coronaviruses is cleaved into S1 and S2 subunits. The S protein is responsible for attachment to cells, hemagglutination, membrane fusion and induction of neutralizing antibodies.
The replicase gene is predicted to encode a protein of ≈740–800 kDa which is co-translationally processed. Several domains within the replicase have predicted functions based on regions of nucleotide homology including two papain-like cysteine proteases, a chymotrypsin-picornaviral 3C-like protease, a cysteine-rich growth factor-related protein, an RNA-dependent RNA polymerase, a nucleoside triphosphate (NTP)-binding/helicase domain and a zinc-finger nucleic acid-binding domain (Siddell, 1995, Enjuanes et al., 2000a, Penzes et al., 2001).
5. Helper dependent expression systems
The coronaviruses have been classified into three groups (1, 2 and 3) (Table 1) based on sequence analysis of a number of coronavirus genes (Siddell, 1995). The helper dependent expression systems have been developed using members of the three groups of coronaviruses (Fig. 3 ), and will be addressed first.
Fig. 3.
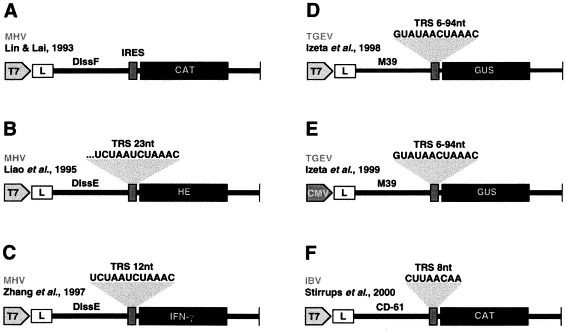
Summary of helper dependent expression systems based on coronavirus derived minigenomes: A–C. Expression modules based on MHV minigenomes DIssF and DIssE cloned under the control of T7 bacteriophage polymerase (T7), used to express chloramphenicol acetyltransferase (CAT), hemagglutinin-esterase (HE) or interferon-γ using either an IRES (A), or TRSs (B–C). D–E. Expression modules based on the TGEV derived minigenome M39 used to express the GUS. The minigenome was cloned either after T7 (D) or the CMV (E) promoters. F. Expression module based on the IBV derived minigenome CD-61 used to express CAT.
5.1. Helper dependent expression systems based on group 1 coronaviruses
Group 1 coronaviruses include porcine, canine, feline and HCoV. Nevertheless, expression systems have been developed for the porcine and HCoV since minigenomes are only available for these two coronaviruses.
Using the TGEV-derived minigenomes (Fig. 3D–E) an expression system has been developed (Méndez et al., 1995, Izeta et al., 1999). The TGEV-derived RNA minigenomes were successfully expressed in vitro using T7 polymerase and amplified after in vivo transfection using a helper virus. To engineer cDNAs encoding TGEV defective RNAs, a deletion mutant of 9.7 kb (DI-C) maintaining the cis-signals required for efficient and stable replication and packaging by helper virus was isolated (Izeta et al., 1999). A collection of 14 DI-C RNA deletion mutants (TGEV minigenomes) was synthetically generated and tested for their ability to be replicated and packaged. The smallest minigenome (M33) that was replicated by the helper virus and efficiently packaged was 3.3 kb in length. TGEV derived minigenomes of 3.3, 3.9 and 5.4 kb (named M33, M39, and M54, respectively) were efficiently used for the expression of heterologous genes.
Using M39 minigenome a two step amplification system was developed similarly to the other amplification system (Herweijer et al., 1995, Dubensky et al., 1996, Berglund et al., 1998), based on the cloning of a cDNA copy of the minigenome after the immediate-early cytomegalovirus promoter (CMV). Minigenome RNAs are first amplified in the nucleus by the cellular RNA pol II and then, the RNAs are translocated into the cytoplasm where they are amplified by the viral replicase of the helper virus. The β-glucuronidase (GUS) and the ORF5 of the PRRSV, a porcine virus with a high impact on animal health (Plana-Durán et al., 1997), have been expressed using this vector (Alonso et al., 2001b). PRRSV ORF5 (603 nt) encodes a surface glycoprotein that is the major PRSSV protective antigen described (Plana-Durán et al., 1997). Maximum expression levels of both GUS and PRRSV ORF5 were detected from passages 3 to 6, although the expression of these genes persisted for at least 10 passages in ST cells.
The HCoV-229E has also been used to express new subgenomic mRNAs although until now it has not been applied to the expression of a foreign protein (Thiel et al., 1998). It was demonstrated that a synthetic RNA comprised of 646 nt from the 5′ end and 1465 from the 3′ end was amplified by the helper virus. Using this minigenome, mRNAs were efficiently expressed under the control of the intergenic region of the HCoV-229E nucleocapsid protein.
5.2. Helper dependent expression systems based on group 2 coronaviruses
Most of the work has been done with MHV defective RNAs (Lin and Lai, 1993, Liao et al., 1995, Zhang et al., 1997). Three heterologous genes have been expressed using the MHV system, chloramphenicol acetyltransferase (CAT), HE, and interferon-γ (Fig. 3A–C). Expression of the reporter gene (CAT) was detected only in passages 0, 1, and 2. The HE was clearly visualized after immunoprecipitation only during the first three passages (Liao and Lai, 1995) and the synthesized protein was incorporated into the virions. When virus vectors expressing CAT and HE were inoculated intracerebrally into mice, HE- or CAT-specific subgenomic mRNAs were detected in the brains at days 1 and 2 p.i. but not later, indicating that the genes in the defective minigenome (DI) vector were expressed only in the early stage of viral infection (Zhang et al., 1998).
A DI RNA of the MHV was also developed as a vector for expressing interferon-γ (IFN-γ). The murine IFN-γ gene was secreted into culture medium as early as 6 h post-transfection and reached a peak level at 12 h post-transfection. The DI-expressed IFN-γ exhibited an antiviral activity comparable to that of recombinant IFN-γ. No inhibition of virus replication was detected when the cells were treated with IFN-γ produced by the DI RNA, but infection of susceptible mice with DI RNA producing IFN-γ caused significantly milder disease, accompanied by less virus replication than that caused by virus containing a control DI vector (Lai et al., 1997, Zhang et al., 1997).
5.3. Helper dependent expression systems based on group 3 coronaviruses
IBV is an avian coronavirus with a single-stranded, positive-sense RNA genome of 27 608 nt (Boursnell et al., 1987). A defective RNA (CD-61) derived from the Beaudette strain of the IBV virus was used as an RNA vector for the expression of two reporter genes, luciferase and CAT (Fig. 3F) (Penzes et al., 1994, Penzes et al., 1996). The defective RNA efficiently expressed the CAT gene but only minimum levels of luciferase (Stirrups et al., 2000).
A helper dependent expression system has recently been described based on arteriviruses (Molenkamp et al., 2000), that belong to the same order as coronaviruses. Also, using equine arteritis virus (EAV) minigenomes of 3.8 kb, the CAT reporter gene has been expressed. The smallest defective RNA obtained (3.0 kb) was replicated by the helper virus but could not be packaged.
5.4. Heterologous gene expression levels in helper dependent expression systems
The expression levels have not been quantified in terms of protein mass for MHV helper dependent expression systems. HCoV-229E mRNA expression levels using engineered expression modules without a heterologous gene, probably are high since the abundance of these RNAs seem to be higher than that of the viral mRNAs within the same cells. Using IBV minigenomes CAT expression levels between 1 and 2 μg/106 cells have been described.
The highest expression levels (2–8 μg of GUS per 106 cells) have been obtained using a two step amplification system based on TGEV derived minigenomes with optimized TRSs (Izeta et al., 1999, Alonso et al., 2001a).
6. Single genome coronavirus vectors
6.1. Vectors constructed by targeted recombination
Reverse genetics were possible by targeted recombination between a helper virus and either non-replicative or replicative coronavirus derived RNAs. This approach was initially developed by Masters’ group (Masters, 1999). First, the engineering of a five nucleotide insertion into the 3′ untranslated region (3′ UTR) of MHV via targeted recombination with an in vitro synthesized RNA was reported (Fig. 4 A) (Koetzner et al., 1992). This approach was facilitated by the availability of an N gene mutant, designated Alb4, that was both temperature sensitive and thermolabile. Alb4 forms tiny plaques at restrictive temperature that are easily distinguishable from wild-type plaques. In addition, incubation of Alb4 virions at non-permissive temperature results in a 100-fold greater loss of titer than for wild-type virions (Koetzner et al., 1992). These phenotypic traits allowed the selection of recombinant viruses generated by a single cross-over event following cotransfection into mouse cells of Alb4 genomic RNA together with a synthetic copy of the smallest subgenomic RNA (RNA7) tagged with a marker in the 3′ UTR.
Fig. 4.
Single genome expression based on the engineering of coronavirus minigenomes by targeted recombination: A. Basic scheme of targeted recombination in MHV. The black box indicates the approximate location of the N gene region (87 nt) that is deleted in the Alb4 mutant. M, insertion of 5 nt used as a genetic marker (Masters, 1999). B. Targeted recombination within the S gene of TGEV and a minigenome carrying the information for an S gene with three nucleotide mutations (Sdmar) that allow escape from neutralization by two mAbs specific for antigenic sub-sites Ac and Aa of S protein (Sola et al., 2001b).
An improvement of the recombination frequency was obtained between the helper virus and replicative defective RNAs as the donor species. Whereas, between replication competent MHV and non-replicative RNAs a recombination frequency of the order of 10−5 was estimated, the use of replicative donor RNA yielded recombinants at a rate of some three orders of magnitude higher (van der Most et al., 1992). This higher efficiency made it possible to screen for recombinants even in the absence of selection. In this manner, the transfer of silent mutation in gene 1a of a minigenome to wild-type MHV at a frequency of about 1% was demonstrated.
Targeted recombination has been applied to the generation of mutants in most of the coronavirus genes. Thus, two silent mutations have been created thus far in gene 1 (van der Most et al., 1992). The S protein has also been modified by targeted recombination. Changes were introduced by one crossover event at the 5′ end of the S gene that modified MHV pathogenicity (Leparc-Goffart et al., 1998). Targeted recombination mediated by two cross-overs allowed the replacement of the S gene of a respiratory strain of TGEV by the S gene of enteric TGEV strain PUR-C11 leading to the isolation of viruses with a modified tropism and virulence (Sánchez et al., 1999). In this case, the recombinants were selected in vivo using their new tropism in piglets. A new strategy for the selection of recombinants within the S gene, after promoting targeting recombination, was based on elimination of the parental replicative TGEV by the simultaneous neutralization with two mAbs (Fig. 4B) (Sola et al., 2001b).
Mutations have been created by targeted mutagenesis within the E and M genes. These mutants provided corroboration for the pivotal role of E protein in coronavirus assembly and identified the carboxyl terminus of the M molecule as crucial to assembly (de Haan et al., 1998, Fisher and Goff, 1998). Targeted recombination was also used to express heterologous genes. For instance, the gene encoding green fluorescent protein (GFP) was inserted into MHV between gene S and E by targeted recombination, resulting in the creation of the largest known RNA viral genome (Fischer et al., 1997).
The frequencies of the targeted recombination event for MHV and TGEV were found to be higher than the standard prediction for the recombination frequency of a multiple crossover. This frequency was expected to be the product of the frequencies of the individual recombination events (Peng et al., 1995, de Haan et al., 1998, Hsue and Masters, 1998, Masters, 1999, Sola et al., 2001b). Nevertheless, the recombinants with several crossovers appear to occur more frequently than would be expected if each cross-over was an independent event. This suggests that the alignment of two templates is the rate-limiting event in recombination, and once this has been achieved, the barrier to multiple crossovers may be only marginally higher than that for single crossovers (Masters, 1999, Sola et al., 2001b).
6.2. Coronavirus vectors derived from infectious cDNA clones
The construction of a full-length genomic cDNA clone could considerably improve the genetic manipulation of coronaviruses. Infectious cDNA clones have now been constructed for members of many positive-stranded RNA virus families (Racaniello and Baltimore, 1981, Ahlquist et al., 1984, Rice et al., 1987, Rice et al., 1989, Liljeström and Garoff, 1991, Satyanarayana et al., 1999), including the Arteriviridae family closely related to coronaviruses (van Dinten et al., 1997, Meulenberg et al., 1998, de Vries et al., 2000). Negative-stranded RNA virus genomes have been generated for Mononegavirales by the simultaneous expression of the ribonucleoprotein containing the N protein, the polymerase cofactor phosphoprotein and the viral RNA polymerase (Schnell et al., 1994). Rescue of engineered RNAs in negative-strand RNA virus with eight genome segments was also possible for influenza virus (Palese, 1998, Fodor et al., 1999, Neumann et al., 1999, Hoffmann et al., 2000a, Hoffmann et al., 2000b).
The enormous length of the coronavirus genome and the instability of plasmids carrying coronavirus replicase sequences have, until recently, hampered the construction of a full-length cDNA clone (Masters, 1999). Now, for the first time, construction of infectious coronavirus cDNA clones is possible (Almazan et al., 2000, Yount et al., 2000, Thiel et al., 2001). Construction of the TGEV full-length cDNA was started from a DI that was stably and efficiently replicated by the helper virus (Méndez et al., 1996, Izeta et al., 1999). Using this DI, the full-length genome was completed and the performance of the enlarged genome was checked after each step. This approach allowed for the identification of a cDNA fragment that was toxic to the bacterial host. This finding was used to advantage by reintroducing the toxic fragment into the cDNA in the last cloning step. In order to express the long coronavirus genome and to add the 5′ cap, a two-step amplification system that couples transcription in the nucleus from the CMV promoter, with a second amplification in the cytoplasm driven by the viral polymerase, was used. In addition, to increase viral cDNA stability within bacteria, the cDNA was cloned as a bacterial artificial chromosome (BAC), that produces only one, maximum two plasmid copies per cell. BACs have been useful to stably clone large DNAs from a variety of complex genomic sources into bacteria (Shizuya et al., 1992), including herpesvirus DNA (Messerle et al., 1997).
A fully functional infectious TGEV cDNA clone (pBAC-TGEVFL), leading to a virulent virus able to infect both the enteric and respiratory tract has been engineered using two BAC plasmids (Fig. 5 A). One plasmid (pBAC-TGEVΔClaI) contained all virus sequences except for a fragment of about 5 kb that was included within a second BAC (pBAC-TGEVClaI) (Almazan et al., 2000). Using this cDNA the GFP gene of 0.72 kb was cloned into the RNA genome by replacing the non-essential 3a and 3b genes (Fig. 5B), leading to an engineered genome with high expression levels (>20 μg/106 cells) and stability (>20 passages in cultured cells) (Sola et al., 2001a). Using the TGEV derived cDNA expression system, the induction of lactogenic immunity in swine has been demonstrated. This immune response led to the acquisition of immunity by newborn piglets (I. Sola and L. Enjuanes, unpublished results).
Fig. 5.
Cloning of the TGEV cDNA in BACs and expression of GFP: A. Plasmid pBAC-TGEVFL (bottom plasmid) was generated using two plasmids, one containing all the virus genome (top plasmid) except a sequence of about 5 kb present between two Cla I sites cloned in a second plasmid (middle plasmid). CMV, cytomegalovirus immediate-early promoter; Poly(A), tail of 24 A residues; HDV, hepatitis delta virus ribozyme; BGH, bovine growth hormone termination and polyadenylation sequences; SC11, S gene of PUR-C11 strain. B. Expression of GFP using an infectious TGEV cDNA clone. Genes 3a and 3b were deleted in the TGEV infectious cDNA, cloned in BAC, leading to a replication competent cDNA (pBAC-TGEV-Δ3ab-GFP). GFP gene (0.72 kb) was inserted within the position of the deleted genes after the TRS of gene 3a. GFP, green fluorescent protein. SC11, S gene of PUR-C11 TGEV strain. An, poly A. HDV, hepatitis delta-virus ribozyme. BGH, bovine growth hormone termination and polyadenylation signals.
These expression levels are similar to those described for vectors based on other positive-strand RNA viruses such as poliovirus and cardiovirus, and one alphavirus, the VEEV (4 μg/106). Nevertheless, these expression levels are still lower than those described for other alphaviruses such as Sindbis virus (50 μg/106 cells) (Frolov et al., 1996, Agapov et al., 1998) and SFV (80–300 μg/106 cells) (Liljeström and Garoff, 1991, Sjoberg et al., 1994, DiCiommo and Bremner, 1998).
DNA based expression systems in general produce high levels of the foreign protein. For instance, vectors based on adenovirus 5 using the major later protein promoter and a tripartite leader may express 90 μg/106 cells and baculovirus expression systems 15–100 μg/106 cells (Kuroda et al., 1989, Sibilia et al., 1995).
A second procedure to assemble a full-length infectious construct of TGEV was based in the in vitro ligation of six adjoining cDNA subclones that span the entire TGEV genome. Each clone was engineered with unique flanking interconnecting junctions which determine a precise assembly with only the adjacent cDNA subclones, resulting in a TGEV cDNA. In vitro transcripts derived from the full-length TGEV construct were infectious (Yount et al., 2000).
More recently, an infectious cDNA clone of HCoV-229 has been reported (Thiel et al., 2001). This system is based upon the in vitro transcription of infectious RNA from a cDNA copy of the human coronavirus 229E genome that has been cloned and propagated in vaccinia virus. In this case, as when the BACs were used to assembly the coronavirus infectious cDNA, the full-length coronavirus genomes can be modified and propagated as single cDNA molecules, in contrast to the infectious cDNA clone derived from six fragments that are in vitro ligated.
The engineered cDNAs will have an important impact on the study of mechanisms of coronavirus replication and transcription and provide an invaluable tool for the experimental investigation of virus-host interactions. These cDNAs may also be the basis for tissue-specific expression systems that may be used in human, porcine, canine and feline species by replacing the S gene included in the cDNA with that of the coronavirus infecting the target species.
6.3. Cloning capacity of coronavirus expression vectors
Coronavirus minigenomes have a theoretical cloning capacity close to 27 kb, since their RNA with a size of about 3 kb is efficiently amplified and packaged by the helper virus and the virus genome has about 30 kb. In contrast, the theoretical cloning capacity for an expression system based on a single coronavirus genome like TGEV may be 3.1 kb taking into account that: (i) the non-essential 3a and 3b genes (1.0 kb) have been deleted; (ii) the standard S gene can be replaced by that of PRCV mutants with a deletion of 0.67 kb; and (iii) both DNA and RNA viruses may accept genomes with sizes up to 105% of the wild-type genome (Bett et al., 1993, Parks and Graham, 1997, Afanasiev et al., 1999). This cloning capacity most likely will be enlarged in the near future when non-essential domains of the replicase gene, with more than 20 kb, could be identified. The present cloning capacity of the coronavirus vectors (around 3 kb) is within the range expected, since other RNA virus vectors, such as those derived from the VEEV, with a genome of around 12 kb, accept inserts of about 1 kb in size that are expressed during ten serial tissue culture passages. Larger genes in the order of 2.5 kb can also be expressed by the vector but are not tolerated as well by these viruses (Caley et al., 1997). Accordingly, in SFV and Sindbis virus expression system recombinants with smaller inserts (e.g. <2 kb) tend to be more stable than those with larger inserts, such as the β-galactosidase gene (about 3 kb) (Bredenbeek and Rice, 1992).
The longest cassette expressed using the Sindbis virus was 3.2 kb in length, which is remarkable for such a small vector (Sindbis genome is roughly 11.8 kb in length) and indicates that, apparently, the Sindbis virion is a rather flexible structure capable of accepting RNA molecules which are at least 30% larger than its own genome. Nevertheless, it should be noted that after the third passage, barely detectable amounts of the expression products were observed in cells infected with all recombinants and after the fifth passage, no heterologous proteins were detected in infected cells (Pugachev et al., 1995).
In VEEV, as well as in the Sindbis system, larger genes have been shown to retard virus growth (Pugachev et al., 1995). The type of gene being expressed may also play an important role in limiting replication, for example, VEEV vectors expressing glycoproteins typically do not replicate to titers as high as those expressing cytoplasmic proteins (Caley et al., 1999).
7. Regulation of transcription
Most of the information on coronavirus transcription has been generated using a helper dependent expression system based on minigenomes encoding new subgenomic mRNAs (Makino et al., 1991, van der Most et al., 1994, van der Most and Spaan, 1995, Krishnan et al., 1996, Penzes et al., 1996, Lai and Cavanagh, 1997, Izeta et al., 1999).
Coronavirus mRNAs have a leader sequence of 65–98 nucleotides at their 5′ ends, which is derived from the 5′ end of the genomic RNA. At the start site of every transcription unit on the viral genomic RNA, there is a CS that is nearly homologous to the 3′ end of the leader RNA. This sequence constitutes part of the signal for subgenomic mRNA transcription.
Coronavirus RNA synthesis occurs in the cytoplasm via a negative-strand RNA intermediate which contains short stretches of oligo(U) at the 5′ end. Both genome-size and subgenomic negative-strand RNAs, which correspond in number of species and size to those of the virus-specific mRNAs have been detected (Fig. 2B). The subgenomic negative-strand RNA sequences appear to be complementary to the positive-strand subgenomic mRNAs.
The common 5′ leader sequence is only found at the very 5′ terminus of the genome, which implies that the synthesis of subgenomic mRNAs involves fusion of non-contiguous sequences (Baric et al., 1983, Spaan et al., 1983, Lai et al., 1984). To explain the synthesis of leader-containing subgenomic mRNAs, two models: the leader-primed transcription (Lai, 1998), and the discontinuous transcription during negative-strand RNA synthesis (Sethna et al., 1989, Sawicki and Sawicki, 1990, van Marle et al., 1999), compatible with most of the experimental data, have been proposed. The leader-primed transcription model proposes that the virion genomic RNA is first transcribed into a genomic-length negative-strand RNA which, in turn, becomes the template for subsequent subgenomic mRNA synthesis. The leader is transcribed from the 3′ end of the negative-strand genomic RNA and dissociates from the template to subsequently associate with the template RNA at the various mRNA start sites serving as a primer for the transcription of the viral subgenomic mRNAs. It is proposed that the discontinuous transcription step takes place during positive-strand RNA synthesis. The discontinuous transcription during negative-strand RNA synthesis model (Fig. 6 A–B) proposes that the discontinuous transcription step occurs during negative-strand RNA synthesis, generating subgenomic negative-strand RNAs, which then serve as templates for subgenomic mRNAs in interrupted transcription. In this model, at the TRS on the genomic RNA the nascent subgenomic negative-strand RNA jumps to the leader RNA sequence at the 5′ end of the genomic RNA to act as a primer for transcription in a process that may be helped by cellular and viral proteins (Fig. 6B).
Fig. 6.
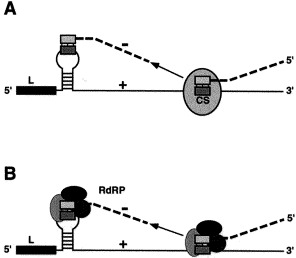
Schematic representation of a coronavirus discontinuous transcription model and the RNA structures involved: A–B. Discontinuous transcription during negative-strand RNA synthesis without (A) and with (B) a schematic representation of protein-RNA complexes potentially involved. During the negative RNA strain synthesis (discontinuous line) the replication complex is detached when the CS sequence is reached, and the complex joins the 3′ end of the leader.
These models are not mutually exclusive, as components of each model may operate at different stages of the viral replication cycle. Nevertheless, recently more high quality experimental evidence is being generated that supports the second model (van Marle et al., 1999, Baric and Yount, 2000).
7.1. Transcription regulatory sequences
The TRSs, including the CS regions, are short sequence elements upstream of the transcription units. Because the leader-mRNA junction occurs within this CS sequence or its minus-sense counterpart (cCS) the CSs are considered to be crucial for mRNA synthesis. The sequence of the cCS probably influences transcription throughout at least two types of recognition events: (i) one related to the potential basepairing between the leader 3′ end, complementary to the cCS, that guides the fusion between the leader and the body of the mRNA (Fig. 6A–B); and (ii) the recognition of the primary or secondary structure within the neighborhood of the cCS, that may cause formation of a bridge between the CS and the 3′ end of the leader, mediated by protein–protein and protein–RNA interaction (Lai, 1998). According to this model, the cCS should act as a classical promoter where transcription is initiated. Alternatively, this sequence may slow down or even detach the transcriptase complex, according to the discontinuous transcription during negative-strand RNA synthesis model (van der Most and Spaan, 1995, Sawicki and Sawicki, 1998, van Marle et al., 1999).
The CS of coronaviruses belonging to groups I (hexameric 5′-CUAAAC-3′) and II (heptameric 5′-UCUAAAC-3′) share homology, whereas the CS of coronaviruses belonging to group III, like that of IBV have the most divergent sequence (5′-CUUAACAA-3′). Also, arterivirus CSs have a sequence (UCAAC) that partially resembles that of IBV. Thus, the CS of different coronaviruses are quite similar though different in length (Fig. 7 A).
Fig. 7.
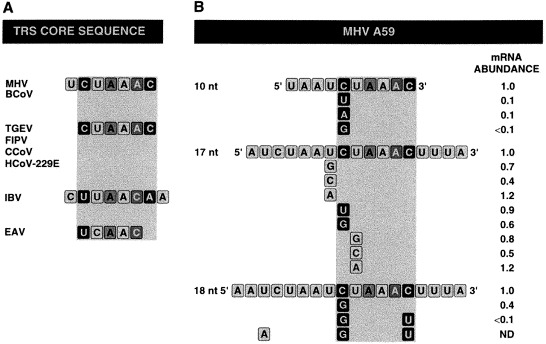
CSs of coronavirus TRSs: A. Alignment of representative CSs of the three groups of coronaviruses and one arterivirus. B. Relative abundance of the mRNAs produced after the mutagenesis of CSs of different lengths (10, 17 and 18 nt) using the MHV-A59 strain derived minigenomes (modified after van der Most et al., 1994).
7.2. The extent of the basepairing could in part determine mRNA levels
The potential basepairing between the 3′ end of the leader and the cCS differs slightly among the different genes of coronaviruses. For MHV, the extent of the basepairing ranges from 9 to 18 bp. Every MHV cCS contains the sequence 3′-AGAUUUG-5′, or a closely related sequence (van der Most and Spaan, 1995). Cloning short oligonucleotides ranging from 10 to 18 nt, comprising the CS sequences in MHV minigenomes, showed that these sequences alone were sufficient to direct subgenomic DI RNA synthesis (van der Most et al., 1994). A series of deletions in the sequences flanking the cCS reduced mRNA production, demonstrating that the sequence flanking the CS 3′-AGAUUUG-5′ affected the efficiency of subgenomic DI RNA transcription (Makino et al., 1991, Joo and Makino, 1992, Makino and Joo, 1993, van der Most et al., 1994). TRS activity became optimal in MHV when a sequence of 18 nt from a region showing full complementarity to the 3′ end of MHV genomic leader was used (Shieh et al., 1987, Makino et al., 1988, La Monica et al., 1992).
In MHV TRS strength is affected only slightly when a single nucleotide is mutated (Fig. 7B) (Joo and Makino, 1992, van der Most et al., 1994). Exceptionally, substitutions in some positions result in a more than 10-fold reduction of transcription. An increase in the TRS length, that leads to an increase in the potential hybridization with the 3′ end of the leader, allows the introduction of more than one mutation without a significant decrease in the mRNA production (Fig. 7B). In TGEV, the presence of the CS 5′-CUAAAC-3′ led to mRNA transcription, but deletion of the ‘U’ or a change in the second ‘C’ led to the complete abrogation of mRNA transcription (Alonso et al., 2001a). These data suggest that transcription initiation require a duplex of certain minimum stability. Once this condition is met, extending the basepairing does not increase TRS strength.
Using TGEV derived RNA minigenomes, we have shown that the CS sequence 5′-CUAAAC-3′ is required and is sufficient for high expression levels providing that it is in the appropriated context (Alonso et al., 2001a). Nevertheless, the mRNA and protein expression levels are highly influenced by the sequences flanking at the 5′ and 3′ sides of this CS sequence. The addition of 5′ upstream sequences from the TGEV N gene to the CS (from 6 to 94 nt) led to an increase in the total mRNA transcription of up to 4-fold. The sequences 3′ downstream of the CS also led to 4-fold higher mRNA levels in TGEV (Alonso et al., 2001a). Similarly, in IBV, expression of the reporter gene was under the canonical octameric IBV CS sequence CU(U/G)AACAA (Stirrups et al., 2000).
In Arteriviruses, it has been shown that discontinuous transcription takes place during the synthesis of the negative RNA strand (van Marle et al., 1999). Using site-directed mutagenesis of an infectious cDNA clone of the EAV it has been shown that subgenomic mRNA (sgmRNA) synthesis requires basepairing interaction between the leader TRS and the complement of a body TRS in the viral negative strand (Fig. 6). EAV TRS core consists of the CS pentanucleotide 5′-UCAAC-3′. It has been shown that mRNA synthesis is probably governed by a direct basepairing interaction between the plus leader TRS and the complement of the TRS. Using TRS mutants with reduced transcriptional activity, evidence was obtained showing that the TRS sequence at the leader-body junction of the sgRNA is derived from the body TRS. This finding supports the idea that sgmRNAs are generated by a mechanism of discontinuous minus strand synthesis (van Marle et al., 1999). The sequences at the 3′ end of the leader complementary to the TRS core are part of a single-strand RNA loop helping its potential interaction with the cCS. In TGEV, the equivalent sequences are also predicted to be mostly within a single-strand RNA loop at the 3′ end of the leader.
7.3. Effect of core sequence copy number on transcription
Studies on coronavirus transcription were performed using more than one CS in order to express the same mRNA. Using BCoV defective RNAs, with one to three heptameric canonical TRS ‘UCAAAC’ separated by 20 nt in the tandem repeats, it was observed that although transcription initiation occurred at each of the three CS sites in the tandem construct, almost all the transcripts were found as a product of the most downstream CS (Krishnan et al., 1996). Nevertheless, the accumulated amounts of subgenomic mRNA remained nearly the same for the three constructs with one to three CSs because the minigenome with three CSs was replicated with lower efficiency than those with two or one CS copy. Similarly in IBV, expression of CAT under the control of TRSs composed of two tandem repeats of the canonical octameric IBV CS sequence ‘CUUAACAA’, showed that either CS sequence can function as acceptor sites for mRNA synthesis but transcription preferentially occurred at the 3′-most TRS (Stirrups et al., 2000).
In other cases, several copies of the CS were inserted within the same minigenome to express more than one RNA. Insertion of two CS copies within an MHV defective RNA resulted in the decrease of the 5′ upstream mRNA if the CSs were separated by 124 nt (Joo and Makino, 1995). In another study (van Marle et al., 1995), in which combinations of up to three CSs separated by 361–761 nt were inserted, it was shown that the position of the CS affected the amounts of mRNAs produced, in spite of the large distance between CSs.
Both sets of experiments resulted in a negative effect on the transcription of upstream CSs by downstream ones. The observation that the most 3′ CS is preferentially used is consistent with the coronavirus discontinuous transcription during the negative-strand synthesis model (Sawicki and Sawicki, 1990). However, this model does not explain all the observations made in coronavirus transcription. In FIPV and TGEV, the shortest mRNAs are produced in lower quantities than the next larger mRNA encoding the nucleocapsid protein (de Groot et al., 1987b, Sethna et al., 1989, Penzes et al., 2001). This result strongly suggests that mRNA abundance is influenced by the presence of additional regulatory signals.
7.4. Expression using an internal ribosomal entry site
Expression under IRES in coronaviruses has been documented for mRNA3 of IBV (Liu and Inglis, 1992, Le et al., 1995) and for mRNA 5 of MHV (Leibowitz et al., 1988, Thiel and Siddell, 1994). CAT expression has been shown using an internal ribosomal entry site (IRES) sequence of the encephalomyocarditis virus in the MHV system (Lin and Lai, 1993) (Fig. 3). CAT activity 20-fold higher than when using control plasmids was detected in passages 0 and 1, but decreased in passage 2. Using the Sindbis virus, CAT expression levels with the IRES of encephalomyocarditis virus are about 5-fold lower than when expression is driven via a second subgenomic promoter (Bredenbeek and Rice, 1992). The combination of coronaviruses TRS and IRES could be useful for the construction of bicistronic vectors.
7.5. Expression system stability and insert size
Expression from MHV defective RNAs of CAT, HE and murine IFN-γ genes was not observed beyond passages 2, 3 and 4, respectively. Using minigenomes derived from TGEV and IBV expression was more stable but highly dependent on the nature of the heterologous gene used. Luciferase expression with TGEV and IBV minigenomes was reduced to almost background levels, while expression of GUS or CAT using TGEV or IBV derived minigenomes, respectively, was observed for about 10 passages (Izeta et al., 1999, Stirrups et al., 2000).
The expression of GUS or PRSSV ORF5 using TGEV minigenomes was increased until passage three, leading to a single new mRNA corresponding to the GUS or the ORF5 mRNAs (Izeta et al., 1999, Alonso et al., 2001b). Expression levels were maintained for 8 passages, but new mRNA bands of a size lower than those of the full-length GUS or ORF5 mRNA were observed at passage 5, steadily decreasing during successive passages (Alonso et al., 2001a, Alonso et al., 2001b).
In general, the insertion of a heterologous gene such as GUS into TGEV derived minigenomes led to a 40–50-fold reduction in the levels of the minigenome RNA (Alonso et al., 2001a). The limited stability of the helper dependent expression systems is most likely due to the foreign gene since TGEV minigenomes of 9.7, 3.9 and 3.3 kb, in the absence of the heterologous gene, are amplified and efficiently packaged for at least 30 passages, without generating new dominant subgenomic RNAs (Méndez et al., 1996, Izeta et al., 1999). The recombination frequency in MHV, TGEV, and IBV seems inversely proportional to the stability of the recombinants expressing a foreign gene. In fact, the MHV based helper dependent expression system has the lowest stability probably because of the higher recombination frequency within this system (Lai, 1996).
The stability of the expression system is also conditioned by the type of polymerases involved in amplification of the minigenome and transcription of the mRNA (Agapov et al., 1998). The accumulation of mutations during the in vitro expression of minigenome RNAs with T7 DNA-dependent RNA-polymerase is 10−4–10−5 consisting mostly of 1-nt insertions and deletions (Boyer et al., 1992, Sooknanan et al., 1994). After transfection of the in vitro produced RNA, synthesis of mRNA by the viral RNA-dependent RNA-polymerase should have an accumulation of mutations with a relatively higher frequency of 10−3–10−4 (Ward et al., 1988, de Mercoyrol et al., 1992). An improvement in expression stability should be observed by using expression systems initiated by DNA transfection, such as those based on the expression of the minigenomes under CMV promoter, since in these cases an eukaryotic RNA polymerase II expresses the minigenome with an estimated error frequency of 5×10−6 on synthetic polynucleotide substrates (de Mercoyrol et al., 1992). In addition, the eukaryotic RNA polymerase II has additional mechanisms to insure even more accurate transcription (Thomas et al., 1998).
8. Modification of coronavirus tropism
Driving vector expression to different tissues may be highly convenient in order to preferentially induce a specific type of immune response, i.e., mucosal immunity by targeting the expression to gut-associated lymph nodes. In addition, it seems useful to change the species-specificity of the vector to expand its use. Both tissue and species-specificity have been modified using coronavirus genomes.
Group 1 coronaviruses attach to host cells through the S glycoprotein (Holmes and Lai, 1996) by interactions with aminopeptidase N (APN) which is the cellular receptor (Delmas et al., 1992, Yeager et al., 1992, Tresnan et al., 1996, Benbacer et al., 1997, Kolb et al., 1997). Interestingly, while porcine and human aminopeptidases show species-specificity, the feline aminopeptidase seems to serve as a receptor for feline, canine, porcine and HCoV (Tresnan et al., 1996, Benbacer et al., 1997). Group 2 coronaviruses use the carcinoembryonic antigen-related cell adhesion molecules (CEACAM) as receptors (Beauchemin et al., 1999). The S protein is also responsible for cell entry of group 2 coronaviruses such as MHV. In these viruses, the S glycoprotein is cleaved into two covalently associated 90-kDa subunits, the amino-terminal S1 and the carboxy-terminal S2 subunits (Frana et al., 1985, Luytjes et al., 1987). It is believed that the S1 subunit forms the membrane-bound stalk portion (de Groot et al., 1987a). A receptor binding activity has been demonstrated in studies using recombinant protein containing the amino-terminal 330 residues of the S1 subunit of MHV-JHM (Kubo et al., 1994).
Engineering the S gene can lead to changes both in the tissue (Ballesteros et al., 1997, Leparc-Goffart et al., 1998, Sánchez et al., 1999) and species-specificity (Kuo et al., 2000). TGEV enteric or respiratory tropism is conditioned by the primary structure of the S gene (Ballesteros et al., 1997). The S glycoprotein domain recognized by the cellular receptor (pAPN) on ST cells is located within the globular domain of the protein encoded between nucleotides 1518 and 2184. This domain is present both in enteric and respiratory porcine coronaviruses, indicating that its presence in a virus is not in itself sufficient to infect the enteric tract. In fact, it has been demonstrated that a second factor mapping in the S gene around nucleotide 655 drastically influences the enteric tropism of the PUR46 strain of TGEV (Ballesteros et al., 1995, Sánchez et al., 1999).
The tissue-specific tropism of TGEV has been modified by constructing recombinant viruses in which part of the S gene from an isolate with an exclusively respiratory tropism has been substituted by the homologous S gene domain of an enteric virus strain. This has been done either by targeted recombination (Sánchez et al., 1999) or by engineering an infectious cDNA clone (Almazan et al., 2000). Studies on the tropism of selected recombinants obtained in these studies confirmed the need for a second S gene domain, distal from the pAPN binding domain, for enteric infection by TGEV.
The species-specificity of coronaviruses has also being modified by targeted RNA recombination. A mutant of the MHV in which the ectodomain of the S glycoprotein was replaced by the high divergent ectodomain of the S protein of FIPV, resulting in a chimeric virus, acquired the ability to infect feline cells and simultaneously lost the ability to infect murine cells in tissue culture (Kuo et al., 2000). The change of tropism opens up the possibility of engineering coronaviruses to target the desired species.
9. Conclusions
Both helper dependent expression systems, based on two components, and single genomes constructed by targeted recombination, or by using infectious cDNAs, have been developed. The sequences that regulate transcription have been characterized mainly using helper dependent expression systems and it will now be possible to validate them using single genome systems. Using helper dependent expression systems, production of high amounts of heterologous antigens (2–8 μg/106 cells) has been achieved, and the synthesis has been maintained for around 10 passages. These amounts have been sufficient to elicit strong immune responses.
The genome of coronaviruses has been engineered either by targeted recombination or by modification of cDNAs encoding infectious coronavirus RNAs. In some cases (TGEV), a foreign gene (GFP of 0.72 kb) has been efficiently expressed during at least 20 passages (Fischer et al., 1997, Sola et al., 2001b). Thus, a new avenue with a great deal of potential has been opened for the coronaviruses with a long genome size and enteric tropism, this makes them of high interest as expression vectors for vaccine development and, possibly in the future, also for gene therapy. The possibility of engineering the tissue and species tropism will make coronaviruses very flexible expression systems, since the same vector can be modified to target expression to different organs and animal species, including humans.
Acknowledgements
This work has been supported by grants from the Comisión Interministerial de Ciencia y Tecnologı́a (CICYT), La Consejerı́a de Educación y Cultura de la Comunidad de Madrid, and Fort Dodge Veterinaria from Spain, and the European Communities (Life Sciences Program, Key Action 2: Infectious Diseases). JMG received a fellowship from the Department of Research and Technology. ISG, FA, JMS, and EC received contracts from the European Union (Biotechnology, FAIR and Key Action 2: Infectious Diseases). AI, SA, and DE received fellowships from the Department of Education, University and Research of the Basque Government. CR received a fellowship from the Health Department (FIS).
References
- Afanasiev B.N., Ward T.W., Beaty B.J., Carlson J.O. Transduction of Aedes aegypti mosquitoes with vectors derived from Aedes densovirus. Virology. 1999;257:62–72. doi: 10.1006/viro.1999.9621. [DOI] [PubMed] [Google Scholar]
- Agapov E.V., Frolov I., Lindenbach B.D., Pragai B.M., Schlesinger S., Rice C.M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P., French R., Janda M., Loesch-Fries L.S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl. Acad. Sci. USA. 1984;81:7066–7070. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., González J.M., Pénzes Z., Izeta A., Calvo E., Plana-Durán J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, S., Izeta, A., Sola, I., Enjuanes, L., 2001a. Transcription regulatory sequences in transmissible gastroenteritis coronavirus. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- Alonso, S., Sola, I., Wege, H., Teifke, J., Enjuanes, L., 2001b. Heterologous gene expression in tissue culture and in vivo using a transmissible gastroenteritis coronavirus helper dependent system. Submitted for publication.
- Andino R., Silvera D., Suggett S.D., Achacoso P.L., Miller C.J., Baltimore D., Feinberg M.B. Engineering poliovirus as a vaccine vector for the expression of diverse antigens. Science. 1994;265:1448–1451. doi: 10.1126/science.8073288. [DOI] [PubMed] [Google Scholar]
- Ballesteros M.L., Sánchez C.M., Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros M.L., Sánchez C.M., Martı́n-Caballero J., Enjuanes L. Molecular bases of tropism in the PUR46 cluster of transmissible gastroenteritis coronaviruses. Adv. Exp. Med. Biol. 1995;380:557–562. doi: 10.1007/978-1-4615-1899-0_89. [DOI] [PubMed] [Google Scholar]
- Baric R.S., Stohlman S.A., Lai M.M.C. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J. Virol. 1983;48:633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Yount B. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 2000;74:4039–4046. doi: 10.1128/jvi.74.9.4039-4046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin N., Draber P., Dveksler G., Gold P., Gray-Owen S., Grunert F., Hammarström S., Holmes K.V., Karlsson A., Kuroki M., Lin S.-H., Lucka L., Najjar S.M., Neumaier M., Öbrink B., Shively J.E., Skubitz K.M., Stanners C.P., Thomas P., Thompson J.A., Virji M., von Kleist S., Wagener C., Watt S., Zimmerman W. Nomenclature announcement. Redefined nomenclature for members of the carcinoembrionic antigen family. Exp. Cell Res. 1999;252:243–249. doi: 10.1006/excr.1999.4610. [DOI] [PubMed] [Google Scholar]
- Benbacer L., Kut E., Besnardeau L., Laude H., Delmas B. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 1997;71:734–737. doi: 10.1128/jvi.71.1.734-737.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund P., Smerdou C., Fleeton M.N., Tubulekas I., Liljestrom P. Enhancing immune responses using suicidal DNA vaccines. Nature Biotech. 1998;16:562–565. doi: 10.1038/nbt0698-562. [DOI] [PubMed] [Google Scholar]
- Bett A.J., Prevec L., Graham F.L. Packaging capacity and stability of human adenovirus type 5 vectors. J. Virol. 1993;67:5911–5921. doi: 10.1128/jvi.67.10.5911-5921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E.G., Brown T.D.K., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Boyer J.C., Bebenek K., Kunkel T.A. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. USA. 1992;89:6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P.J., Rice C.M. Animal RNA virus expression systems. Semin. Virol. 1992;3:297–310. [Google Scholar]
- Brian, D.A., 2001. Nidovirus genome replication and subgenomic mRNA synthesis. Pathways followed and cis-acting elements required. In: Lavi E., Weiss S., Hingley S.T. (Eds.), Nidoviruses, Plenum Press, New York, in press. [DOI] [PubMed]
- Caley I.J., Betts M.R., Davis N.L., Swanstrom R., Frelinger J.A., Johnston R.E. Venezuelan equine encephalitis virus vectors expressing HIV-1 proteins: vector design strategies for improved vaccine efficacy. Vaccine. 1999;17:3124–3135. doi: 10.1016/s0264-410x(99)00142-5. [DOI] [PubMed] [Google Scholar]
- Caley I.J., Betts M.R., Irlebeck D.M., Davis N.L., Swanstrom R., Frelinger J.A., Johnston R.E. Humoral, mucosal, and cellular immunity in response to a human immunodeficiency virus type 1 immunogen expressed by a Venezuelan equine encephalitis virus vaccine vector. J. Virol. 1997;71:3031–3038. doi: 10.1128/jvi.71.4.3031-3038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T.J., Nestorowicz A., Mason P.W., Rice C.M. Yellow fever/Japanese encephalitis chimeric viruses: construction and biological properties. J. Virol. 1999;73:3095–3101. doi: 10.1128/jvi.73.4.3095-3101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- de Groot R.J., Luytjes W., Horzinek M.C., van der Zeijst B.A.M., Spaan W.J.M., Lenstra J.A. Evidence for a coiled-coil structure in the spike protein of coronaviruses. J. Mol. Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Ter Haar R.J., Horzinek M.C., van der Zeijst B.A.M. Intracellular RNAs of the feline infectious peritonitis coronavirus strain 79–1146. J. Gen. Virol. 1987;68:995–1002. doi: 10.1099/0022-1317-68-4-995. [DOI] [PubMed] [Google Scholar]
- de Haan C.A.M., Kuo L., Masters P.S., Vennema H., Rottier P.J.M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mercoyrol L., Corda Y., Job C., Job D. Accuracy of wheat-germ RNA polymerase II. General enzymatic properties and effect of template conformational transition from right-handed B-DNA to left-handed Z-DNA. Eur. J. Biochem. 1992;206:49–58. doi: 10.1111/j.1432-1033.1992.tb16900.x. [DOI] [PubMed] [Google Scholar]
- de Vries A.A.F., Glaser A.L., Raamsman M.J.B., de Haan C.A.M., Sarnataro S., Godeke G.J., Rottier P.J.M. Genetic manipulation of equine arteritis virus using full-length cDNA clones: separation of overlapping genes and expression of a foreign epitope. Virology. 2000;270:84–97. doi: 10.1006/viro.2000.0245. [DOI] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L'Haridon R., Vogel L.K., Norén O., Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R. In: Dolin R., Wringht P.F., editors. Vol. 127. Marcel Dekker; New York (Basel): 1999. The common cold. Rhinoviruses and coronaviruses; pp. 253–280. (Viral Infections of the Respiratory Tract). [Google Scholar]
- DiCiommo D.P., Bremner R. Rapid, high level protein production using DNA-based Semliki Forest virus vectors. J. Biol. Chem. 1998;17:18060–18066. doi: 10.1074/jbc.273.29.18060. [DOI] [PubMed] [Google Scholar]
- Dubensky T.W., Driver D.A., Polo J.M., Belli B.A., Latham E.M., Ibanez C.E., Chada S., Brumm D., Banks T.A., Mento S.J., Jolly D.J., Chang S.M.W. Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J. Virol. 1996;70:508–519. doi: 10.1128/jvi.70.1.508-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P., Rottier P., Siddell S.G., Spaan W.J.M., Taguchi F., Talbot P. In: Virus Taxonomy. Classification and Nomenclature of Viruses. van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carsten E.B., Estes M.K., Lemon S.M., McGeoch D.J., Maniloff J., Mayo M.A., Pringle C.R., Wickner R.B., editors. Academic Press; New York: 2000. Coronaviridae; pp. 835–849. [Google Scholar]
- Enjuanes L., Spaan W., Snijder E., Cavanagh D. In: Virus Taxonomy. Classification and Nomenclature of Viruses. van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carsten E.B., Estes M.K., Lemon S.M., McGeoch D.J., Maniloff J., Mayo M.A., Pringle C.R., Wickner R.B., editors. Academic Press; New York: 2000. Nidovirales; pp. 827–834. [Google Scholar]
- Enjuanes L., Van der Zeijst B.A.M. In: The Coronaviridae. Siddell S.G., editor. Plenum Press; New York: 1995. Molecular basis of transmissible gastroenteritis coronavirus epidemiology; pp. 337–376. [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Stegen C.F., Koetzner C.A., Masters P.S. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 1997;71:5148–5160. doi: 10.1128/jvi.71.7.5148-5160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J., Goff S.P. Mutational analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- Fodor E., Devenish L., Engelhardt O.G., Palese P., Brownlee G.G., Garcı́a-Sastre A. Rescue of influenza A virus from recombinant DNA. J. Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frana M.F., Behnke J.N., Sturman L.S., Holmes K.V. Proteolytic cleavage of the E2 polyprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 1985;56:912. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I., Frolova E., Schlesinger S. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 1997;71:2819–2829. doi: 10.1128/jvi.71.4.2819-2829.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I., Hoffman T.A., Prágai B.M., Dryga S.A., Huang H.V., Schlesinger S., Rice C.M. Alphavirus-based expression vectors: strategies and applications. Proc. Natl. Acad. Sci. USA. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herweijer H., Latendresse J.S., Williams P., Zhang G., Danko I., Schlesinger S., Woff J.A. A plasmid-based self-amplifying Sindbis virus vector. Hum. Gene Ther. 1995;6:1161–1167. doi: 10.1089/hum.1995.6.9-1161. [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Neumann G., Hobom G., Webster R.G., Kawaoka Y. ‘Ambisense’ approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology. 2000;267:310–317. doi: 10.1006/viro.1999.0140. [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Neumann G., Kawaoka Y., Hobom G., Webster R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V., Lai M.M.C. In: Fundamental Virology. third ed. Fields B.N., Knipe D.M., Howley P.M., editors. Lippincott-Raven; Philadelphia, PA: 1996. Coronaviridae: the viruses and their replication; pp. 541–559. [Google Scholar]
- Hsue B., Masters P.S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B., Masters P.S. An essential secondary structure in the 3′ untranslated region of the mouse hepatitis virus genome. Adv. Exp. Med. Biol. 1998;440:297–302. doi: 10.1007/978-1-4615-5331-1_39. [DOI] [PubMed] [Google Scholar]
- Izeta A., Smerdou C., Alonso S., Penzes Z., Méndez A., Plana-Durán J., Enjuanes L. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 1999;73:1535–1545. doi: 10.1128/jvi.73.2.1535-1545.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M., Makino S. Mutagenic analysis of the coronavirus intergenic consensus sequence. J. Virol. 1992;66:6330–6337. doi: 10.1128/jvi.66.11.6330-6337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M., Makino S. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 1995;69:272–280. doi: 10.1128/jvi.69.1.272-280.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Westaway E.G. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J. Virol. 1994;68:4580–4588. doi: 10.1128/jvi.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Westaway E.G. Subgenomic replicons of the flavivirus kunjin: construction and applications. J. Virol. 1997;71:1479–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetzner C.A., Parker M.M., Ricard C.S., Sturman L.S., Masters P.S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A.F., Hegui A., Siddell S.G. Identification of residues critical for the human coronavirus 229E receptor function of human aminopeptidase N. J. Gen. Virol. 1997;78:2795–2802. doi: 10.1099/0022-1317-78-11-2795. [DOI] [PubMed] [Google Scholar]
- Krishnan R., Chang R.Y., Brian D.A. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology. 1996;218:400–405. doi: 10.1006/viro.1996.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.-J., Raamsman M.J.B., Masters P.S., Rottier P.J.M. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda K., Groner A., Frese K., Drenckhahn D., Hauser C., Rott R., Doerfler W., Klenk H.D. Synthesis of biologically active influenza virus hemagglutinin in insect larvae. J. Virol. 1989;63:1677–1685. doi: 10.1128/jvi.63.4.1677-1685.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Monica N., Yokomori K., Lai M.M.C. Coronavirus mRNA synthesis: identification of novel transcription initiation signals which are differentially regulated by different leader sequences. Virology. 1992;188:402–407. doi: 10.1016/0042-6822(92)90774-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 1996;7:381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- Lai M.M.C., Baric R.S., Brayton P.R., Stohlman S.A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc. Natl. Acad. Sci. USA. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Zhang X., Hinton D., Stohlman S. Modulation of mouse hepatitis virus infection by defective-interfering RNA-mediated expression of viral proteins and cytokines. J. Neurovirol. 1997;3(Supp. 1):S33–S34. [PubMed] [Google Scholar]
- Lapps W., Hogue B.G., Brian D.A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.Y., Sonenberg N., Maizel J.V. Distinct structural elements and internal entry of ribosomes in mRNA 3 encode by infectious bronchitis virus. Virology. 1995;198:405–411. doi: 10.1006/viro.1994.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J.L., Perlman S., Weinstck G., DeVries J.R., Budzilowicz C., Weissemann J.M., Weiss S.R. Detection of a murine coronavirus nonstructural protein encoded in a downstream open reading frame. Virology. 1988;164:156–164. doi: 10.1016/0042-6822(88)90631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leparc-Goffart I., Hingley S.T., Chua M.M., Phillips J., Lavi E., Weiss S.R. Targeted recombination within the spike gene of murine coronavirus mouse hepatitis virus-A59: Q159 is a determinant of hepatotropism. J. Virol. 1998;72:9628–9636. doi: 10.1128/jvi.72.12.9628-9636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.-L., Lai M.M.C. A cis-acting viral protein is not required for the replication of a coronavirus defective-interfering RNA. Virology. 1995;209:428–436. doi: 10.1006/viro.1995.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.L., Zhang X., Lai M.M.C. Coronavirus defective-interfering RNA as an expression vector: the generation of a pseudorecombinant mouse hepatitis virus expressing hemagglutinin-esterase. Virology. 1995;208:319–327. doi: 10.1006/viro.1995.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeström P. Alphavirus expression systems. Curr. Opin. Biotech. 1994;5:495–500. doi: 10.1016/0958-1669(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Liljeström P., Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- Lin Y.J., Lai M.M.C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontinuous sequence for replication. J. Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Identification of two new polypeptides encoded by messenger RNA5 of the coronavirus infectious bronchitis virus. Virology. 1992;186:342–347. doi: 10.1016/0042-6822(92)90094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby J.P., Clinton R., Kurtz S. Adaptation of human enteric coronavirus to growth in cell lines. J. Clin. Virol. 1999;12:43–51. doi: 10.1016/S0928-0197(98)00067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Bredenbeek P., Noten A., Horzinek M., Spaan W. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Sturman L., Bredenbeek P., Charite J., van der Zeijst B., Horzinek M., Spaan W. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology. 1987;161:479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J. Virol. 1993;67:3304–3311. doi: 10.1128/jvi.67.6.3304-3311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M., Makino J.K. A system for study of coronavirus messenger RNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C.-K., Soe L.H., Baker S.C., Lai M.C. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988;166:550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Reverse genetics of the largest RNA viruses. Adv. Virus Res. 1999;53:245–264. doi: 10.1016/S0065-3527(08)60351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Kapikian A.Z., Hardison K.A., Hartley J.W., Chanock R.M. Antigenic relationships among the coronaviruses of man and between human and animal coronaviruses. J. Immunol. 1969;102:1109–1118. [PubMed] [Google Scholar]
- Méndez A., Smerdou C., Gebauer F., Izeta A., Enjuanes L. Structure and encapsidation of transmissible gastroenteritis coronavirus (TGEV) defective interfering genomes. Adv. Exp. Med. Biol. 1995;380:583–589. doi: 10.1007/978-1-4615-1899-0_93. [DOI] [PubMed] [Google Scholar]
- Méndez A., Smerdou C., Izeta A., Gebauer F., Enjuanes L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerle M., Crnkovic I., Hammerschmidt W., Ziegler H., Koszinowski U.H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg J.J.M., Bos-de-Ruijter J.N.A., Wenswoort G., Moormann R.J.M. An infectious cDNA clone of porcine reproductive and respiratory syndrome virus. Adv. Exp. Med. Biol. 1998;440:199–206. doi: 10.1007/978-1-4615-5331-1_24. [DOI] [PubMed] [Google Scholar]
- Molenkamp R., Rozier B.C.D., Greve S., Spaan W.J.M., Snijder E.J. Isolation and characterization of an arterivirus defective interfering RNA genome. J. Virol. 2000;74:3156–3165. doi: 10.1128/jvi.74.7.3156-3165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Watanabe T., Ito H., Watanabe S., Goto H., Gao P., Hughes M., Perez D.R., Donis R., Hoffmann E., Hobom G., Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor J.B., Brian D.A. Downstream ribosomal entry for translation of coronavirus TGEV gene 3b. Virology. 2000;269:172–182. doi: 10.1006/viro.2000.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P. RNA virus vectors: where are we and where do we need to go? Proc. Natl. Acad. Sci. USA. 1998;95:12750–12752. doi: 10.1073/pnas.95.22.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks R.J., Graham F.L. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D., Koetzner C.A., Masters P.S. Analysis of second-site revertants of a murine coronavirus nucleocapsid protein deletion mutant and construction of nucleocapsid protein mutants by targeted RNA recombination. J. Virol. 1995;69:3449–3457. doi: 10.1128/jvi.69.6.3449-3457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z., González J.M., Calvo E., Izeta A., Smerdou C., Sánchez C.M., Sola I., Almazán F., Enjuanes L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the purdue virus cluster. Virus Genes. 2001;23:101–114. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z., Tibbles K., Shaw K., Britton P., Brown T.D.K., Cavanagh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z., Wroe C., Brown T.D.K., Britton P., Cavanagh D. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J. Virol. 1996;70:8660–8668. doi: 10.1128/jvi.70.12.8660-8668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plana-Durán J., Climent I., Sarraseca J., Urniza A., Cortes E., Vela C., Casal J.I. Baculovirus expression of proteins of porcine reproductive and respiratory syndrome virus strain Olot/91. Involvement of ORF3 and ORF5 protein in protection. Virus Genes. 1997;14:19–29. doi: 10.1023/a:1007931322271. [DOI] [PubMed] [Google Scholar]
- Pugachev K.V., Mason R.W., Shope R.E., Frey T.K. Double-subgenomic Sindbis virus recombinants expressing immunogenic proteins of Japanese encephalitis virus induce significant protection in mice against lethal JEV infection. Virology. 1995;212:587–594. doi: 10.1006/viro.1995.1516. [DOI] [PubMed] [Google Scholar]
- Pushko P., Parker M., Ludwing G.V., Davis N.L., Johnston R.E., Smith J.F. Replication-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- Racaniello V.R., Baltimore D. Cloned poliovirus cDNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- Resta S., Luby J.P., Rosenfeld C.D., Siegel J.D. Isolation and propagation of a human enteric coronavirus. Science. 1985;229:978–981. doi: 10.1126/science.2992091. [DOI] [PubMed] [Google Scholar]
- Rice C.M., Grakoui A., Galler R., Chambers T.J. Transcription of infectious yellow fever RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989;1:285–296. [PubMed] [Google Scholar]
- Rice C.M., Levis R., Strauss J.H., Huang H.V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Antón I.M., Suñé C., Pedregosa A.M., Martı́n-Alonso J.M., Parra F., Carrascosa J.L., Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J. Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C.M., Gebauer F., Suñé C., Méndez A., Dopazo J., Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190:92–105. doi: 10.1016/0042-6822(92)91195-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C.M., Izeta A., Sánchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Durán J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana T., Gowda S., Boyko V.P., Albiach-Marti M.R., Mawassi M., Navas-Castillo J., Karasev A.V., Dolja V., Hilf M.E., Lewandowski D.J., Moreno P., Bar-Joseph M., Garnsey S.M., Dawson W.O. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc. Natl. Acad. Sci. USA. 1999;96:7433–7438. doi: 10.1073/pnas.96.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 1998;440:215–220. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- Schnell M.J., Mebatsion T., Conzelmann K.K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P.B., Hung S.-L., Brian D.A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh C.-k., Soe L.H., Makino S., Chang M.-F., Stohlman S.A., Lai M.M.C. The 5′-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology. 1987;156:320–330. doi: 10.1016/0042-6822(87)90412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya H., Birren B., Kim U.J., Mancino V., Slepak T., Tachiiri Y., Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilia M., Boniotti M.B., Angoscini P., Capucci L., Rossi C. Two independent pathways of expression lead to self-assembly of the rabbit hemorrhagic disease virus capsid protein. J. Virol. 1995;69:5812–5815. doi: 10.1128/jvi.69.9.5812-5815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S.G. In: The Viruses. Fraenkel-Conrat H., Wagner R.R., editors. Plenum Press; New York: 1995. The Coronaviridae; p. 418. [Google Scholar]
- Sjoberg E.M., Suomalainen M., Garoff H. A significantly improved Semliki Forest virus expression system based on translation enhancer segments from the viral capsid gene. Bio/Technol. 1994;12:1127–1131. doi: 10.1038/nbt1194-1127. [DOI] [PubMed] [Google Scholar]
- Sola, I., Alonso, S., Plana-Durán, J., Enjuanes, L., 2001a. Heterologous gene expression with a single genome derived from transmissible gastroenteritis coronavirus, submitted for publication.
- Sola, I., Izeta, A., Enjuanes, L., 2001b. Tissue specific targeting of coronavirus neutralizing antibodies using vectors based on RNA minigenomes, submitted for publication.
- Sooknanan R., Howes M., Read L., Malek L.T. Fidelity of nucleic acid amplification with avian myeloblastosis virus reverse transcriptase and T7 RNA polymerase. Biotechniques. 1994;17:1077–1085. [PubMed] [Google Scholar]
- Spaan W.H., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B.A.M., Siddell S.G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirrups K., Shaw K., Evans S., Dalton K., Casais R., Cavanagh D., Britton P. Expression of reporter genes from the defective RNA CD-61 of the coronavirus infectious bronchitis virus. J. Gen. Virol. 2000;81:1687–1698. doi: 10.1099/0022-1317-81-7-1687. [DOI] [PubMed] [Google Scholar]
- Tahara S.M., Dietlin T.A., Nelson G.W., Stohlman S.A., Manno D.J. Mouse hepatitis virus nucleocapsid protein as a translational effector of viral mRNAs. Adv. Exp. Med. Biol. 1998;440:313–318. doi: 10.1007/978-1-4615-5331-1_41. [DOI] [PubMed] [Google Scholar]
- Talbot P.J. Virus-induced autoimmunity in multiple sclerosis: the coronavirus paradigm. Adv. Clin. Neurosci. 1997;7:215–233. [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S. Infectious RNA transcribed in vivo from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Thiel V., Siddell S.G. Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J. Gen. Virol. 1994;75:3041–3046. doi: 10.1099/0022-1317-75-11-3041. [DOI] [PubMed] [Google Scholar]
- Thiel V., Siddell S.G., Herold J. Replication and transcription of HCV 229E replicons. Adv. Exp. Med. Biol. 1998;440:109–114. doi: 10.1007/978-1-4615-5331-1_14. [DOI] [PubMed] [Google Scholar]
- Thomas M.J., Platas A.A., Hawley D.K. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- Tresnan D.B., Levis R., Holmes K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R.G., De Groot R.J., Spaan W.J.M. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 1994;68:3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R.G., Heijnen L., Spaan W.J.M., Degroot R.J. Homologous RNA recombination allows efficient introduction of site-specific mutations into the genome of coronavirus MHV-A59 via synthetic co-replicating RNAs. Nucleic Acids Res. 1992;20:3375–3381. doi: 10.1093/nar/20.13.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R.G., Spaan W.J.M. In: The Coronaviridae. Siddell S.G., editor. Plenum Press; New York: 1995. Coronavirus replication, transcription, and RNA recombination; pp. 11–31. [Google Scholar]
- van Dinten L.C., den Boon J.A., Wassenaar A.L.M., Spaan W.J.M., Snijder E.J. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., Dobbe J.C., Gultyaev A.P., Luytjes W., Spaan W.J.M., Snijder E.J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Nat. Acad. Sci. USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., Luytjes W., Van der Most R.G., van der Straaten T., Spaan W.J.M. Regulation of Coronavirus mRNA transcription. J. Virol. 1995;69:7851–7856. doi: 10.1128/jvi.69.12.7851-7856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.D., Stokes M.A.M., Flanagan J.B. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J. Virol. 1988;62:558–562. doi: 10.1128/jvi.62.2.558-562.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.G., Chang R.-Y., Brian D.A. In: Corona and Related Viruses. Talbot P.J., Levy G.A., editors. Plenum Press; New York: 1995. Evidence for a pseudoknot in the 3′-untranslated region of the bovine coronavirus genome; pp. 511–514. [DOI] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: the transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hinton D.R., Cua D.J., Stohlman S.A., Lai M.M.C. Expression of interferon-γ by a coronavirus defective-interfering RNA vector and its effect on viral replication, spread, and pathogenicity. Virology. 1997;233:327–338. doi: 10.1006/viro.1997.8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hinton D.R., Park S., Parra B., Liao C.-L., Lai M.M.C. Expression of hemagglutinin/esterase by a mouse hepatitis virus coronavirus defective-interfering RNA alters viral pathogenesis. Virology. 1998;242:170–183. doi: 10.1006/viro.1997.8993. [DOI] [PMC free article] [PubMed] [Google Scholar]


