Abstract
During dengue outbreaks, acute diagnosis at the patient's point of need followed by appropriate supportive therapy reduces morbidity and mortality. To facilitate needed diagnosis, we developed and optimized a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay that detects all 4 serotypes of dengue virus (DENV). We used a quencher to reduce nonspecific amplification. The assay does not require expensive thermocyclers, utilizing a simple water bath to maintain the reaction at 63 °C. Results can be visualized using UV fluorescence, handheld readers, or lateral flow immunochromatographic tests. We report a sensitivity of 86.3% (95% confidence interval [CI], 72.7–94.8%) and specificity of 93.0% (95% CI, 83.0–98.1%) using a panel of clinical specimens characterized by DENV quantitative reverse transcription–polymerase chain reaction. This pan-serotype DENV RT-LAMP can be adapted to field-expedient formats where it can provide actionable diagnosis near the patient's point of need.
Keywords: Dengue, Loop-mediated isothermal amplification, RT-LAMP
Highlights
-
•
We developed a pan-serotype DENV RT-LAMP assay.
-
•
The assay demonstrates a sensitivity of 86.3% and specificity of 93.0% using a panel of clinical samples.
-
•
Feasibility of pairing this reaction with different visualization methods was demonstrated.
1. Introduction
Dengue is one of the fastest growing global health problems today, caused by a single-stranded, positive-sense enveloped RNA virus that belongs to the Flaviviridae family (Bhatt et al., 2013, Clopper and Pearson, 1934). The human immune system recognizes 4 antigenically distinct serotypes of dengue virus (DENV) (i.e., DENV1-4) (Curtis et al., 2008, Curtis et al., 2009). Symptoms can range from mild fever, headaches, myalgia, and rashes (Dineva et al., 2007, Ding et al., 2010) to severe disease characterized by plasma leakage and hemorrhage. Severe dengue can be fatal, especially without intensive medical care. The virus is transmitted by Aedes aegypti and Aedes albopictus mosquitoes and is endemic in most tropical and subtropical areas where these vectors are found. An estimated 390 million DENV infections are believed to occur each year, resulting in 96 million symptomatic cases (Bhatt et al., 2013). Neither vaccines nor antiviral treatments are available to reduce DENV-associated morbidity or mortality. The only available treatment option is supportive bed rest, fluids, and symptomatic relief with analgesics.
Diagnosing dengue is important for guiding appropriate supportive care and for alerting the physician to disease-specific warning signs that may require hospitalization; however, it is not possible to make an accurate differential diagnosis of dengue based on clinical features alone, as many symptoms of dengue resemble those of other diseases, such as malaria, chikungunya, measles, influenza, and rickettsial infections (Francois et al., 2011, Fredricks and Relman, 1998). Traditional laboratory techniques for dengue diagnosis include viral isolation followed by indirect immunofluorescence assay or serological assays such as plaque reduction neutralization test, hemagglutination inhibition, and IgM antibody capture enzyme-linked immunosorbent assay (Francois et al., 2011, Fredricks and Relman, 1998, George and Lum, 1997). Some of these techniques can take days or weeks to complete. Furthermore, serological methods require convalescent samples demonstrating an increase in antibody titer from pre-exposure samples in order to make a definitive diagnosis, making it impossible to confirm a diagnosis in the acute stage, limiting the value of serological assays for informing patient management.
Reverse transcription–polymerase chain reaction (RT-PCR) and quantitative RT-PCR (qRT-PCR) can be used to identify DENV RNA, which enables diagnosis during the acute phase of dengue infection. This can be an advantage because it facilitates earlier detection of dengue (within 1–7 days post onset of symptoms), before an immune response has been mounted and serology-based diagnostics can detect IgM or IgG levels (typically detectable 5–14 days post onset of symptoms). However, PCR equipment is expensive, requires trained personnel, and is best suited to diagnostic reference laboratories. Other nucleic acid detection strategies based on isothermal amplification such as nucleic acid sequence–based amplification (NASBA) and reverse transcription loop-mediated isothermal amplification (RT-LAMP) have been developed to address these challenges and move molecular assays closer to point-of-care use (Gubler, 1998, Hayasaka et al., 2013).
Here, we have explored a DENV diagnostic solution using the RT-LAMP technology. The technology was originally developed by Eiken Chemical Company (Notomi et al., 2000) and is based on the principle of strand displacement. Briefly, Bst polymerase has both polymerase and strand displacement activity and can be used to amplify nucleic acids using a series of primers that initiate a specific stem loop structure following binding to a target sequence (Henchal et al., 1983, Hong et al., 2004). This structure allows the enzyme to polymerize nucleic acids continuously at permissive temperatures beyond 57–65 °C. A variety of methods can be used to visualize the amplified DNA product or the magnesium pyrophosphate by-product. The DNA amplicon can be labeled with an intercalating agent such as SYBR Green and visualized using the naked eye or under UV light. Alternatively, the magnesium pyrophosphate causes turbidity, which can be visualized by the naked eye or using a turbidimeter (Mori et al., 2001). Another benefit of this assay is that it does not require expensive instrumentation and takes less than an hour to perform, making LAMP and RT-LAMP suitable for diagnostic applications in low-resource settings. The technology has been adapted to detect a variety of microorganisms ranging from bacteria such as Mycobacterium (Iwamoto et al., 2003); parasites such as Plasmodium (Sattabongkot et al., 2014); and viruses such as human immunodeficiency virus (Kaneko et al., 2007, Kao et al., 2005), severe acute respiratory syndrome-coronavirus (Hong et al., 2004), and hepatitis B virus (Kwallah et al., 2013, Hong et al., 2004). RT-LAMP has also been evaluated for diagnosing other members of the Flaviviridae family, including yellow fever virus (Kwallah et al., 2013), Japanese encephalitis virus (Li et al., 2011, McAvin et al., 2005), West Nile virus (Parida et al., 2004), and tick-borne encephalitis virus (Hayasaka et al., 2013).
In this study, we developed an RT-LAMP assay to detect all 4 serotypes of DENV in a single reaction. Other groups have explored this concept, with several assays detecting the 4 serotypes of DENV in separate reactions (Hayasaka et al., 2013, Mori et al., 2004). Some groups have developed pan-serotype DENV RT-LAMP assays (Teoh et al., 2013), and others have expanded this concept to a pan-flaviviral assay that detects DENV1-4, Japanese encephalitis virus, and West Nile virus in a single reaction (Li et al., 2011). We report a pan-serotype DENV RT-LAMP assay that has been optimized and evaluated with an extensive panel of clinical samples obtained from febrile patients in South America. We have additionally explored a method of reducing assay variability in a DENV quantitative RT-LAMP (qRT-LAMP) that uses a quencher to reduce false positives. Finally, we have also examined methods for streamlining sample preparation and evaluated visualization of the qRT-LAMP product using lateral flow immunochromatographic tests (ICTs) and a handheld fluorescence reader. Together, this represents a complete assay, from sample to result, which can be used to identify DENV with high accuracy in low-resource settings.
2. Methods
2.1. Human use statement
The procedures applied in this study were done in accordance with the ethical standards of the Naval Medical Research Center (NMRC) Institutional Review Board and with the Helsinki Declaration of 1975, as revised in 1983. Study protocols were approved by the NMRC Institutional Review Board (NMRCD.2000.0006 and NMRC.2005.0007) in compliance with all applicable federal regulations governing the protection of human subjects. Clinical serum samples of DENV viremic patients or those determined to have other febrile illness were collected during ongoing febrile surveillance studies (years 1996-2005) at the Naval Medical Research Unit-6 Peru at regional sites in Piura, Tumbes, Madre de Dios, and Iquitos. The samples were deidentified and shipped to the NMRC. Additional serum samples from DENV-negative individuals were obtained from a dengue vaccine trial at the NMRC and a blood bank at the Walter Reed Army Medical Center, both in the United States. All of these specimens were obtained with institutional review board approval.
2.2. Reference methods
Tissue culture–derived laboratory virus stocks propagated in Vero African green monkey kidney cells at the NMRC were used as positive controls. The following DENV strains were used to cover all 4 serotypes: DENV1, WP74; DENV2, 16803; DENV3, CH53489; and DENV4, 341750. RNA extraction for these DENV stocks as well as clinical serum samples was performed using QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol. RNA was eluted in 60 μL of nuclease-free water. Samples were characterized as positive or negative for DENV using a qRT-PCR assay, modified from one previously described (McAvin et al., 2005). Briefly, the sequences of the qRT-PCR primers used were as follows: DENV forward: 5′-GGTTAGAGGAGACCCCTC-3′, DENV reverse: 5′-CAGAGATCCTGCTGTCTC-3′, probe: 5′FAM-CAGCATATTGACGCTGGGA-TAMRA3′. We utilized the TaqMan® EZ RT-PCR core reagents kit (Life Technologies, Carlsbad, CA, USA) with a final volume of 25 μL, with the following reaction concentrations: 0.7 μmol/L forward primer, 0.7 μmol/L reverse primer, 0.27 μmol/L probe, 0.2 mmol/L dNTP, 4 mmol/L Mn(OAc)2, 1X ABI Buffer, 2.5 U rTth Polymerase, and 0.25 U AmpErase. The reaction was held at 50 °C for 2 minutes and 60 °C for 30 minutes, followed by 40 cycles of denaturation at 94 °C for 20 seconds and polymerization at 61 °C for 45 seconds. To quantify RNA copies, the DENV 3′UTR was cloned into the PCR_Script AMP vector (Stratagene, La Jolla, CA, USA), and the transcript was transcribed in vitro using the Riboprobe T7 Transcription kit (Promega, Madison, WI, USA). RNA was quantified using a spectrophotometer, and the RNA copy number was calculated. This RNA standard with known copy numbers was serially diluted and run alongside all qRT-PCR reactions to enable quantification of RNA copies in the reaction.
2.3. RT-LAMP design and reaction conditions
RT-LAMP primers were designed against the conserved regions of DENV using Eiken's PrimerExplorer version 4 (http://primerexplorer.jp/e/) utilizing DENV2 genome (GenBank accession number FM210242.2) as the initial template. These primers were then aligned with DENV1, 3, and 4, and degenerate primers were manually created to improve sequence consensus. These included 2 outer primers (F3 and B3), 2 inner primers (forward inner primer [FIP] and backward inner primer [BIP]), and 1 loop primer (LoopB). These primers were then aligned with up to 36 published strains each of DENV1, DENV3, and DENV4. Six additional primers were added to broaden detection of these DENV strains: FIPdegen1/3, FIPdegen4, BIPdegen1/3, BIPdegen4, B3degen1/3, and B3degen4.
The RT-LAMP reactions were carried out in a 25-μL volume with the following concentrations: 20 pmol/L FIP, FIPdegen4, BIP, and BIPdegen4; 30 pmol/L FIPdegen1/3, BIPdegen1/3, and LoopB; 2.5 pmol/L F3 and B3 (Integrated DNA Technologies, Coralville, IA, USA); 1X ThermoPol Buffer (New England BioLabs, Ipswich, MA, USA); 0.4 mol/L Betaine (Sigma-Aldrich, St. Louis, MO, USA); 1.4 mmol/L each dNTP (Life Technologies, Grand Island, NY, USA); 8 mmol/L total MgSO4 (Sigma-Aldrich); 16 U Bst DNA Polymerase (New England BioLabs); 2.4 U AMV RT (Life Technologies, Grand Island, NY); and 3 μL of template. Reactions were held at 63 °C for 60 minutes unless otherwise indicated. A positive control template corresponding to 106 copies/reaction DENV RNA was used in RT-LAMP reactions, unless otherwise stated.
2.4. UV fluorescence-based readout
Completed reactions were run on a 2% agarose gel containing 1X SybrSafe (Life Technologies). The RT-LAMP product banding pattern was also compared against those from true positives. To visualize RT-LAMP product in the tubes, 400X SybrSafe was added. The agarose gels and tubes were visualized using an E-Gel Imager system (Life Technologies). Fluorescence in reaction tubes was also measured using the PicoflourTM Model number 8000-004 (Turner Biosystems, Sunnyvale, CA, USA).
2.5. Lateral flow immunochromatographic tests
The method of visualizing RT-LAMP product on lateral flow ICTs was adapted from previous reports (Notomi et al., 2000, Parida et al., 2004). A home-made ICT strip, which had streptavidin on the detection zone and the mouse anti-FITC antibody conjugated with colloidal gold in the sample pad of the strip, was used for detection of the LAMP product. For visualizing the RT-LAMP product on this system, the reaction was performed as described above except with the addition of Biotin-11-dUTP (Life Technologies) and FITC-12-dUTP both to a final concentration of 10 μmol/L. Following a LAMP reaction, the mixture was heated to 100 °C for 10 minutes and then cooled on ice for 5 minutes. Approximately 0.5 μL of the biotinylated RT-LAMP product was applied to the binding pad of the lateral flow ICT. Running buffer (200 μL of 0.1% Tween 20) was added to facilitate capillary flow, and the lateral flow ICTs were visually inspected after 30 minutes. The presence of a visible purple line was considered reactive.
2.6. Sequence-specific detection
Sequence-specific detection was performed as above, but incorporating a FAM-labeled–LoopB primer instead of the unlabeled primer. To reduce visualization of nonspecific amplification, 60 pmol/L of Black Hole Quencher (BHQ) 1–labeled Antisense LoopB primer was added to the RT-LAMP reaction tube following isothermal amplification. Both the FAM- and BHQ-labeled primers were obtained from Integrated DNA Technologies. The tubes were quenched for approximately 2 hours at room temperature, followed by visualization using a UV source as described above.
3. Results
3.1. Assay characteristics
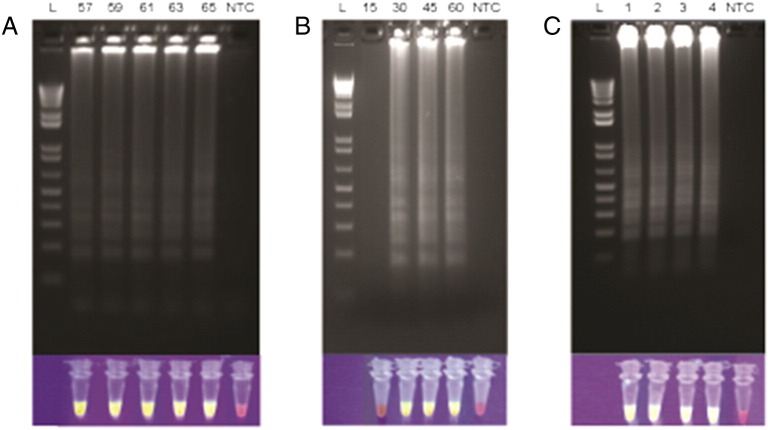
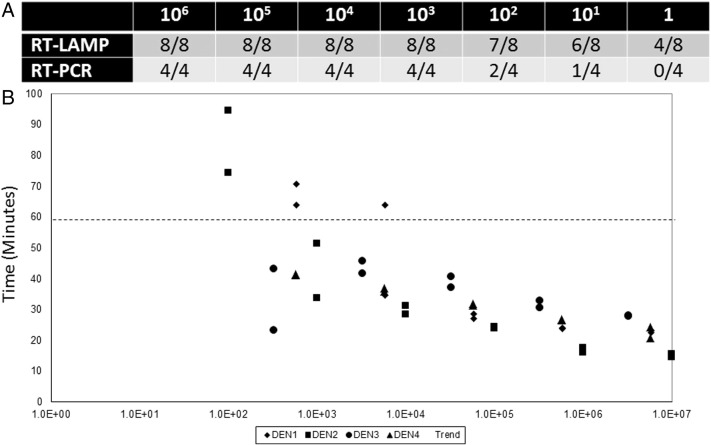
We designed four sets of LAMP primers to detect all serotypes and multiple strains of DENV in a single reaction. Among these, 1 set of primers was down-selected based on faster time to positivity in a pilot experiment (data not shown). Table 1 summarizes the primers used. The primers target a 3′ untranslated region of DENV that is highly conserved between all 4 serotypes and multiple strains. Unlike traditional LAMP assays, which require 4–6 primers, this pan-dengue RT-LAMP uses 11 primers, including 3 FIP, 3 BIP, and 3 B3 degenerate primers. Using a positive control template, the RT-LAMP reaction was tested within a range of temperatures (57–65 °C, Fig. 1A) using a simple water bath as the heat source. Nucleic acid amplification took place at all the temperatures tested, and subsequent experiments were performed at 63 °C. Next, various assay characteristics were systematically optimized, including primer, enzyme, and betaine concentrations, in order to achieve the fastest time to result (data not shown). Under optimized conditions, 106 copies/reaction of DENV RNA was detectable in approximately 30 minutes when using gel electrophoresis or UV as a readout (Fig. 1B). The assay was designed to be reactive to all 4 serotypes of DENV, and this was confirmed using tissue culture–derived positive strains (Fig. 1C). Using a serial dilution of DENV2, we determined a limit of detection of 103 copies/reaction where samples were detected reliably a majority (>50%) of the time, comparable to qRT-PCR (Fig. 2A). Fig. 2B illustrates how time to positivity is affected by the number of copies of DENV template present in the reaction. Based on these results, we selected a reaction time of 60 minutes to enable reliable detection of >103 copies/reaction for all 4 DENV serotypes while minimizing nonspecific amplification. We also saw positive RT-LAMP reactions using DENV-positive samples that had been heat treated for 10 minutes at 100 °C. Boiling samples can eliminate the need for formal RNA extraction steps as has been previously reported with DNA-based pathogen targets (Parida et al., 2005, Parida et al., 2006).
Table 1.
RT-LAMP assay primers used for amplification of DENV.
| Primer name | Type | Length | Sequence |
|---|---|---|---|
| FIP DENV1/3 | Forward inner | 49 | GGTTATTCATCAGAGATCTGCTCTCTTTTTTTCGAATCGGAAGCTTCGT |
| FIP DENV2 | Forward inner | 49 | GGTTATTCATCAGAGATCTGCTCTCTTTTATTCTTTGAGGGAGCTAAGC |
| FIP DENV4 | Forward inner | 49 | TTCATTTTTCCAGAGATCTGCTCTCTTTTTTTCGAATCGGAAGCTTCGT |
| BIP DENV1/3 | Reverse inner | 49 | AACGGAAAAAGACGGGTCAACCGTTTTTCTTTGTCAGCTGTTGCACAGT |
| BIP DENV2 | Reverse inner | 49 | AACGGAAAAAGGCGAGAAATACGCTTTTCTTTGTCAGCTGTTGCACAGT |
| BIP DENV4 | Reverse inner | 49 | AACGAAAAAAGGTGGTTAGACCACTTTTCTTCACCAACCCTTGAGGGGT |
| F3 | Forward outer | 18 | GTGGACCGACAAAGACAG |
| B3 DENV1/3 | Reverse outer | 18 | GTGAGCAATCCTTTTGAG |
| B3 DENV2 | Reverse outer | 18 | TGCAGCATTCCAAGTGAG |
| B3 DENV4 | Reverse outer | 18 | GAAAAAAGTCCGGTTGAG |
| LoopB | Reverse loop | 19 | GCGAGAGAAACCGCGTGTC |
| FAM-LoopB | Reverse loop | 20 | [Fam]a-CGCGAGAGAAACCGCGTGTC |
| Antisense LoopB | Reverse loop | 20 | GACACGCGGTTTCTCTCGCG-[Bhq] b |
Final sequences for the optimized primer combination are given. The forward inner primers and backward inner primers contain a TTTT spacer as suggested in previous literature.
Oligo labeled with FAM (5 carboxy fluorescein).
Oligo labeled with BHQ.
Fig. 1.
Characteristics of a pan-dengue RT-LAMP assay. Amplicons generated via RT-LAMP were visualized via the incorporation of SybrSafe using agarose gel electrophoresis or UV illumination. A) 106 copies/reaction DENV1 at 57-65 °C, B) 106 copies/reaction DENV1 held at 63 °C for 15–60 min, C) 105 copies/reaction DENV1, 2, 3, or 4 held at 63 °C for 60 min. NTC = no template control.
Fig. 2.
Comparison of limit of detection between pan-serotype RT-LAMP and RT-PCR. Ten-fold limiting dilutions of DENV2 beginning at 106 copies/reaction were used to compare the limit of detection of our optimized pan-dengue RT-LAMP to an in-house RT-PCR assay.
3.2. Clinical performance
A panel of 44 clinical specimens PCR confirmed as DENV positive was used to determine the clinical sensitivity of our optimized RT-LAMP reaction. The panel included DENV1, 2, and 3 naturally circulating in northern Peru at the time of sample collection. DENV-negative samples, including 17 human serum samples from individuals without fever symptoms as well as 40 samples from individuals presenting fever symptoms of unknown origin that tested negative for DENV by qRT-PCR, were used to determine clinical specificity (Fig. 3 ). Our optimized RT-LAMP assay demonstrated 86.3% sensitivity (95% confidence interval [CI], 72.7–94.8% using binomial confidence interval (Clopper and Pearson, 1934)) and 93.0% specificity (95% CI, 83.0–98.1%). The 6 false-negative samples consisted of 2 DENV1 and 4 DENV3 specimens. Overall, the assay demonstrated a 90.1% agreement with DENV qRT-PCR, underscoring the potential clinical utility of this assay.
Fig. 3.
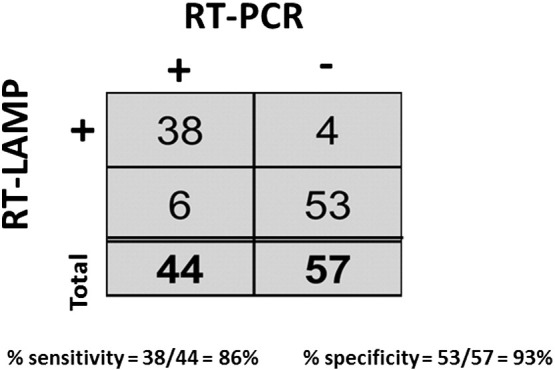
Clinical sensitivity and specificity of RT-LAMP. Clinical specimens from subjects suspected of having dengue were tested in triplicate via RT-LAMP and RT-PCR. If 2 out of 3 reactions were positive, the sample was considered positive.
3.3. Sequence-specific detection
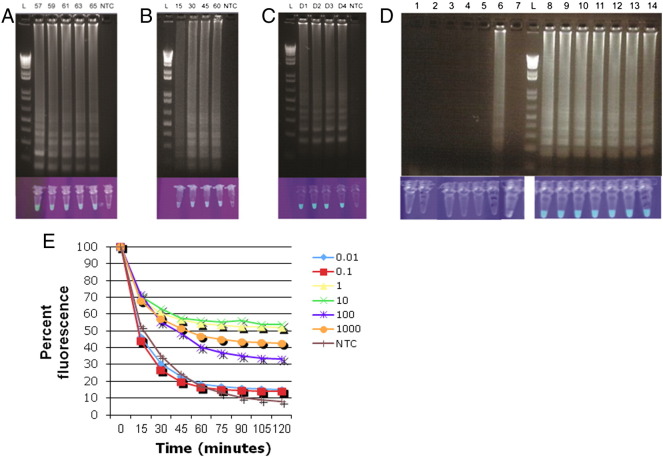
As has been reported by others, we occasionally observed false-positive results when using our DENV RT-LAMP assay (Curtis et al., 2009). Precautionary measures were implemented to reduce risk of contamination from template, and these substantially reduced initial false-positive results. However, another kind of nonspecific amplification was also infrequently observed, which was distinguishable from contamination-derived false-positive results based on a different banding pattern on agarose gels (Fig. 5 ). It has been hypothesized that this is due to non–template-driven self-priming. Other groups were able to limit this phenomenon by attaching a fluorescent label to their loop primer, followed by the addition of a probe with a fluorescence quencher (Curtis et al., 2009). As the increased number of primers in our pan-serotype assay may facilitate nonspecific amplification, we examined whether sequence-specific detection was a feasible solution. At the end of the RT-LAMP reaction, any unbound FAM-loop probe was quenched by BHQ-labeled probe (which consists of the reverse complement sequence of the FAM-loop probe with a BHQ modification.) We optimized this formulation and found no difference in the permissible temperature range, time to positivity, or number of serotypes detected when including a fluorescently labeled loop primer (Fig. 5A–C). In addition, this formulation was able to reduce the nonspecific amplification (Fig. 5D).
Fig. 5.
Sequence-specific detection. Amplicons generated via sequence-specific RT-LAMP were visualized via agarose gel electrophoresis or UV illumination 90 minutes after the addition of a reverse complement loop quencher. A) 106 copies/reaction DENV1 at 57–65 °C, B) 106 copies/reaction DENV1 held at 63 °C for 15–60 minutes, C) 106 copies/reaction DENV1, 2, 3, or 4 held at 63 °C for 60 minutes, D) 7 replicates each of NTC (lanes 1–7) or DENV3 (lanes 8–14) after 60 minutes at 63 °C. Reactions were quenched for 2 hours prior to UV illumination. E. Quenching reaction measured using Picflour shows detection of RT-LAMP product following quenching. NTC = no template control.
3.4. LAMP visualization using lateral flow ICTs and fluorescent readers
In our hands, RT-LAMP readout was not prone to operator error, and the positives and negatives were always clearly distinguishable. However, various methods to visualize RT-LAMP results including real-time PCR thermocyclers, agarose gel electrophoresis, turbidity measurements, and UV illumination are not practical in resource-limited settings. We thus explored the feasibility of determining RT-LAMP positivity via an inexpensive lateral flow ICT. We found 100% correlation between our agarose gel and ICT results when using 24 DENV1-, DENV2-, or DENV3-positive specimens. One specimen that was false negative when visualized via agarose gel electrophoresis was also negative via ICT (Fig. 4), suggesting that ICT has sensitivity comparable to other methods. Our data demonstrate that RT-LAMP results can be reliably determined without the aid of expensive or bulky equipment. We further evaluated the ability of our RT-LAMP assay to be visualized using a battery-operated handheld fluorescent reader and observed the kinetics of quenching over time. True-positive samples remain above a fluorescent threshold of 30 units following 1 hour of quenching.
Fig. 4.
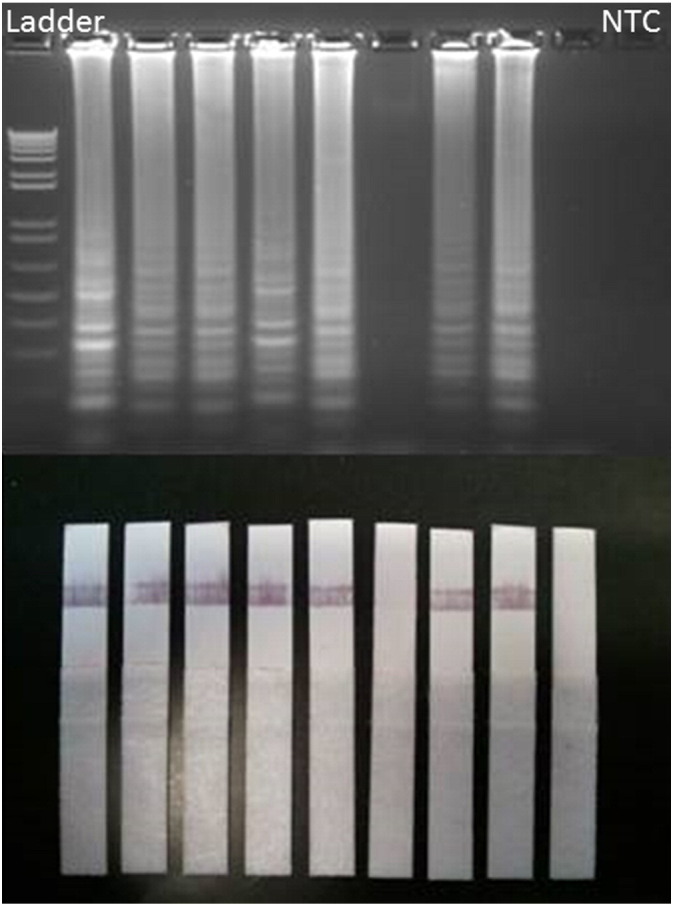
Evaluation of pan-dengue RT-LAMP ICT. Eight DENV-positive clinical specimens each for DENV1, 2, and 3 were amplified via RT-LAMP and the product run on an agarose gel or applied to an immunochromatographic test. DENV2 results are shown. NTC = no template control.
4. Discussion
Diagnosing dengue accurately during acute infection is critical for informing patient management decisions. Lateral flow ICTs targeting the DENV nonstructural protein 1 (NS1) antigen have had some success in enabling early diagnosis at the point of need; however, nucleic acid tests (NATs) may detect DENV RNA earlier than NS1 antigen, and NATs can be more sensitive and specific. The primary disadvantage with PCR-based NATs involves the need for thermocyclers, which are often bulky and too expensive to be used in resource-limited settings. Commercially available PCR kits cost $50–100/test and are generally unsuitable for high-throughput usage in developing economies (Rico-Hesse, 2003, Sattabongkot et al., 2014). In addition, thermocyclers can cost $20,000–$120,000, which may be prohibitive in many dengue-endemic countries (Dineva et al., 2007). Training personnel to operate the instrument and interpret results also requires resources typically only found in a reference laboratory. PCR is also subject to inhibition by substances such as heme compounds, heparin, and EDTA often present in a clinical samples and blood collection tubes, leading to false-negative results (Shu and Huang, 2004, Tanner et al., 2012); removing PCR inhibitors can lead to increased overhead costs and turnaround time to results. Conversely, PCR is also often subject to reporting false positives from the presence of contaminating nucleic acids (Fredricks and Relman, 1998), necessitating the use of template-free “clean rooms” for preparing a master mix, which is impractical in the field.
To develop a diagnostic solution that overcomes these limitations, we developed a pan-DENV RT-LAMP assay. We demonstrate that the reaction can proceed continuously within a wide temperature range 57–65 °C, making it suitable for a water bath or electricity-free heater (LaBarre et al., 2011). In addition, we have explored various methods of visualizing the assay results. Electrophoresis gel confirmation of reaction products may be performed in a reference laboratory, but in the field, the reaction can be visualized in real time by measuring an increase in turbidity from magnesium pyrophosphate by-products (Mori et al., 2001). We propose a method involving the measurement of color change by the incorporation of SYBR Green adapted from a previous report, which was easier to read with the naked eye (Parida et al., 2005). We also explored visualization using lateral flow ICTs and demonstrated 100% concordance between RT-LAMP visualized on a gel versus those visualized using ICTs. This format is likely to be easier to read in resource-limited settings where a UV light source is not available. Finally, we explored the feasibility of using a handheld UV fluorescence reader, which when coupled with a quencher, can provide higher specificity detection of DENV. The handheld UV reader that we used was approximately $500, and such readers could improve accuracy of readout as well as provide some level of quantitation to RT-LAMP results. LAMP has several other advantages over traditional PCR, including less sensitivity to inhibitors such as ethanol, isopropanol, EDTA, and sodium acetate (Teoh et al., 2013, Toriniwa and Komiya, 2006). The use of loop-specific primers can reduce assay times to provide faster sample to result than PCR, and other groups have reported higher sensitivity (Nagamine et al., 2002). LAMP also does not require the template to be denatured (Nagamine et al., 2001), and contrasted with NASBA (a similar isothermal NAT), LAMP does not require a separate nucleic acid extraction step (Francois et al., 2011). This robustness further reduces the cost as well as time to result.
Like qPCR, qLAMP is amenable to multiplexing (Tanner et al., 2012), and we adapted it to detect all 4 serotypes of DENV in 1 reaction. Our data demonstrate that the use of degenerate primers is a feasible method by which to design assays for targets of interest even when sequence conservation is limited. The assay was able to detect a majority of DENV PCR–positive clinical samples obtained from South America (sensitivity of 86.3%), representing multiple circulating strains of DENV1–3; however, the clinical performance is limited with only 44 dengue-positive clinical specimens from 1 region and lack of inclusion of DENV4-positive specimens. The assay was also specific to DENV and did not react to other pathogens that cause similar fever-like symptoms. This demonstrates the assay's effectiveness for enabling differential diagnosis in a clinical situation with multiple strains and serotypes. Since this is a molecular assay, it will likely be able to detect DENV in mosquito samples and have further applications for vector surveillance. Due to the robustness of LAMP, it may be used without explicit sample preparation in applications where having false negatives for low copy number samples are acceptable and with integrated sample preparation where it is not. Acute DENV viremic patients have 103–1012 plaque-forming units/mL (Vaughn et al., 2000), and such high titers are detectable by LAMP without explicit sample preparation. The assay reagents can also be lyophilized for field expediency and provided in a single tube to which one may add hydration buffer and test sample. The assay can be run at a range of temperatures and can be performed using a water bath or other available heat source. We also envision the assay readout adapted to specific applications and present UV visualization, handheld fluorescence readers, and lateral flow ICTs as viable alternatives.
In conclusion, this work adds to the growing body of isothermal amplification assays available to detect DENV. More than previous reports, we have explored all aspects of DENV detection from sample to result, with an emphasis on visualizing the RT-LAMP product, as readout is critical for ensuring accuracy. The assay can be adapted to use in dengue-endemic countries where it is most needed to reduce morbidity and mortality associated with this widespread disease.
Disclaimer
The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, or the US Government.
Copyright statement
Dr Allison Dauner, Ms Indrani Mitra, Mr Theron Gilliland Jr, Ms Sajeewane Seales, and Dr Subhamoy Pal are (or were) employed by the Henry M. Jackson Foundation for the Advancement of Military Medicine and were funded to do this work by the US Government. Dr Shuenn-Jue Wu and Dr Tadeusz Kochel are employees of the US Government or members of the US Military. This work was prepared as part of their official duties. Title 17 U.S.C. § 105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. § 101 defines a US Government work as a work prepared by military service members or employees of the US Government as part of those persons' official duties.
Funding statement
This work was funded by the Military Infectious Diseases Research Program, L0205_11_NM. The work was supported by Work Unit Number 6000.RAD1.L.A0311.
Acknowledgments
This work would not have been possible without the generous support of the Ministry of Health Peru, Direccion General de Epidemiologia and DIRESA Piura, Tumbes, Madre de Dios, and Iquitos.
References
- Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clopper C.J., Pearson E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- Curtis K.A., Rudolph D.L., Owen S.M. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP) J Virol Methods. 2008;151(2):264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Curtis K.A., Rudolph D.L., Owen S.M. Sequence-specific detection method for reverse transcription, loop-mediated isothermal amplification of HIV-1. J Med Virol. 2009;81(6):966–972. doi: 10.1002/jmv.21490. [DOI] [PubMed] [Google Scholar]
- Dineva M.A., MahiLum-Tapay L., Lee H. Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. Analyst. 2007;132(12):1193–1199. doi: 10.1039/b705672a. [DOI] [PubMed] [Google Scholar]
- Ding W.C., Chen J., Shi Y.H., Lu X.J., Li M.Y. Rapid and sensitive detection of infectious spleen and kidney necrosis virus by loop-mediated isothermal amplification combined with a lateral flow dipstick. Arch Virol. 2010;155(3):385–389. doi: 10.1007/s00705-010-0593-4. [DOI] [PubMed] [Google Scholar]
- Fiscus S.A., Cheng B., Crowe S.M., Demeter L., Jennings C., Miller V. HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3(10):e417. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois P., Tangomo M., Hibbs J., Bonetti E.J., Boehme C.C., Notomi T. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol. 2011;62(1):41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- Fredricks D.N., Relman D.A. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 1998;36(10):2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R., Lum L. CAB International; Wallingford: 1997. Clinical spectrum of dengue infection. [Google Scholar]
- Gubler D.J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11(3):480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka D., Aoki K., Morita K. Development of simple and rapid assay to detect viral RNA of tick-borne encephalitis virus by reverse transcription-loop-mediated isothermal amplification. Virol J. 2013;10:68. doi: 10.1186/1743-422X-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchal E.A., M. J.M., S. M.C., G. M.K., B. W. E. Rapid identification of dengue virus isolates by using monoclonal antibodies in an indirect immunofluorescence assay. Am J Trop Med Hyg. 1983;32:164–169. doi: 10.4269/ajtmh.1983.32.164. [DOI] [PubMed] [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42(5):1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins H., Gonzalez I.J., Polley S.D., Angutoko P., Ategeka J., Asiimwe C. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J Infect Dis. 2013;208(4):645–652. doi: 10.1093/infdis/jit184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T., Sonobe T., Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41(6):2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jittmittraphap A., Thammapalo S., Ratanasetyuth N., Wongba N., Mammen M.P., Jampangern W. Rapid detection of dengue viral RNA in mosquitoes by nucleic acid-sequence based amplification (NASBA) Southeast Asian J Trop Med Public Health. 2006;37(6):1117–1124. [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods. 2007;70(3):499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kao C.L., King C.C., Chao D.Y., Wu H.L., Chang G.J. Laboratory diagnosis of dengue virus infection: current and future perspectives in clinical diagnosis and public health. J Microbiol Immunol Infect. 2005;38(1):5–16. [PubMed] [Google Scholar]
- Klein A., Barsuk R., Dagan S., Nusbaum O., Shouval D., Galun E. Comparison of methods for extraction of nucleic acid from hemolytic serum for PCR amplification of hepatitis B virus DNA sequences. J Clin Microbiol. 1997;35(7):1897–1899. doi: 10.1128/jcm.35.7.1897-1899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwallah A., Inoue S., Muigai A.W., Kubo T., Sang R., Morita K. A real-time reverse transcription loop-mediated isothermal amplification assay for the rapid detection of yellow fever virus. J Virol Methods. 2013;193(1):23–27. doi: 10.1016/j.jviromet.2013.05.004. [DOI] [PubMed] [Google Scholar]
- LaBarre P., Hawkins K.R., Gerlach J., Wilmoth J., Beddoe A., Singleton J. A simple, inexpensive device for nucleic acid amplification without electricity-toward instrument-free molecular diagnostics in low-resource settings. PLoS One. 2011;6(5) doi: 10.1371/journal.pone.0019738. e19738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Fang M., Zhou B., Ni H., Shen Q., Zhang H. Simultaneous detection and differentiation of dengue virus serotypes 1-4, Japanese encephalitis virus, and West Nile virus by a combined reverse-transcription loop-mediated isothermal amplification assay. Virol J. 2011;8:360–369. doi: 10.1186/1743-422X-8-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAvin J.C., Escamilla E.M., Blow J.A., Turell M.J., Quintana M., Bowles D.E. Rapid identification of dengue virus by reverse transcription-polymerase chain reaction using field-deployable instrumentation. Mil Med. 2005;170(12):1053–1059. doi: 10.7205/milmed.170.12.1053. [DOI] [PubMed] [Google Scholar]
- Mikita K., Maeda T., Yoshikawa S., Ono T., Miyahira Y., Kawana A. The Direct Boil-LAMP method: a simple and rapid diagnostic method for cutaneous leishmaniasis. Parasitol Int. 2014;63(6):785–789. doi: 10.1016/j.parint.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289(1):150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Mori Y., Kitao M., Tomita N., Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004;59(2):145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Nagamine K., Watanabe K., Ohtsuka K., Hase T., Notomi T. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin Chem. 2001;47(9):1742–1743. [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16(3):223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Neeraja M., Lakshmi V., Lavanya V., Priyanka E.N., Parida M.M., Dash P.K. Rapid detection and differentiation of dengue virus serotypes by NS1 specific reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay in patients presenting to a tertiary care hospital in Hyderabad, India. J Virol Methods. 2015;211:22–31. doi: 10.1016/j.jviromet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42(1):257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Horioke K., Ishida H., Dash P.K., Saxena P., Jana A.M. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43(6):2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.M., Santhosh S.R., Dash P.K., Tripathi N.K., Saxena P., Ambuj S. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid and real-time detection of Japanese encephalitis virus. J Clin Microbiol. 2006;44(11):4172–4178. doi: 10.1128/JCM.01487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthawibool T., Senapin S., Flegel T.W., Kiatpathomchai W. Rapid and sensitive detection of Macrobrachium rosenbergii nodavirus in giant freshwater prawns by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. Mol Cell Probes. 2010;24(5):244–249. doi: 10.1016/j.mcp.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–341. doi: 10.1016/s0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattabongkot J., Tsuboi T., Han E.T., Bantuchai S., Buates S. Loop-mediated isothermal amplification assay for rapid diagnosis of malaria infections in an area of endemicity in Thailand. J Clin Microbiol. 2014;52(5):1471–1477. doi: 10.1128/JCM.03313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu P.Y., Huang J.H. Current advances in dengue diagnosis. Clin Diagn Lab Immunol. 2004;11(4):642–650. doi: 10.1128/CDLI.11.4.642-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N.A., Zhang Y., Evans T.C., Jr. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques. 2012;53(2):81–89. doi: 10.2144/0000113902. [DOI] [PubMed] [Google Scholar]
- Teles F.R., Prazeres D.M., Lima-Filho J.L. Trends in dengue diagnosis. Rev Med Virol. 2005;15(5):287–302. doi: 10.1002/rmv.461. [DOI] [PubMed] [Google Scholar]
- Teoh B.T., Sam S.S., Tan K.K., Johari J., Danlami M.B., Hooi P.S. Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification. BMC Infect Dis. 2013;13:387–396. doi: 10.1186/1471-2334-13-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriniwa H., Komiya T. Rapid detection and quantification of Japanese encephalitis virus by real-time reverse transcription loop-mediated isothermal amplification. Microbiol Immunol. 2006;50(5):379–387. doi: 10.1111/j.1348-0421.2006.tb03804.x. [DOI] [PubMed] [Google Scholar]
- Trent D.W., Manske C.L., Fox G.E., Chu M.C., Kliks S.C., Monath T.P. The molecular epidemiology of Dengue viruses, genetic variation and microevolution. In: Kurstk E., editor. Applied Virology Research, Virus Variation and epidemiology. Plenum Press; Plenum, NY: 1989. p. 2. [Google Scholar]
- Trent D.W., Manske C.L., Fox G.E., Chu M.C., Kliks S.C., Monath T.P. The molecular epidemiology of dengue viruses, genetic variation and microevolution. Appl Virol Res. 1990;2:293–315. [Google Scholar]
- Vaughn D.W., Green S., Kalayanarooj S., Innis B.L., Nimmannitya S., Suntayakorn S. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181(1):2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]