Abstract
The unclassified bovine enteric calicivirus (BEC) is a new bovine enteric calicivirus that is different from bovine norovirus, and causes diarrhea and pathologies in the small intestine of calves. This virus includes Nebraska (NB)- and Newbury agent 1 (NA1)-like strains. The prevalence of this BEC and its genetic characterization has only been reported in the UK and the USA. This study examined the prevalence and genetic diversity of these BECs in diarrheic calves in South Korea. Among a total of 645 diarrheic fecal specimens obtained from 629 cattle herds, these unclassified BECs were detected in 59 (9.1%) diarrheic fecal samples from 57 herds (9.3%) by either RT-PCR or nested PCR. Sequence and phylogenetic analyses of the partial RdRp gene showed that all the Korean BECs clustered together and were closely related to the NB-like viruses (80.9–88.1% nucleotide and 84.5–98.4% amino acid) but not to the NA1-like viruses (75.8–78.4% nucleotide and 79.7–82.8% amino acid). Although these viruses could not be classified into NA1- and NB-like viruses from the sequence and phylogenetic data of the entire capsid gene, all the Korean BECs clustered together on a branch separate from the other known BECs. These results show that these BEC infections are endemic in diarrheic calves in South Korea. The infecting strains are genetically closer to the NB-like viruses but have a distinct evolutionary pathway.
Keywords: Unclassified bovine enteric calicivirus, Calves, Prevalence, Genetic diversity
1. Introduction
Caliciviruses are small, non-enveloped viruses that are 27–38 nm in diameter and possess a single-stranded, plus-sense RNA genome of 7.3–8.3 kb in length and a major structural protein (Green et al., 2001, Sosnovtsev et al., 1998). The family Caliciviridae is classified into four genera: Norovirus (NoV), Sapovirus (SaV), Vesivirus and Lagovirus (Green et al., 2000) with a new proposed genus, Nebraska (NB)-like (Smiley et al., 2002). Caliciviruses have been identified in a number of animal species in association with respiratory, vesicular and hemorrhagic and enteric diseases. In humans, NoVs and SaVs have been associated with enteric diseases only, and are the most common cause of food- and waterborne, acute, non-bacterial gastroenteritis in humans worldwide (Green et al., 2001). Animal SaVs and NoVs also cause gastroenteritis in swine, calves and mink (Bridger, 1990, Guo et al., 2001, Saif et al., 1980).
In addition to bovine noroviruses (BNoV), another bovine enteric calicivirus, Newbury agent-1 (NA1), was reported to be associated with calf diarrhea in the UK in 1978 (Woode and Bridger, 1978). Electron microscopy, animal cross-protection experiments and solid-phase immune electron microscopy (Bridger et al., 1984, Dastjerdi et al., 1999, Woode and Bridger, 1978) revealed NA1 to be unrelated to the bovine enteric calicivirus, Newbury agent-2, which is the prototype BNoV. Genomic data were recently obtained from two virus strains, Bo/Nebraska/80/US and Bo/Newbury1/76/UK (Oliver et al., 2006, Smiley et al., 2002). Smiley et al. (2002) reported that the NB-like bovine enteric calicivirus is genetically most similar to SaVs and lagoviruses, and causes pathology in the small intestine of gnotobiotic calves. Oliver et al. (2006) showed that NB and NA1 viruses form a distinct clade that is independent of the 4 recognized genera. The evidence for classifying the NA1 and NB viruses into the same genus was their identical genome organization, identical composition and location of predicted ORF1 protease cleavage sites as well as the almost identical capsid proteins (Oliver et al., 2006). Until now, this potentially new genus, including NB- and NA1-like unclassified bovine enteric caliciviruses (BECs), has not been named by the International Committee on Viral Taxonomy. In this study, therefore, we use the name of these viruses as “unclassified BECs” with “NB- and NA1-like viruses” to designate the strains.
Determination of the antigenic relationship between genogroups and genotypes of enteric caliciviruses is difficult because most viruses, with the exception of Cowden porcine SaV, do not grow in a cell culture (Flynn and Saif, 1988, Han et al., 2004, Parwani et al., 1991). Therefore, molecular studies have relied upon the availability of stool samples collected during outbreaks or from human volunteers and experimental animal studies. Similar to the genomes of lagoviruses and SaVs, unclassified BECs have two predicted open reading frames (ORFs) that encode non-structural and contiguous capsid proteins (ORF1), as well as a basic protein that is characteristic of the family Caliciviridae (Oliver et al., 2006, Smiley et al., 2002). The NA1 and NB viruses can be assigned into two separate genotypes, based on the genetic divergence in the RNA-dependent RNA polymerase (RdRp) gene (Oliver et al., 2006).
The prevalence of the unclassified BECs and its genetic characterization has only been reported in the UK (Oliver et al., 2006, Woode and Bridger, 1978) and the USA (Han et al., 2004, Smiley et al., 2002). Therefore, this study examined the prevalence and genetic diversity of unclassified BECs in diarrheic calves in South Korea.
2. Materials and methods
2.1. Specimens
Local veterinary clinicians sampled one or two diarrheic fecal samples from each farm and immediately submitted them to the College of Veterinary Medicine, Chonnam National University. Between January 2004 and December 2005, a total of 645 diarrheic fecal specimens were collected from 629 Korean native beef (Hanwoo) calf herds (aged 3–70 days) during the spring (407 samples/406 herds), summer (107 samples/98 herds), autumn (73 samples/69 herds) and winter (58 samples/56 herds). As the fecal samples had arrived, they were examined immediately for common enteric viral, bacterial and protozoan pathogens including unclassified BECs, groups A, B and C bovine rotaviruses (BRV A–C), bovine torovirus (BToV), bovine coronavirus (BCoV), BNoV, bovine viral diarrhea virus (BVDV), Salmonella spp., Clostridium spp., Campylobacter spp., shiga-toxin-producing Escherichia coli, Coccidium spp. and Cryptosporidium spp. (Asakura et al., 1998, Park et al., 2007a, Park et al., 2007b, Park et al., 2008). The results of the BNoV, BToV and BCoV are reported elsewhere (Park et al., 2007a, Park et al., 2007b, Park et al., 2008). Detailed data on the other enteric viruses will be reported elsewhere.
2.2. RNA extraction
For RNA extraction, fecal suspensions of each sample were prepared by diluting the feces 1:10 in 0.01 M phosphate-buffered saline, pH 7.2. The suspensions were then vortexed for 30 s and centrifuged (1200 × g for 20 min). The 200 μl supernatant was used for RNA extraction using the Trizol-LS (Gibco-BRL, Life Tech, Grand Island, NY) procedure. The total RNA recovered was suspended in 50 μl of RNase-free water and stored at −80 °C until needed.
2.3. RT-PCR and nested PCR
The oligonucleotide primers as well as the RT-PCR and nested PCR conditions used to detect unclassified BEC, BToV, BCoV, BRV A-C, BNoV and BVDV are described elsewhere (Cho et al., 2001, Park et al., 2006, Park et al., 2007a, Park et al., 2007b, Park et al., 2008). As a negative control for all RT-PCR assays, RNA was extracted from the normal feces of colostrum-deprived calves that had been inoculated with 50 ml sterile PBS. In order to check for cross-contamination between RT-PCR and nested PCR, 5 μl of the RT-PCR product with the RNA extracted from the normal mock-infected calves was used for all nested PCR assays. The amplification products were analyzed by 1.5 or 2% agarose gel electrophoresis and visualized by irradiating the ethidium bromide stained samples with UV.
2.4. DNA sequencing
The RT-PCR products of a portion of the capsid gene (1692 bp), and the nested PCR products of a portion of the RdRp gene (194 bp) were selected from different test reactions and sequenced to confirm the specificity of the reaction as well as to obtain the genomic data for phylogenetic analysis. The RT-PCR products were purified using a GenClean II kit (BIO 101, Inc., La Jolla, CA) according to the manufacture's instructions. DNA sequencing was carried out using an automated DNA sequencer (ABI system 3700, Applied Biosystem Inc., Foster City, CA).
2.5. Molecular analysis
Using the DNA Basic module (DNAsis MAX, Alameda, CA), the nucleotide (nt) and deduced amino acid (aa) sequences of the capsid gene and a portion of the RdRp gene were compared with those selected from other known caliciviruses (Table 1 ). The variation in the nt sequences within the capsid gene was analyzed using the sliding-window genetic diversity plot (SimPlot, version 3.5.1; http://sray.med.som.jhmi.edu/RaySoft).
Table 1.
Name and Genbank accession number of the reference unclassified bovine enteric caliciviruses (BEC) and other caliciviruses used in phylogenetic analysis
| Viruses | Strains | Source | Viruses | Strains | Source |
|---|---|---|---|---|---|
| BEC | Bo/MA16/04/Korea | DQ984640 | BEC | Bo/MA474/05/Korea | EF528566 |
| BEC | Bo/MA56/04/Korea | DQ984641 | BEC | Bo/MA567-1/05/Korea | EF528567 |
| BEC | Bo/SA68/04/Korea | DQ984642 | BEC | Bo/MA567-2/05/Korea | EF528568 |
| BEC | Bo/MA91/04/Korea | DQ984643 | BEC | Bo/MA729/05/Korea | EF528569 |
| BEC | Bo/MA95/04/Korea | DQ984644 | BEC | Bo/NB/80/US | AY082891 |
| BEC | Bo/MA99/04/Korea | DQ984645 | BEC | Bo/CV23-OH/02/US | AY082890 |
| BEC | Bo/MA120/04/Korea | DQ984646 | BEC | Bo/CV504-OH/02/US | AY549168 |
| BEC | Bo/MA129/04/Korea | DQ984647 | BEC | Bo/CV519-OH/02/US | AY549169 |
| BEC | Bo/MA137/04/Korea | DQ984648 | BEC | Bo/CV526-OH/02/US | AY549170 |
| BEC | Bo/MA164/04/Korea | DQ984649 | BEC | Bo/CV531-OH/02/US | AY549171 |
| BEC | Bo/HP167/04/Korea | DQ984650 | BEC | Bo/CV548-OH/02/US | AY549172 |
| BEC | Bo/MA176/04/Korea | DQ984651 | BEC | Bo/CV562-OH/02/US | AY549173 |
| BEC | Bo/SA181/04/Korea | DQ984652 | BEC | Bo/Newbury1/76/UK | DQ013304 |
| BEC | Bo/MA216/04/Korea | DQ984653 | BEC | Bo/PenrithC39/00/UK | DQ228162 |
| BEC | Bo/MA236/04/Korea | DQ984654 | BEC | Bo/Penrith142/00/UK | DQ228160 |
| BEC | Bo/MA360/04/Korea | DQ984655 | BEC | Bo/Penrith143/00/UK | DQ228161 |
| BEC | Bo/MA39/04/Korea | EF528558 | BEC | Bo/Penrith150/00/UK | DQ228157 |
| BEC | Bo/SA63/04/Korea | EF528559 | BEC | Bo/Penrith151/00/UK | DQ228158 |
| BEC | Bo/MA271/04/Korea | EF528560 | BEC | Bo/Starcross93/00/UK | DQ228165 |
| BEC | Bo/MA274/04/Korea | EF528561 | BEC | Bo/Starcross117/00/UK | DQ228164 |
| BEC | Bo/MA278/04/Korea | EF528562 | Norovirus | Bo/Newbury2/76/UK | AF097917 |
| BEC | Bo/MA298/04/Korea | EF528563 | Sapovirus | Hu/Sapporo/82/JP | U65427 |
| BEC | Bo/MA362/04/Korea | EF528564 | Lagovirus | Ra/RHDV/GH/88/GE | M67473 |
| BEC | Bo/MA415/05/Korea | EF528565 | Vesivirus | Fe/FCV/F9/58/US | M86379 |
Phylogenetic and bootstrap (1000 replicates) analyses based on the nt sequences of the capsid gene (1650 bp without primer sequence) and a portion of the RdRp gene (154 bp without primer sequence) was carried out using the neighbor-joining method and the unweighted-pair group method of Molecular Evolutionary Genetics analysis (MEGA version 3.1) with a pairwise distance (Kumar et al., 2004). A sequence similarity search was performed for the unclassified BEC capsid and RdRp genes using the LALIGN Query program of the GENESTREAM network server at Institut de Génétque Humaine, Montpellier, FRANCE (http://www.eng.uiowa.edu/∼tscheetz/sequence-analysis/examples/LALIGN/lalign-guess.html).
3. Results and discussion
A total of 645 diarrheic fecal samples from 629 calf herds were screened by RT-PCR and nested PCR. Nine positive fecal samples from 9 herds were detected using the 1-step RT-PCR assay, which targeted a 594 bp fragment of the RdRp region in the unclassified BECs. Fifty-nine fecal samples (9.2%) from 57 herds (9.3%) were found positive by the nested PCR assay, which targeted a 194 bp fragment of the RdRp region. Of the 59 unclassified BECs-positive fecal specimens, 19 fecal samples (2.9%) from 18 herds (2.8%) tested positive to this virus alone. The other 40 fecal samples (6.2%) from the 39 herds (6.2%) also tested positive to other enteric pathogens including BRV A-C, BCoV, BNoV, BVDV, BToV and E. coli. In addition, 402 fecal specimens from the 401 herds that tested negative for unclassified BECs tested positive for other enteric pathogens. No enteric pathogens were detected in the 184 fecal samples from 171 herds. The reported fecal prevalence of the BECs in calf diarrhea ranged from 8.4% in the UK (Oliver et al., 2006) to 28.0% in the US (Smiley et al., 2003). These results suggest that BEC infections are endemic in diarrheic calves in South Korea in a similar manner to the prevalence of BEC infections in the UK (Oliver et al., 2006).
Seasonally, BEC infections were less prevalent in diarrheic fecal samples of calves obtained in spring than in the other seasons; 30 (7.4%) out of 407 fecal samples [13 (6.6%) out of 198 samples in 2004 and 17 (8.1%) out of 209 samples in 2005] tested positive in spring; 14 (13.1%) out of 107 fecal samples [6 (11.5%) out of 52 samples in 2004 and 8 (14.5%) out of 55 samples in 2005] tested positive in summer; 8 (11.0%) out of 73 fecal samples [4 (10.5%) out of 38 samples in 2004 and 4 (11.4%) out of 35 samples in 2005] tested positive in autumn; and 8 (13.8%) out of 58 fecal samples [4 (12.9%) out of 31 samples in 2004 and 3 (11.1%) out of 27 samples in 2005] tested positive in winter. To our knowledge, there is no report showing a clear seasonal distribution of BEC infections in calves. Therefore, more epidemiological studies throughout the world will be needed to properly understand the seasonal pattern of BEC infections and to establish BEC surveillance programs to prevent BEC infections.
Molecular analysis was performed by sequencing 194 nt of the nt 5698 to nt 6454 region of the RdRp gene from 16 BECs, which were sequenced from the 59 positive samples for the RdRp region by nested PCR. The paired comparisons of these viruses revealed the nt and aa sequences of the Korean viruses RdRp gene to be more homologous to each other (96.4–100% nt and 95.3–100% aa) and more closely related to the NB-like viruses (80.9–88.1% nt and 84.5–98.4% aa) than to the NA1-like viruses (75.8–78.4% nt and 79.7–82.8% aa) (Table 2 ). Phylogenetic analysis of the nt sequences of the partial RdRp gene from the unclassified BECs also revealed the 16 Korean RdRp gene to cluster together and closely in a branch with the NB-like strains, particularly with the PenrithC39 and Starcross93 strains (Fig. 1 ). The NA1-like viruses including the NA1, Penrith142, Starcross117, Penrith151, Penrith143 and Penrith150 strains clustered on a separate major branch (Fig. 1). These results suggest that the Korean BECs are closely related to NB-like but distantly related to the NA1-like viruses. A previous study reported that unclassified BECs can be divided into two potential genotypes, NA1-like and NB-like, according to phylogenetic analysis of the RdRp gene (Oliver et al., 2006). It was reported that both genotypes are circulating in the UK but the US has only NB-like viruses (Oliver et al., 2006). The genetic similarity of the Korean BECs to NB-like viruses from the US can be explained by the fact that from the 1960s, South Korea imports live cows mainly from countries in North American and Oceania, not from European countries. However, the possibility of NA1-like virus infections in South Korea, even with a very low infection status, cannot be excluded. Therefore, continuous monitoring and intensive study of the BECs with a larger number of bovine fecal samples will be needed.
Table 2.
Amino acid and nucleotide identity of the partial polymerase and the full length capsid genes of the Korean unclassified bovine enteric caliciviruses compared with Bo/Newbury1/1976/UK and Bo/NB/1980/US
| Isolate | Nucleotide (nt) and amino acid (aa) identity to |
|||||||
|---|---|---|---|---|---|---|---|---|
| Bo/Newbury1/1976/UK |
Bo/NB/1980/US |
|||||||
| Polymerase |
Capsid |
Polymerase |
Capsid |
|||||
| nt | aa | nt | aa | nt | aa | nt | aa | |
| Bo/MA16/04/KOR | 78.4 | 82.8 | NA | 87.1 | 93.8 | NA | ||
| Bo/MA56/04/KOR | 78.4 | 82.8 | NA | 87.1 | 93.8 | NA | ||
| Bo/SA68/04/KOR | 77.8 | 81.3 | NA | 85.6 | 92.2 | NA | ||
| Bo/MA91/04/KOR | 77.8 | 82.8 | NA | 86.6 | 93.8 | NA | ||
| Bo/MA95/04/KOR | 78.4 | 82.8 | NA | 87.1 | 93.8 | NA | ||
| Bo/MA99/04/KOR | 77.3 | 79.7 | NA | 85.1 | 90.6 | NA | ||
| Bo/MA120/04/KOR | 76.8 | 81.3 | NA | 86.6 | 92.2 | NA | ||
| Bo/MA129/04/KOR | 77.8 | 81.3 | NA | 85.6 | 92.2 | NA | ||
| Bo/MA137/04/KOR | 76.8 | 81.3 | NA | 86.6 | 92.2 | NA | ||
| Bo/MA164/04/KOR | 77.3 | 81.3 | NA | 86.1 | 92.2 | NA | ||
| Bo/HP167/04/KOR | 78.4 | 82.8 | NA | 87.6 | 93.8 | NA | ||
| Bo/MA176/04/KOR | 77.8 | 81.3 | NA | 86.1 | 92.2 | NA | ||
| Bo/SA181/04/KOR | 78.4 | 82.8 | NA | 87.1 | 93.8 | NA | ||
| Bo/MA216/04/KOR | 78.4 | 82.8 | NA | 86.1 | 93.8 | NA | ||
| Bo/MA236/04/KOR | 77.3 | 81.3 | NA | 86.1 | 92.2 | NA | ||
| Bo/MA360/04/KOR | 77.8 | 81.3 | NA | 85.6 | 92.2 | NA | ||
| Bo/MA39/04/KO | NA | 86.1 | 92.3 | NA | 87 | 92.9 | ||
| Bo/SA63/04/KOR | NA | 86.7 | 93.3 | NA | 87.6 | 93.8 | ||
| Bo/MA271/04/KOR | NA | 86.5 | 93.1 | NA | 87.5 | 93.6 | ||
| Bo/MA274/04/KOR | NA | 85.7 | 92.7 | NA | 86.9 | 93.1 | ||
| Bo/MA278/04/KOR | NA | 86.2 | 93.1 | NA | 87.4 | 93.6 | ||
| Bo/MA298/04/KOR | NA | 86.1 | 93.3 | NA | 87.3 | 93.8 | ||
| Bo/MA362/04/KOR | NA | 86.1 | 93.3 | NA | 87.2 | 93.6 | ||
| Bo/MA415/04/KOR | NA | 86.3 | 93.3 | NA | 87.3 | 93.8 | ||
| Bo/MA474/05/KOR | NA | 85.9 | 93.3 | NA | 87 | 93.8 | ||
| Bo/MA567-1/05/KOR | NA | 86.3 | 92.9 | NA | 87.4 | 93.4 | ||
| Bo/MA567-2/05/KOR | NA | 86.2 | 92.7 | NA | 87.3 | 93.3 | ||
| Bo/MA729/05/KOR | NA | 85.8 | 92.3 | NA | 86.8 | 92.9 | ||
| Bo/Newbury1/76/UK | NA | NA | 81.4 | 84.4 | 92.9 | 98.9 | ||
| Bo/Penrith142/00/UK | 94.3 | 96.6 | NA | 79.4 | 84.4 | NA | ||
| Bo/Penrith143/00/UK | 93.8 | 96.6 | NA | 78.9 | 84.4 | NA | ||
| Bo/Penrith150/00/UK | 92.8 | 95.3 | NA | 77.8 | 82.8 | NA | ||
| Bo/Penrith151/00/UK | 93.3 | 96.9 | NA | 78.4 | 84.4 | NA | ||
| Bo/Starcross93/00/UK | 80.9 | 84.4 | NA | 88.1 | 95.3 | NA | ||
| Bo/Starcross117/00/UK | 93.3 | 96.9 | NA | 78.4 | 84.4 | NA | ||
| Bo/NB/80/US | 81.4 | 84.4 | 92.9 | 98.9 | NA | NA | ||
| Bo/CV23-OH/02/US | 79.9 | 82.8 | NA | 85.6 | 92.2 | NA | ||
| Bo/CV504-OH/02/US | NA | 89.8 | 98 | NA | 92.3 | 98.5 | ||
| Bo/CV519-OH/02/US | NA | 86.9 | 93.8 | NA | 87.8 | 93.4 | ||
| Bo/CV526-OH/02/US | NA | 89.8 | 98 | NA | 92.2 | 98.5 | ||
| Bo/CV531-OH/02/US | NA | 89.9 | 98.4 | NA | 92 | 98.5 | ||
| Bo/CV548-OH/02/US | NA | 89.3 | 98.2 | NA | 91.4 | 98.7 | ||
| Bo/CV562-OH/02/US | NA | 89.6 | 98.4 | NA | 91.7 | 98.7 | ||
NA, genomic sequence for region not available.
Fig. 1.
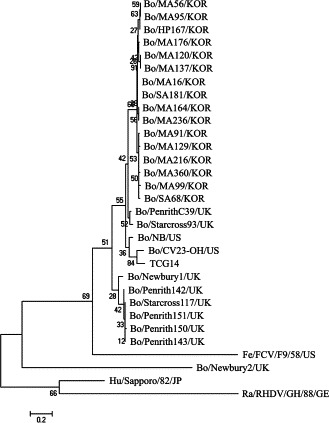
The phylogenetic trees of the nucleotide sequences of the partial RdRp gene of caliciviruses were constructed using the neighbor-joining method of Molecular Evolutionary Genetics Analysis (Kumar et al., 2004). The names of the viruses used are listed in Table 1.
The RT-PCR assay with the primer pair specific to the whole capsid gene (1692 nt of the nt 5035 to nt 6726) carried out using the 59 fecal samples testing positive to the RdRp gene by nested PCR. Among the positive amplicons, 12 amplicons that had been amplified strongly by RT-PCR were selected and sequenced. The Korean viruses shared high nt and aa identity with both NB-like (86.8–87.7% nt and 92.9–93.8 aa) and NA1-like (85.7–86.7% nt and 92.3–93.3% aa) viruses for the entire capsid gene (Table 2). In the S domain, which is the most conserved region in the capsid gene (Green et al., 2001), the Korean viruses showed high nt and aa identity with both NB-like (92.9–93.9% nt and 98.2–99.1% aa) and NA1-like (93.1–94.0% nt and 97.7–98.6% aa) viruses (Table 3 ). However, the Korean BECs had lower nt and aa identity with the NB (76.9–78.1% nt and 82.2–84.0% aa) and NA1 (75.3–76.7% nt and 81.7–82.8% aa) viruses in the P2 domain, which is the putative receptor binding and the most hypervariable region in the capsid gene (Green et al., 2001) (Table 3). The SimPlot results were equivalent to those of the nt and aa identities of each domain of the capsid gene, where the most hypervariable region was observed in the P2 region (Fig. 2 ). In addition, an analysis of the nt and aa sequences of the Korean BEC capsid as a whole, S and P regions were more homologous to each other (Fig. 2).
Table 3.
Amino acid and nucleotide identity of the various capsid domains of the South Korean unclassified bovine calicivirus strains to the Bo/Newbury1/1976/UK and Bo/NB/1980/US
| Virus | Amino acid [nucleotide] identity of capsid genea |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bo/Newbury 1/1976/UK |
Bo/NB/1980/US |
|||||||||||
| Complete | S-domain | P1A-domain | P2-domain | P1B-domain | P-domain | Complete | S-domain | P1A-domain | P2-domain | P1B-domain | P-domain | |
| Bo/MA39/KOR | 92.3 [86.1] | 97.7 [93.4] | 92.9 [88.7] | 82.2 [75.9] | 97.1 [85.4] | 88.7 [81.1] | 92.9 [87.0] | 98.2 [93.2] | 92.9 [89.9] | 83.4 [76.9] | 97.1 [88.3] | 89.3 [82.7] |
| Bo/SA63/KOR | 93.3 [86.7] | 98.2 [94.0] | 98.2 [88.7] | 82.8 [76.7] | 97.1 [85.8] | 89.9 [81.6] | 93.8 [87.6] | 98.6 [93.5] | 98.2 [89.9] | 84.0 [78.1] | 97.1 [89.3] | 90.5 [83.6] |
| Bo/MA271/KOR | 93.1 [86.5] | 98.6 [94.0] | 98.2 [88.7] | 81.7 [76.3] | 97.1 [85.8] | 89.3 [81.4] | 93.6 [87.5] | 99.1 [93.5] | 98.2 [89.9] | 82.8 [77.7] | 97.1 [89.3] | 99.9 [83.4] |
| Bo/MA274/KOR | 92.7 [85.7] | 98.2 [93.1] | 98.2 [86.9] | 81.7 [75.3] | 96.1 [85.8] | 89.0 [80.6] | 93.1 [86.9] | 98.6 [92.9] | 98.2 [88.1] | 82.2 [77.5] | 96.1 [88.7] | 89.3 [82.8] |
| Bo/MA278/KOR | 93.1 [86.2] | 98.2 [93.4] | 98.2 [88.1] | 82.2 [76.2] | 96.1 [85.4] | 89.3 [81.2] | 93.6 [87.4] | 98.6 [93.9] | 98.2 [89.3] | 83.4 [77.8] | 96.1 [88.3] | 99.9 [83.1] |
| Bo/MA298/KOR | 93.3 [86.1] | 98.6 [93.2] | 98.2 [88.1] | 82.8 [76.1] | 96.1 [85.4] | 99.6 [81.1] | 93.8 [87.3] | 99.1 [93.7] | 98.2 [89.3] | 84.0 [77.5] | 96.1 [88.3] | 90.2 [82.9] |
| Bo/MA362/KOR | 93.3 [86.1] | 98.6 [93.4] | 98.2 [88.7] | 82.8 [76.1] | 96.1 [85.0] | 99.6 [81.1] | 93.6 [87.2] | 99.1 [93.2] | 98.2 [89.9] | 83.4 [77.5] | 96.1 [88.6] | 99.9 [83.1] |
| Bo/MA415/KOR | 93.3 [86.3] | 98.6 [93.7] | 98.2 [88.1] | 82.8 [76.3] | 96.1 [85.4] | 99.6 [81.2] | 93.8 [87.3] | 99.1 [93.5] | 98.2 [89.3] | 84.0 [77.7] | 96.1 [88.3] | 90.2 [83.0] |
| Bo/MA474/KOR | 93.3 [85.9] | 98.6 [93.2] | 98.2 [88.1] | 82.8 [75.7] | 96.1 [85.4] | 99.6 [80.9] | 93.8 [87.0] | 99.1 [93.1] | 98.2 [89.3] | 84.0 [77.5] | 96.1 [88.3] | 90.2 [82.9] |
| Bo/MA567-1/KOR | 92.9 [86.3] | 97.7 [93.4] | 98.2 [88.7] | 82.2 [76.3] | 97.1 [85.8] | 99.6 [81.4] | 93.4 [87.4] | 98.2 [93.2] | 98.2 [89.9] | 83.4 [77.7] | 97.1 [89.3] | 90.2 [83.4] |
| Bo/MA567-2/KOR | 92.7 [86.2] | 98.2 [93.5] | 96.4 [88.1] | 81.7 [76.1] | 97.1 [85.8] | 89.0 [81.2] | 93.3 [87.3] | 98.6 [93.4] | 96.4 [89.3] | 82.8 [77.5] | 97.1?? [89.3] | 99.6 [83.2] |
| Bo/MA729/KOR | 92.3 [85.8] | 97.7 [93.1] | 98.2 [88.7] | 81.7 [75.5] | 95.1 [84.8] | 88.7 [80.7] | 92.9 [86.8] | 98.2 [92.9] | 98.2 [89.9] | 82.8 [77.3] | 95.1 [87.7] | 89.3 [82.7] |
Each domain is based on the amino acid and nucleotide sequences of S (1-217 nt and 1-651 aa), P1A (218–273 nt and 652–819 aa), P2 (274–442 nt and 820–1326 aa) and P1B (443–545 nt and 1327–1635 aa) domains for the Parkville virus (PV; genus Sapovirus) capsid protein, as determined by X-ray crystallography (Chen et al., 2004).
Fig. 2.
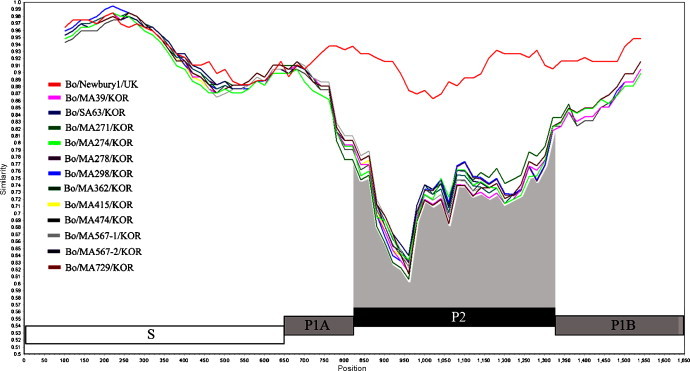
The nucleotide identity plot of the Newbury1 and all Korean unclassified bovine calicivirus strains was generated in comparison with the Nebraska strain using Simplot with a 200-nucleotide window moving along in 20 nucleotide steps. The capsid gene was divided into the S (white box), P1A and P1B (gray box) and P2 (black box) domains (Chen et al., 2004).
Unlike the phylogenetic data of RdRp region, the phylogenetic tree of the nt sequences of the whole capsid gene did not show any characteristic clustering to the NB-like or NA1-like viruses. Moreover, all Korean BECs clustered on a separate branch. The NB-like and NA1-like viruses were on a different branch (Fig. 3 ). In addition, the Korean BECs in the phylogenetic analysis of the RdRp and capsid genes clustered together, and the other known BECs clustered on a separate branch. In contrast to genotyping by the RdRp gene, it was reported that there are no capsid genotypes (Oliver et al., 2006). Therefore, this data can support the above suggestion (Oliver et al., 2006) and suggests that the Korean BECs may have a distinct evolutionary pathway from the NB- and NA1-like BECs.
Fig. 3.
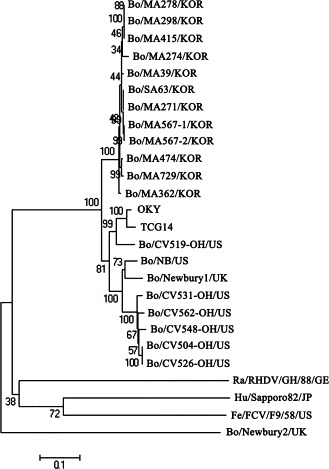
The phylogenetic trees of the nucleotide sequences of the entire capsid gene of the caliciviruses were constructed using the neighbor-joining method of Molecular Evolutionary Genetics Analysis (Kumar et al., 2004). The names of viruses used are listed in Table 1.
In summary, unclassified BEC infections are endemic in diarrheic calves in South Korea. The infecting strains are genetically closer to NB-like viruses but may have a distinct evolutionary pathway from the NB- and NA1-like BECs.
Acknowledgments
This study was supported by the funds from the National Veterinary Research and Quarantine Service (NVRQS), Ministry of Agriculture and Forestry, and the Regional Technology Innovation Program (RTI05-01-01) of the Ministry of Commerce, Industry and Energy (MOCIE), Republic of Korea. The authors acknowledge a graduate fellowship provided by the Korean Ministry of Education and Human Resources Development through the Brain Korea 21 project.
References
- Asakura H., Makino S., Shirahata T., Tsukamoto T., Kurazono H., Ikeda T., Takeshi K. Detection and genetical characterization of Shiga toxin-producing Escherichia coli from wild deer. Microbiol. Immunol. 1998;42:815–822. doi: 10.1111/j.1348-0421.1998.tb02356.x. [DOI] [PubMed] [Google Scholar]
- Bridger J.C. Small viruses associated with gastroenteritis in animals. In: Saif L.J., Theil K.W., editors. Viral Diarrheas of Man and Animals. CRC Press; Boca Raton: 1990. pp. 161–182. [Google Scholar]
- Bridger J.C., Hall G.A., Brown J.F. Characterization of calici-like virus (Newbury agent) found in association with astrovirus in bovine diarrhea. Infect. Immun. 1984;43:133–138. doi: 10.1128/iai.43.1.133-138.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Neill J.D., Noel J.S., Huston A.M., Glass R.I., Estes M.K., Venkataran Prasad B.V. Inter- and intragenus structural variations in caliciviruses and their functional implications. Virology. 2004;78:6469–6479. doi: 10.1128/JVI.78.12.6469-6479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.O., Hasoksuz M., Nielsen P.R., Chang K.O., Lathrop S., Saif L.J. Cross-protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection. Arch. Virol. 2001;146:2401–2419. doi: 10.1007/s007050170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastjerdi M.M., Green J., Callimore C.I., Brown D.W.G., Bridger J.C. The bovine Newbury agent-2 is genetically more closely related to human SRSVs than to animal caliciviruses. Virology. 1999;254:1–5. doi: 10.1006/viro.1998.9514. [DOI] [PubMed] [Google Scholar]
- Flynn W.T., Saif L.J. Serial propagation of porcine enteric calicivirus-like virus in primary porcine kidney cell cultures. J. Clin. Microbiol. 1988;26:206–212. doi: 10.1128/jcm.26.2.206-212.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.Y., Chanock R.M., Kapikian A.Z. Human caliciviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. fourth ed. Lippinocott Williams & Wilkins; Philadelphia: 2001. pp. 841–874. [Google Scholar]
- Green K.Y., Ando T., Balayan M.S., Berke T., Clarke I.N., Estes M.K., Matson D.O., Nakata S., Neill J.D., Studdert M.J., Thiel H.-J. Taxonomy of the caliciviruses. J. Infect. Dis. 2000;181:S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- Guo M., Evermann J.E., Saif L.J. Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Arch. Virol. 2001;146:479–493. doi: 10.1007/s007050170157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.G., Smiley J.R., Thomas C., Saif L.J. Genetic recombination between two genotypes of genogroup III bovine noroviruses (BoNVs) and capsid sequence diversity among BoNVs and Nebraska-like bovine enteric caliciviruses. J. Clin. Microbiol. 2004;42:5214–5224. doi: 10.1128/JCM.42.11.5214-5224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Oliver S.L., Asobayire E., Dastjerdi A.M., Bridger J.C. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology. 2006;350:240–250. doi: 10.1016/j.virol.2006.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Lim G.K., Park S.I., Kim H.H., Cho K.O. Detection and molecular characterization of calf diarrhea bovine coronaviruses circulating in South Korea during 2004–2005. Zoonoses Publ. Health. 2007;54:223–230. doi: 10.1111/j.1863-2378.2007.01045.x. [DOI] [PubMed] [Google Scholar]
- Park S.I., Jeong C., Kim H.H., Park S.H., Park S.J., Hyun B.H., Yang D.K., Kim S.K., Kang M.I., Cho K.O. Molecular epidemiology of bovine noroviruses in South Korea. Vet. Microbiol. 2007;124:125–133. doi: 10.1016/j.vetmic.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Oh E.H., Park S.I., Kim H.H., Jeong Y.J., Lim G.K., Hyun B.H., Cho K.O. Molecular epidemiology of bovine torovirus circulating in South Korea. Vet. Microbiol. 2008;126:364–371. doi: 10.1016/j.vetmic.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jeong C., Yoon S.S., Choy H.E., Saif L.J., Park S.H., Kim Y.J., Jeong J.H., Park S.I., Kim H.H., Lee B.J., Cho H.S., Kim S.K., Kang M.I., Cho K.O. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J. Clin. Microbiol. 2006;44:3178–3188. doi: 10.1128/JCM.02667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parwani A.V., Flynn W.T., Gadfied K.L., Saif L.J. Serial propagation of porcine enteric calicivirus in a continuous cell line. Effect of medium supplementation with intestinal contents or enzymes. Arch. Virol. 1991;120:115–122. doi: 10.1007/BF01310954. [DOI] [PubMed] [Google Scholar]
- Saif L.J., Bohl E.H., Theil K.W., Cross R.F., House J.A. Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. J. Clin. Microbiol. 1980;12:105–111. doi: 10.1128/jcm.12.1.105-111.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley J.R., Hoet A.E., Tråvén M., Tsunemitsu H., Saif L.J. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J. Clin. Microbiol. 2003;41:3089–3099. doi: 10.1128/JCM.41.7.3089-3099.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley J.R., Chang K.O., Hayes J., Vinjé J., Saif L.J. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J. Virol. 2002;76:10089–10098. doi: 10.1128/JVI.76.20.10089-10098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Sosnovtseva S.A., Green K.Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 1998;72:3051–3059. doi: 10.1128/jvi.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woode G.N., Bridger J.C. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J. Med. Microbiol. 1978;11:441–452. doi: 10.1099/00222615-11-4-441. [DOI] [PubMed] [Google Scholar]


