Abstract
Fibrillarin, one of the major proteins of the nucleolus, has methyltransferase activity directing 2′-O-ribose methylation of rRNA and snRNAs and is required for rRNA processing. The ability of the plant umbravirus, groundnut rosette virus, to move long distances through the phloem, the specialized plant vascular system, has been shown to strictly depend on the interaction of one of its proteins, the ORF3 protein (protein encoded by open reading frame 3), with fibrillarin. This interaction is essential for several stages in the groundnut rosette virus life cycle such as nucleolar import of the ORF3 protein via Cajal bodies, relocalization of some fibrillarin from the nucleolus to cytoplasm, and assembly of cytoplasmic umbraviral ribonucleoprotein particles that are themselves required for the long-distance spread of the virus and systemic infection. Here, using atomic force microscopy, we determine the architecture of these complexes as single-layered ringlike structures with a diameter of 18–22 nm and a height of 2.0 ± 0.4 nm, which consist of several (n = 6–8) distinct protein granules. We also estimate the molar ratio of fibrillarin to ORF3 protein in the complexes as approximately 1:1. Based on these data, we propose a model of the structural organization of fibrillarin–ORF3 protein complexes and discuss potential mechanistic and functional implications that may also apply to other viruses.
Abbreviations used: ORF3 protein, protein encoded by open reading frame 3; RNP, ribonucleoprotein; CB, Cajal body; GRV, groundnut rosette virus; CP, coat protein; AFM, atomic force microscopy; EM, electron microscopy; GAR domain, glycine- and arginine-rich domain
Keywords: atomic force microscopy, fibrillarin, umbravirus, virus movement, protein complexes
Fibrillarin, one of the major proteins of the nucleolus, is a core component of box C/D small nucleolar ribonucleoprotein (RNP) particles and is required for rRNA processing.1 Fibrillarin has methyltransferase activity directing 2′-O-ribose methylation of rRNA and snRNAs.2, 3, 4 It is also localized to another class of subnuclear body or domain, the Cajal body (CB). CBs are involved in the maturation of small nuclear RNPs and small nucleolar RNPs, which traffic through CBs before accumulating in splicing speckles and the nucleolus, respectively.5 Although interaction of some animal viruses with fibrillarin has been reported,6, 7, 8, 9 the specific role of these interactions in virus life cycles remains largely uncharacterized.
Recently, we have shown that the ability of the plant umbravirus, groundnut rosette virus (GRV), to move long distances through the phloem, the specialized vascular system used by plants for the transport of assimilates and macromolecules, strictly depends on the interaction of one of its proteins, the ORF3 protein (protein encoded by open reading frame 3), with fibrillarin.10 , 11 Umbraviruses are single-stranded RNA-containing viruses. Their genomes are relatively small, comprising slightly more than 4000 nucleotides with three to four open reading frames. Umbraviruses differ from most other viruses in that they do not encode a coat protein (CP) such that conventional virus particles are not formed in infected plants.12 Nevertheless, they accumulate and spread efficiently within the infected plant since their lack of a CP is compensated for by the ORF3 protein (26–29 kDa). The ORF3 protein fulfils umbraviral functions that are normally provided by the CPs of other plant viruses, such as long-distance movement of viral RNA through the phloem.13 , 14
Our recent studies have shown that, upon GRV infection, the ORF3 protein produced in the cytoplasm enters the nucleus and is targeted to CBs. The CBs are reorganized and move to and fuse with the nucleolus by an unknown mechanism.10 Finally, the ORF3 protein is exported from the nucleus leading to the formation of viral RNP particles in cytoplasmic inclusions. This transport pathway is absolutely required for the formation of these particles that are themselves essential for the long-distance spread of the virus and systemic infection. The purpose of this nuclear/nucleolar trafficking of the ORF3 protein is to recruit and relocalize some of the nuclear pool of fibrillarin to the cytoplasm (fibrillarin normally does not accumulate in cytoplasm).10 , 11 Our studies have also demonstrated a direct physical interaction between fibrillarin and ORF3 protein that is involved in at least two stages in the GRV life cycle: (1) CB fusion with the nucleolus and (2) the formation of viral RNP particles that are capable of long-distance movement, causing systemic viral infection. This model demonstrates completely novel functions for fibrillarin in triggering nucleolar import of the ORF3 protein via CBs and mediating assembly of umbraviral RNPs. Thus, the interaction of the GRV ORF3 protein with fibrillarin in CBs triggers all the molecular and cellular events necessary to establish a systemic infection, and hence, formation of the fibrillarin–ORF3 protein complexes appears to be the key prerequisite for both these processes. Here, we determine the architecture of these complexes using atomic force microscopy (AFM) as a high-resolution technique, which has been shown to be successful in studies of proteins, nucleic acids, and their complexes.15, 16, 17, 18 We also estimate the molar ratio of fibrillarin to ORF3 protein in the complexes as approximately 1:1. Based on these data, we propose a model of the structural organization of fibrillarin–ORF3 protein complexes and discuss the potential implications that may also apply to other virus life cycles.
In preliminary experiments, purified recombinant ORF3 protein and fibrillarin were visualized separately. The ORF3 protein tagged with six histidine residues (ORF3–His) was expressed from a tobacco mosaic virus vector in tobacco plants and isolated as described previously.14 Recombinant fibrillarin [Arabidopsis fibrillarin 2 (Fib2);2 ∼ 35 kDa] was expressed in and purified from Escherichia coli either as a fusion with glutathione S-transferase with a consequent removal of glutathione S-transferase from fibrillarin by thrombin treatment as described by Kim et al. 11 or as a fusion with six histidine residues (fibrillarin–His). Most of the fibrillarin molecules were observed in AFM as randomly distributed small granules with a height of 1.9 ± 0.3 nm (data here and below represent means ± SD for 50 different granules or complexes) and an apparent diameter measured at half of the height of about 10.0 ± 1.1 nm. It should be noted, however, that in AFM images, horizontal dimensions of objects are usually greatly overestimated due to the well-known effect of tip convolution. The lateral dimension of the fibrillarin molecule was estimated as 3.2 ± 0.4 nm, using the method of Stemmer and Engel.19 Given the uniform size of these molecules, they probably represent monomers of fibrillarin. The height of ORF3 protein molecules was not uniform and varied from 1.6 to 4.2 nm, representing at least three classes of particles of 1.6 ± 0.2, 3.1 ± 0.3, and 4.0 ± 0.3 nm. This confirms our previous biochemical data that the ORF3 protein preparations were not homogeneous and can form dimers and higher-order associations.14 This suggests that the smaller particles (with a height of 1.6 ± 0.2 nm) correspond to monomers, whereas the bigger ones (with a height of 3.1 ± 0.3 and 4.0 ± 0.3 nm) represent dimers and trimers of the ORF3 protein. About 60% of the preparation was represented by the putative monomers.
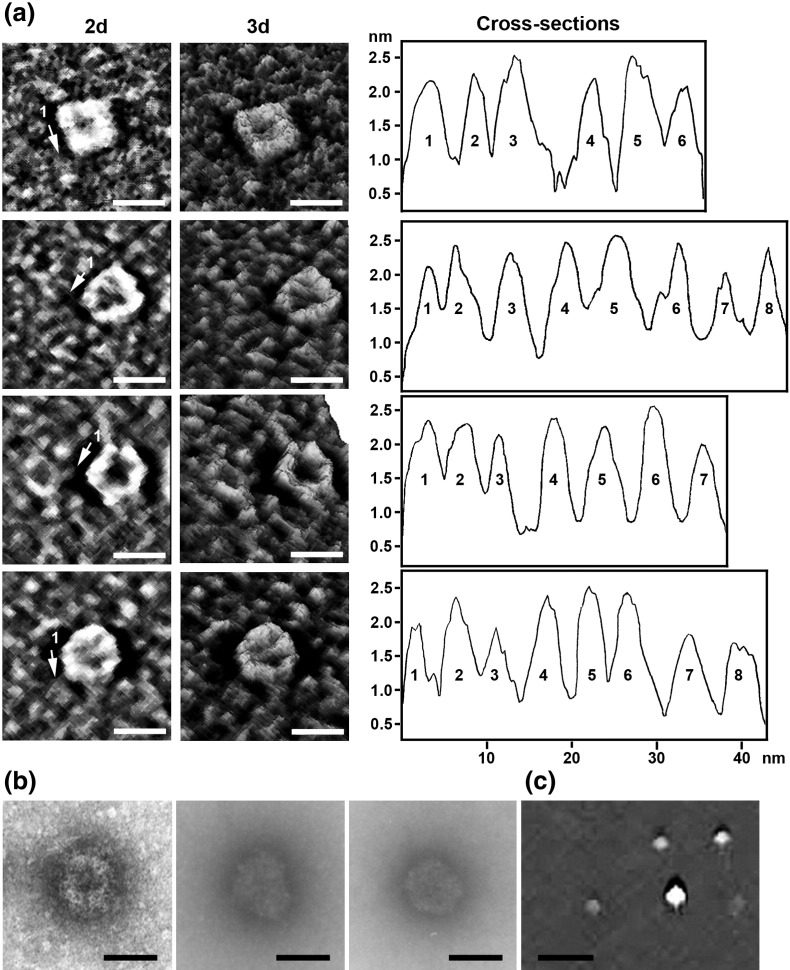
Mixing of the ORF3 protein and fibrillarin at equal (1:1) or unequal (1:2 and 2:1) molar ratios gave rise to ringlike complexes with a diameter of 18–22 nm consisting of several (n = 6–8) distinct protein granules arranged in ringlike structures with inner holes (Fig. 1a). The height of the granules (2.0 ± 0.4 nm) approximately corresponded to the heights of fibrillarin and ORF3 monomers, suggesting that the rings formed by these granules represent single-layered chains of the molecules. Given that neither the ORF3 protein nor fibrillarin alone can form such rings, it is likely that the rings consist of both the ORF3 protein and fibrillarin. While the height of the granules forming the rings was comparable with the heights of ORF3 protein and fibrillarin monomers, their width (5.5 ± 0.6 nm) could possibly correspond to protein dimers formed either by two ORF3 protein molecules or by an ORF3 and fibrillarin protein molecule (fibrillarin does not form dimers or oligomers as shown above). Electron microscopy (EM) detected similar rings formed by mixing the ORF3 protein and fibrillarin in vitro (Fig. 1b).11 Rings showing apparent granular structures in EM also exhibited six, seven, or eight granules per ring in approximately equal ratio (of 50 ringlike complexes, 16 complexes contained six granules, 18 complexes contained seven granules, and 16 complexes contained eight granules).
Fig. 1.
AFM (a and c) and EM (b) images of ORF3 protein–fibrillarin complexes formed in vitro. (a and b) Mixtures of fibrillarin with wild-type ORF3 protein. (c) Mixture of fibrillarin with the ORF3 L149A mutant, which does not interact with fibrillarin [similar images were obtained when wild-type ORF3 protein was mixed with fibrillarin mutant lacking the GAR domain (Fib2ΔGAR),11 which does not interact with the ORF3 protein]. The recombinant ORF3 protein (ORF3–His; 10 ng) and fibrillarin were mixed in 15 μl of buffer A (10 mM Hepes–KOH, pH 7.6, 100 mM KCl) in a 1:1 molar ratio (15 ng/μl) and incubated at room temperature for 30 min. (a and c) For AFM, mixtures of fibrillarin with ORF3 protein were diluted to ∼ 5 ng/μl in deionized water and 5–10 μl was placed onto freshly cleaved mica strips for 5–15 min. The strips were rinsed with water and dried at room temperature. Imaging of complexes was done in tapping mode by using a Nanowizard® BioAFM (JPK, Berlin, Germany). Silicon beam cantilevers (Veeco Instruments Ltd., Cambridge, UK) with a nominal spring constant of 40 N/m and a resonant frequency of 300 kHz were used. All the AFM experiments were performed in air at room temperature, and the images were captured in constant height mode by using a scan speed of 0.5 Hz. Images, including two-dimensional (2d) and three-dimensional (3d) representations, were processed using JPK software and transferred to Adobe Photoshop for layout. Sample heights and lengths were measured automatically using the JPK software. Cross sections were made around the ring structures along lines connecting centers of granules in the anticlockwise direction starting from granule 1 (as indicated by arrows) to illustrate the heights of the complexes. Periodical height variations represent complexes containing six to eight granules arranged into ringlike structures. (b) For EM, complexes of the ORF3 protein and fibrillarin were negatively stained with 2% sodium phosphotungstate (pH 7.0). The specimens were examined and photographed in a Phillips CM 10 transmission electron microscope. Scale bars represent 20 nm.
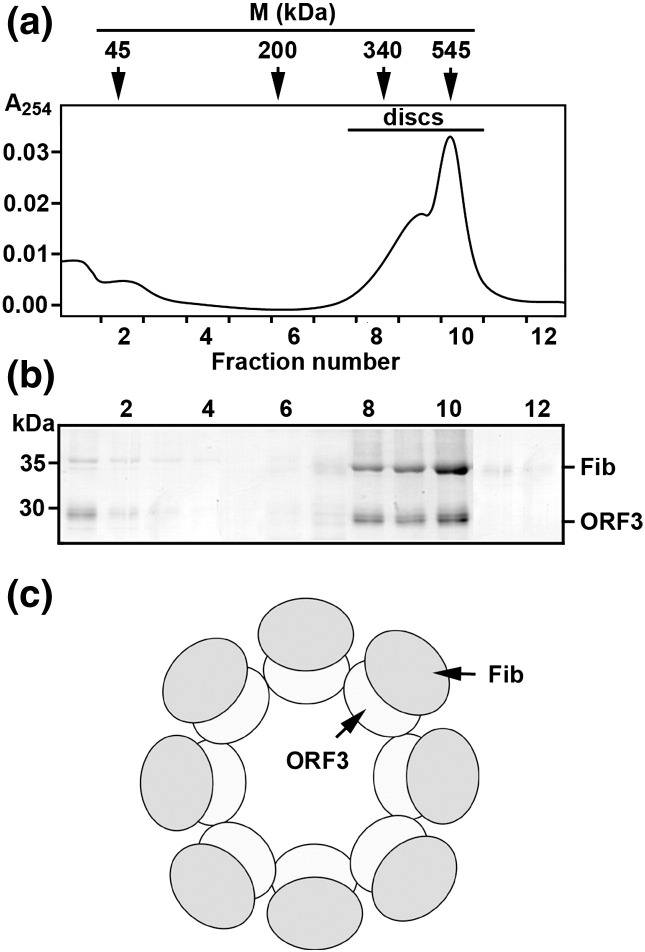
The complexes formed at 1:1, 1:2, and 2:1 molar ratios were analyzed using centrifugation in a sucrose concentration gradient to analyze the composition of the ORF3 protein–fibrillarin complexes in more detail. Figure 2a shows a relatively broad sedimentation distribution of the complexes formed at a molar ratio of 1:1. The complexes were present in 3 of 12 gradient fractions (8–10), which corresponded to a sedimentation zone flanked by molecular mass protein markers of 340 kDa (fraction 8) and 545 kDa (fraction 10). EM analysis confirmed the presence of the ringlike complexes only in these fractions of the gradient (data not shown). No other types of macromolecular aggregates were found in any of the other gradient fractions.
Fig. 2.
Analysis of the oligomeric state of the ORF3 protein–fibrillarin complexes. (a) Sedimentation distribution of the complexes. Samples of the ORF3–His and fibrillarin–His complexes were loaded onto the top of 10–30% sucrose gradients and centrifuged for 20 h in a Beckman SW41 rotor at 150,000g at 4 °C. Gradients were monitored for absorbance at 254 nm (A254) during collection of the fractions from the top (fraction 1). The positions in the gradient of molecular mass markers (M), 45 kDa (albumin from chicken egg white), 200 kDa (β-amylase), 340 kDa (fibrinogen from bovine plasma), 545 kDa (urease from Jack bean), are shown. The presence of ringlike complexes in fractions 8–10 was confirmed by EM. (b) Distribution of the ORF3 protein and fibrillarin over gradient fractions determined by SDS-PAGE. The gels were stained with Coomassie brilliant blue, and protein bands were analyzed by densitometry using ImageJ software20 showing the equimolar ORF3 protein–fibrillarin ratio in the complexes (gradient fractions 8–10). Positions of the ORF3–His (ORF3) and fibrillarin–His (Fib) are shown on the right, and those of molecular mass markers are on the left. (c) Schematic representation of ORF3 protein–fibrillarin complex containing eight granules. Each single granule consists of one molecule of the ORF3 protein (ORF3) and one molecule of fibrillarin (Fib). The rings are formed by ORF3 protein–ORF3 protein association. Shapes and dimensions of molecules do not reflect real proportions.
Fractions obtained after centrifugation in sucrose gradient (Fig. 2a) were subjected to SDS-PAGE to estimate the ORF3 protein–fibrillarin ratio in the complexes. Figure 2b shows that the vast majority of both the ORF3 protein and fibrillarin are present in fractions 8–10 presumably in the form of the complexes. Furthermore, densitometry of the ORF3 protein- and fibrillarin-specific bands on the gel (Fig. 2b) showed that the molar ORF3 protein–fibrillarin ratios in all these fractions are about 1:1, implicating the equimolar nature of the ORF3 protein–fibrillarin complexes. Some ORF3 and fibrillarin, apparently uncomplexed, were also detected in the top fractions of the gradient (fractions 1 and 2) (Fig. 2a and b). Analysis of the ORF3 protein–fibrillarin complexes formed at 1:2 or 2:1 molar ratios did not reveal any differences in sedimentation profiles (in fractions 8–10), shape, or dimensions of the complexes (data not shown).
Taking into account the molecular mass of the ringlike complexes in fractions 8–10 (340–545 kDa) and the number of granules in the complexes (n = 6–8), we expect the molecular mass of individual granules to lie in a range of approximately 57–68 kDa. This suggests that granules in the ORF3–fibrillarin complexes correspond to either heterologous dimers of ORF3 protein (26–29 kDa) and fibrillarin (35 kDa) molecules or potentially homologous dimers of ORF3 protein molecules. However, we have previously shown that fibrillarin and ORF3 interact with one another through single domains on each protein: the glycine- and arginine-rich domain (GAR domain) of fibrillarin and the leucine-rich domain of ORF3.11 In line with these observations, no ringlike complexes were formed with the ORF3 L149A mutant (Fig. 1c). This mutant has the leucine at position 149 replaced with alanine and is unable to interact with fibrillarin.11 A fibrillarin mutant lacking the GAR domain (Fib2ΔGAR)11 was also unable to form ringlike complexes. For both of these mutants, only smaller particles apparently corresponding to individual granules of the ORF3 protein (monomers and oligomers of higher order) and fibrillarin were observed (Fig. 1c). Therefore, the most likely interpretation, given the equimolar ratio of ORF3 and fibrillarin in the ringlike complexes, is that the ORF3–fibrillarin interaction leads to formation of individual heterodimers (“granules”) with a molecular mass of ∼ 63 kDa. The higher-order ring complexes are formed by additional ORF3–ORF3 association, which has been demonstrated previously14 (Fig. 2c).
ORF3 targets CBs and causes their fusion with the nucleolus prior to relocalization of fibrillarin to the cytoplasm and formation of RNP particles.10 That fibrillarin is concentrated in CBs and the nucleolus suggests that the initial interaction between the ORF3 protein and fibrillarin, leading to the formation of dimers and higher-order complexes, is likely to occur in CBs.11 The physical association of the nucleolus and CBs is well documented and is controlled by molecular interactions among CB and nucleolar proteins.3 Therefore, the fusion of CBs with the nucleolus, which is mediated by the ORF3–fibrillarin interaction,11 probably reflects a disruption or interference of normal protein–protein interactions and processes required for CB integrity. Thus, the ringlike complexes may occur in the CBs and move to the nucleolus by virtue of the fusion of the CBs with the nucleolus. However, it is also possible that complexes could move from CBs to the nucleolus independently of CB fusion, or that some of the ORF3 protein pool enters the nucleolus via CBs as uncomplexed molecules, and interact with fibrillarin in the nucleolus to form ORF3–fibrillarin complexes for export to cytoplasm.
The biological relevance of the ringlike complexes is demonstrated by two observations. Firstly, such structures were observed when ORF3 and fibrillarin were mixed in vitro and the addition of GRV RNA to the mixture led to the formation of infectious regular filamentous structures similar to viral RNP particles formed in vivo following GRV infection.11 Secondly, ringlike structures containing the ORF3 protein with sizes similar to those of the ORF3 protein–fibrillarin complexes described here have also been found in plants infected with GRV.14 This observation also indicates that potential posttranslational modifications of fibrillarin in plants (such as, e.g., the arginine dimethylation of mammalian and yeast fibrillarin orthologues21) are probably not essential for production of the ringlike complexes because, in our work, fibrillarin was expressed in and purified from E. coli and would therefore not be modified. Moreover, it has been reported that the lack of posttranslational modifications in bacterially expressed mammalian fibrillarin does not prevent it from interactions with other proteins.7 , 21 Importantly, the height and diameter of the rings formed by ORF3 protein and fibrillarin in vitro are about 2.0 ± 0.4 nm and 18–22 nm, respectively, and therefore, they could be exported from the nucleus through the nuclear pore complex. For example, intact nucleocapsids of human hepatitis B virus with diameters of 32 and 36 nm are able to cross the nuclear pore without disassembly.22 This suggests that the ORF3 protein and fibrillarin can move from the nucleus to the cytoplasm as ringlike complexes. Furthermore, in the cytoplasm, fibrillarin, along with the ORF3 protein and viral RNA, takes part in the assembly of infectious long-distance movement-competent RNP particles.11 , 14 These particles have a filamentous structure with regular helical elements but not the uniformity typical of virus particles. The diameter of these filamentous particles is ∼ 18–20 nm, which correlates with that of the ORF3 protein–fibrillarin rings (18–22 nm). Thus, the rings formed by these proteins interact with viral RNA, encapsidating it and reorganizing it into helical structures, and thereby play a key role in the assembly of umbraviral RNP complexes. Whether the rings stack on the RNA or nucleate, the addition and assembly of the RNP are unknown.
Collectively, these results demonstrate that, in addition to traditional functions in rRNA processing and modification (e.g., methyltransferase), fibrillarin possesses completely novel functions in triggering nucleolar import of the ORF3 protein via CBs and mediating assembly of umbraviral RNPs. These functions are presumably based on the ability of the ORF3 protein to interact and form ringlike complexes with fibrillarin such that the virus alters and exploits the properties of fibrillarin (e.g., RNA binding) for successful virus propagation. Other viral proteins, such as the nucleoproteins encoded by porcine arterivirus7 or infectious bronchitis coronavirus,6 are also able to interact with fibrillarin. Although the structure and architecture of these complexes and how they impact the viral life cycle remain unknown, mechanisms that lead to formation of the GRV ORF3 protein–fibrillarin complexes may also apply to other viruses.
Acknowledgements
The work was supported by the Scottish Government and the Royal Society (J.W.S.B., N.O.K., and M.T.). We also thank the University of Abertay—Dundee for the award of a postdoctoral research assistantship (E.C.), the Scottish Higher Education Funding Council for the SRIF II Grant to purchase the AFM (A.K.A.), and the Russian Foundation for Basic Research (N.O.K.).
Edited by J. Karn
References
- 1.Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Barneche F., Steinmetz F., Echeverria M. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 2000;275:27212–27220. doi: 10.1074/jbc.M002996200. [DOI] [PubMed] [Google Scholar]
- 3.Cioce M., Lamond A.I. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 4.Matera A.G., Shpargel K.B. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Sleeman J.E., Ajuh P., Lamond A.I. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J. Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- 6.Chen H., Wurm T., Britton P., Brooks G., Hiscox J.A. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002;76:5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo D., Wootton S.K., Li G., Song C., Rowland R.R. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J. Virol. 2003;77:12173–12183. doi: 10.1128/JVI.77.22.12173-12183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiscox J.A. The nucleolus—a gateway to viral infection? Arch. Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiscox J.A. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev., Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S.H., Ryabov E., Kalinina N.O., Rakitina D., Gillespie T., MacFarlane S. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007;26:2169–2179. doi: 10.1038/sj.emboj.7601674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S.H., MacFarlane S., Kalinina N.O., Rakitina D.V., Ryabov E.V., Gillespie T. Interaction between a plant virus-encoded protein and the major nucleolar protein, fibrillarin, is required for virus systemic infection. Proc. Natl Acad. Sci. USA. 2007;104:11115–11120. doi: 10.1073/pnas.0704632104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taliansky M.E., Robinson D.J. Molecular biology of umbraviruses: phantom warriors. J. Gen. Virol. 2003;84:1951–1960. doi: 10.1099/vir.0.19219-0. [DOI] [PubMed] [Google Scholar]
- 13.Ryabov E.V., Robinson D.J., Taliansky M.E. A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proc. Natl Acad. Sci. USA. 1999;96:1212–1217. doi: 10.1073/pnas.96.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taliansky M.E., Roberts I.M., Kalinina N., Ryabov E.V., Raj S.K., Robinson D.J., Oparka K.J. An umbraviral protein, involved in long-distance RNA movement, binds viral RNA and forms unique, protective ribonucleoprotein complexes. J. Virol. 2003;77:3031–3040. doi: 10.1128/JVI.77.5.3031-3040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansma H.G., Kim K.J., Laney D.E., Garcia R.A., Argaman M., Allen M.J., Parsons S.M. Properties of biomolecules measured from atomic force microscope images: a review. J. Struct. Biol. 1997;119:99–108. doi: 10.1006/jsbi.1997.3855. [DOI] [PubMed] [Google Scholar]
- 16.Kiselyova O.I., Yaminsky I.V., Karpova O.V., Rodionova N.P., Kozlovsky S.V., Arkhipenko M.V., Atabekov J.G. AFM study of potato virus X disassembly induced by movement protein. J. Mol. Biol. 2003;332:321–325. doi: 10.1016/s0022-2836(03)00835-0. [DOI] [PubMed] [Google Scholar]
- 17.Skabkin M.A., Kiselyova O.I., Chernov K.G., Sorokin A.V., Dubrovin E.V., Yaminsky I.V. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 2004;32:5621–5635. doi: 10.1093/nar/gkh889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torrance L., Andreev I.A., Gabrenaite-Verhovskaya R., Cowan G., Mäkinen K., Taliansky M.E. An unusual structure at one end of potato potyvirus particles. J. Mol. Biol. 2006;357:1–8. doi: 10.1016/j.jmb.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Stemmer A., Engel A. Imaging biological macromolecules by STM: quantitative interpretation of topographs. Ultramicroscopy. 1990;34:129–140. doi: 10.1016/0304-3991(90)90067-v. [DOI] [PubMed] [Google Scholar]
- 20.Abramoff M.D., Magelhaes P.J., Ram S.J. Image processing with ImageJ. Biophoton. Int. 2004;11:36–42. [Google Scholar]
- 21.Jones K.W., Gorzynski K., Hales C.M., Fischer U., Badbanchi F., Terns R.M., Terns M.P. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- 22.Pante N., Kann M. Nuclear pore complex is able to transport macromolecules with diameters of ∼ 39 nm. Mol. Biol. Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]