Abstract
RNA interference (RNAi) mediated by double stranded small interfering RNA (siRNA) is a novel mechanism of post-transcriptional gene silencing. It is projected as a potential tool to inhibit viral replication. In the present paper, we demonstrate the suppression of replication of an avian herpes virus (Anatid Herpes Virus-1, AHV-1) by siRNA mediated gene silencing in avian cells. The UL-6 gene of AHV-1 that codes for a protein involved in viral packaging was targeted. Both cocktail and unique siRNAs were attempted to evaluate the inhibitory potential of AHV-1 replication in duck embryo fibroblast (DEF) cell line. DEF cells were chemically transfected with different siRNAs in separate experiments followed by viral infection. The observed reduction in virus replication was evaluated by cytopathic effect, viral titration and quantitative real time PCR (QRT-PCR). Among the three siRNA targets used the unique siRNA UL-B sequence was found to be more potent in antiviral activity than the cocktail and UL6-A-siRNA sequences.
Keywords: siRNA, Anatid Herpes Virus, UL-6, Duck, DEF
1. Introduction
The RNA interference phenomenon is known to exist in diverse organisms ranging from plants to man. Application of this phenomenon is wide open, from endogenous gene knock down to inhibition of viral replication in cells. RNA interference has widely been proven as an effective mechanism to suppress the replication of different viruses viz. Poliovirus, human corona virus, human immunodeficiency virus-1 (HIV-1), Hepatitis-B virus, Hepatitis-C virus, foot and mouth disease virus (FMDV), etc. (Gitlin et al., 2002, Wang et al., 2004, Lee et al., 2002, McCaffrey et al., 2003, Wilson et al., 2003, Chen et al., 2004). The phenomenon operates at the post transcription level, thereby, hindering the expression of some essential proteins required for viral replication. RNAi is presumed to be an evolutionarily conserved host defense mechanism against viruses. Evidences also suggest that viruses also have evolved proteins that suppress the RNA silencing pathway as an adaptive mechanism (Mourrain et al., 2000, Dalmay et al., 2001, Gitlin and Andino, 2003). The mechanism has been demonstrated for suppression of viral replication in a wide range of organisms starting from plants, insects, mammals, etc. (Waterhouse et al., 2001, Adelman et al., 2002, Gitlin and Andino, 2003). The present report demonstrates the amenability of AHV1 virus to siRNA mediated viral suppression which may have relevance in developing alternate strategies for control of this virus in ducks and water fowls.
AHV-1 infects birds of the group Anseriformes and causes severe epidemic called duck virus enteritis alternatively known as duck plague (Dardiri, 1975, Kaleta, 1990, Foulon, 1992). The virus belongs to the family Herpesviridae and has not been placed in any of the subfamilies of Herpesviridae (Roizman and Pellett, 2001). The genome of AHV-1 is a linear double stranded DNA (dsDNA) molecule of approximately 180 kb with high G + C content. The genome organization is similar to known alpha herpes viruses (Fukuchi et al., 1984, Gardner et al., 1993). The virus is transmitted by contact of susceptible birds and epidemic may occur within a week. Like other herpes viruses, AHV-1 can also establish latency after primary infection and survivors may act as carriers, shedding virus at regular intervals causing epidemics (Burgess et al., 1979). The RNAi could be a very effective method of controlling emerging viral infections against which vaccines are not available or not so effective. This mechanism appears to be more specific than any antiviral since only the viral genes are targeted. In this paper, we report the inhibition of AHV-1 replication using the siRNA approach targeting the UL6 gene of the virus.
2. Materials and methods
2.1. Cells and viruses
Duck embryo fibroblast (DEF) cells (ATCC CCL-141) were propagated in Eagle's minimum essential medium (EMEM, Sigma) supplemented with 10% fetal bovine serum (FBS, Hyclone) and antibiotics at 37 °C with 5% CO2. DEF monolayer was infected with chick embryo fibroblast (CEF) adapted AHV-1, Ranipet strain. Cytopathic effects (CPE) were observed after 48 h.
2.2. Selection of siRNA target sequences and generation of siRNAs
UL-6 gene of AHV-1 whose sequence is available in the database (Accession Number: AF043730, NCBI) was selected as target for silencing. This gene codes for an essential protein needed for the cleavage and packaging of viral DNA to form viable virions (Plummer et al., 1998). SiRNA targets were identified in this gene using siRNA target finder programme of Ambion (http://www.ambion.com/sirnatargetfinder) which identified a region between 708 and 1032 bp of the UL-6 mRNA. The selected sequence was subjected to global blast against non-redundant nucleotide sequences. Cocktail siRNAs were generated using Silencer siRNA cocktail kit (Ambion Inc, USA) as per manufacturer's protocol. In brief, primers were designed using primer select program of DNA Star® software to amplify 325 bp region extending from 708 to 1032 of UL-6 gene (Table 1 ). T7 promoter sequences were appended to 5′ end of both forward and reverse primers (Table 1) and are used to amplify the same 325 bp region of the UL-6 gene. The resultant 371 bp amplicon was used as template in in vitro transcription reaction. In vitro transcription was carried out using T7 RNA polymerase and rNTPs to generate both sense and anti-sense RNAs, which were annealed to form dsRNA. A cocktail of siRNAs was generated by digesting the dsRNA with RNase-III enzyme. Unique siRNA targets in the UL-6 gene sequence were identified using siRNA target finder programme of Ambion®. Two siRNA target sequences showing least homology with other genes were identified and were synthesized using Silencer™ siRNA construction kit (Ambion Inc, USA) as per manufacturers recommended protocol. Essentially, oligomers synthesized were annealed with T7 promoter primers and extended by Klenow enzyme to get a 48 bp DNA oligonucleotide template. In vitro transcription was carried out from this template to generate sense and anti-sense siRNA strands that were annealed to produce double stranded siRNAs. All the primer sequences and the oligonucleotide sequences required for the experiment were got synthesized from Invitrogen®.
Table 1.
List of primers and siRNA target sequences used
| Name | Primer sequence | Location | Length |
|---|---|---|---|
| AHV-UL-6B-F | 5′-ATAATCAGGGTCAAGAGGTAGCACT-3′ | 708–1032 | 325 |
| AHV-UL-6B-R | 5′-GGTTCATATCCATCTTCCGCAAATC-3′ | 708–1032 | 325 |
| T7 promoter appended UL-6 primersa | 5′-TAATACGACTCACTATAGGGAGAATAATCAGGGTCAAGAGGTAGCACT-3′ | ||
| 5′-TAATACGACTCACTATAGGGAGAGGTTCATATCCATCTTCCGCAAATC-3′ | |||
| AHV-UL6-AsiRNA | 5′-UUUAGCAAAGGCGCGAAUGUU-3′ | 395 | |
| 3′-UUAAAUCGUUUCCGCGCUUAC-5′ | |||
| AHV-UL6-BsiRNA | 5′-UAACUCUCCUCGUUGCACCUU-3′ | 751 | |
| 3′-UUAUUGAGAGGAGCAACGUGG-5′ | |||
For the T7 promoter appended UL-6 primers, promoter sequences are marked in italics, the purine sequences highlighted and the primer sequences underlined.
2.3. Transient transfection of DEF cells with siRNAs
The siRNAs, both cocktail and unique, were introduced into DEF cells using the transfection agent siPORT Amine™ (Ambion Inc, USA). 75 nM final concentration of each siRNA was used for transfecting 70–80% confluent DEF cells. The cells were incubated for 6 h with siRNA-transfection agent complex. After removing the siRNA-transfection agent complex and washing with serum free EMEM, cells were re-incubated in normal growth medium (EMEM with 10% FCS) for another 12 h. The cells were infected with one multiplicity of infection (m.o.i.) final concentration of AHV-1 in maintenance medium (EMEM with 2% FCS). Controls were treated in the same way except for the addition of siRNA. The cells were analyzed for CPE and viral load, 48 h post-infection. Images of cells in control and experimental groups were collected with an Olympus® CK40 microscope fitted with Olympus® SC35 video camera at a magnification of 100×.
2.4. Titration of AHV-1
The chick embryo adapted vaccine strain of AHV-1 was adapted in DEF cells and was titrated in confluent DEF monolayer. Transfection plates were harvested 48 h post-infection and freeze thawed three times. Ten fold dilutions of the sample were used for titration and reading was taken 48 h post-infection. The TCID50 was calculated using Reed and Muench (1938) method.
2.5. Quantitative Real Time-PCR for evaluation of virus load
The standardization of the Real Time-PCR for AHV-1 was carried out using a pair of primers for amplifying a 325 bp long sequence from the UL-6 gene (Table 1). Real Time-PCR was performed from the viral genomic DNA isolated from experimental and control cells using Brilliant SYBR® Green Q-PCR master mix (Stratagene®, USA). Real Time-PCR was performed in an Mx3000P™ instrument from Stratagene®. The reaction was performed in 25 μl volume containing 12.5 μl of Brilliant SYBR® Green Q-PCR master mix, 2.5 μl of genomic DNA isolated from cell culture supernatants and 0.5 μM each of forward and reverse primers. The PCR conditions included 30 cycles, with denaturation at 94 °C for 30 s, primer annealing at 57 °C for 1 min and primer extension at 72 °C for 1 min. An initial denaturation at 94 °C for 10 min and a final extension at 72 °C for 10 min were also included.
3. Results
3.1. Silencing of AHV-1 by cocktail siRNAs
The objective of the present study was to demonstrate the utility of RNAi mechanism to control an avian herpes virus, AHV-1. The chick embryo adapted AHV-1 readily adapted to DEF. The susceptibility of DEF cells for transfection with nucleic acid was also assessed using a functional β-galactosidase construct pSV β-galactosidase (data not shown).
Two approaches, both targeting the UL-6 gene of AHV-1, which is endowed with the function of packaging of mature viral particles (Plummer et al., 1998), were used. In the first instance, to evaluate the amenability of the target gene selected for RNAi, a cocktail siRNA approach spanning 325 bp long region of the genomic sequence was used. This region contained 21 potential siRNA target sequences as identified using a target finder programme of M/s. Ambion. The cocktail siRNA generated for this region as per the method described earlier was used for transfection of DEF followed by viral infection. The CPE observed at 48 h post-infection in control and experimental group is shown in Fig. 1 . Apparently, there is a marked reduction in the CPE, especially in vacuolation and rounding of cells. This was further corroborated by titration of the virus yield between the experimental and control groups. There was 0.88 log difference, which is equivalent to 7.5 fold difference of virions in the cocktail siRNA transfected group in comparison to the control group (average of four individual wells).
Fig. 1.
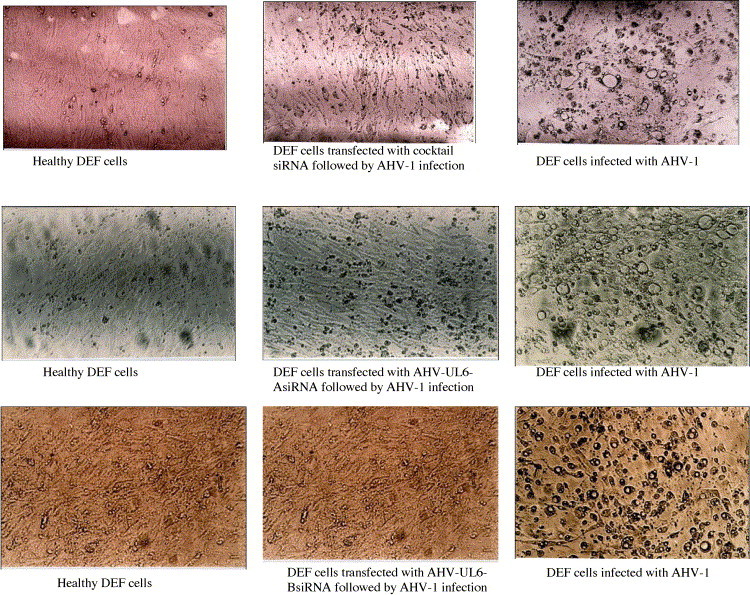
Transfection of DEF cells with cocktail and unique siRNAs followed by AHV-1 infection (48 h post-infection) (100×).
3.2. Silencing of AHV-1 by unique siRNAs
3.2.1. CPE and virus titration
After observing the existence of RNAi mechanism directly by reduction in CPE and virus titre using the cocktail approach, two unique siRNA sequences were also synthesized (Table 1), targeting the same UL-6 gene of AHV-1. siRNAs were generated for this region regions using a different approach as described in the materials and methods. Transfection of 75 nM of individual siRNAs per well followed by infection with ten m.o.i produced marked reduction in CPE, after 48 h of infection. The reduction in CPE between the control and experimental groups were more marked than the cocktail siRNA transfection experiments (Fig. 1). Titration of the virus load, also brought out clear reduction in the experimental groups compared to control groups, confirming the interferences achieved with these siRNAs. The AHV-UL6-AsiRNA brought out a log difference of 1.21 with that of control whereas AHV-UL6-BsiRNA resulted in a log difference of 1.33 (Average value of four independent observations). The log difference brought out by AHV-UL6-AsiRNA and AHV-UL6-BsiRNA corresponds to 16.2 and 21.3 fold differences with control, respectively. This indicates the superiority of AHV-UL6-BsiRNA in eliciting a higher interference effect.
3.2.2. Real Time-PCR
The effect of RNAi using unique siRNAs was also proved by estimating quantitatively the virus DNA concentration present in the experimental and control groups, 48 h post-infection, using Real Time-PCR. SYBR® Green based Q-PCR method was used for this purpose. The threshold cycle (C t), which indicates the significant increase in fluorescent level, is compared between the control and experimental groups. The difference in the threshold cycle (ΔC t) between the groups will characterize the difference in the initial virus load in the samples. The AHV-UL6-AsiRNA transfected sample attained threshold at 16.70th cycle (average of four independent PCR reactions), and control group attained C t at 12.44th cycle (average of four independent PCR reactions). ΔC t between the two was found to be 4.26 cycles, which is equivalent to a reduction of 19 fold difference in virus concentration, 48 h post-infection, in the presence of AHV-UL6-AsiRNA (Fig. 2A). On the other hand, AHV-UL6-BsiRNA transfected sample attained C t at 18.28th cycle (average of four independent PCR reactions), and control group attained C t at 22.79th cycle (average of four independent PCR reactions), yielding a ΔC t of 4.51 cycles (Fig. 2B). This is equivalent to about 23 fold difference in reduction in the virus load in the AHV-UL6-BsiRNA transfected cells, 48 h post-infection. Comparison of ΔC t between AHV-UL6-AsiRNA and AHV-UL6-BsiRNA reveals that the latter produced 3.5 folds more reduction in virus titre in comparison to the former. Thus AHV-UL6-BsiRNA appears to be the choicest siRNA for controlling AHV-1 replication.
Fig. 2.
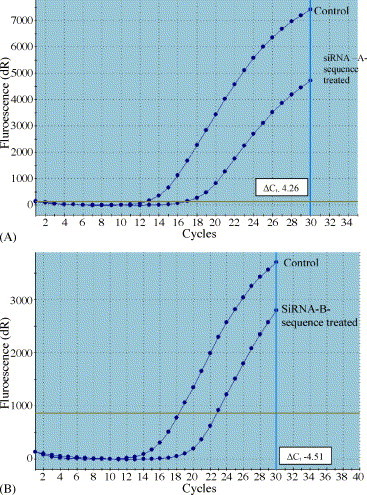
Real Time-PCR amplification plot of AHV-1 genomic DNA isolated from cells transfected with AHV-UL6-AsiRNA and virus control cells 48 h post-infection (A) and with AHV-UL6-BsiRNA and virus control cells 48 h post-infection (B).
4. Discussion
Post-transcriptional gene silencing (PTGS) mediated by siRNA has been a well-proven method for controlling replication of different viruses of plant and animal origin. Even though, initially there were apprehensions about siRNA mediated interference mechanism operating in mammalian systems owing to dsRNA induced global shutdown, Elbashir et al. (2001) had proven that direct introduction of siRNAs can induce RNAi in mammalian cells also. Consequently, many mammalian viruses were demonstrated to be amenable to siRNA mediated suppression both in cell culture system as well as in in vivo system (Chen et al., 2004).
Duck virus enteritis, alternatively called as duck plague, caused by an Herpesvirus, is a major problem for the duck rearing regions of the world, as well as free ranging water fowls (Devos et al., 1964, Jansen, 1968, Hall and Simmons, 1972, Erickson et al., 1974, Montali et al., 1976, Pritchard et al., 1999). Even though, a CEF adapted vaccine is available, alternate methods of viral control is essential as the vaccines is less effective and since this virus can go into latency and survivors of outbreak can act as carriers (Burgess et al., 1979).
Hepatitis-B virus, Hepatitis-C virus, HIV, Poliovirus, SARS virus, Dengue virus, etc. are some of the human viruses that have been shown to be under check with siRNA (McCaffrey et al., 2003, Wilson et al., 2003, Capodici et al., 2002, Coburn and Cullen, 2002, Hu et al., 2002, Jacque et al., 2002, Lee et al., 2002, Novina et al., 2002, Park et al., 2002, Surabhi and Gaynor, 2002, Gitlin et al., 2002, Wang et al., 2004, Adelman et al., 2002). This method offers a promising alternative method for control of animal viruses affecting livestock industry. Consequently, Foot and Mouth Disease (FMD), one of the major scourges of cattle farming world over has also been found to be responsive to siRNA (Chen et al., 2004). Attempts are also being made to control Peste des Petits Ruminants (PPR) virus, a major scourge of small ruminants by siRNA (Sahay et al., 2004, personal communication). One of the major applications of siRNA mediated viral suppression in animal husbandry sector would be in poultry production. Owing to the small generation interval and the facility of in ovo administration, siRNA has much promise in poultry production. Unfortunately, existence of RNAi mechanism has not yet demonstrated in birds. The present study demonstrates the existence of RNAi mechanism in birds by suppressing an avian virus, AHV-1.
Various approaches are used for generating siRNA that include cocktail siRNA, synthetic siRNA, siRNA expressing plasmids, viral vectors, etc. (Byrom et al., 2003, Kronke et al., 2004, Elbashir et al., 2002, Brummelkamp et al., 2002, Lee et al., 2002, Miyagishi and Taira, 2002, Paddison et al., 2002, Sui et al., 2002, Yu et al., 2002, Barton and Medzhitov, 2002, Shen et al., 2003, Tiscornia et al., 2003). Kronke et al. (2004) introduced esiRNA cocktail approach to control Hepatitis-C virus replication. Long dsRNA generated by in vitro transcription of PCR products were digested with RNase-III to generate a cocktail of siRNAs. This approach leads to the formation of a large number of siRNAs against same targeted gene, which provides a sure shot inhibition of viral target even if the region is hyper mutable. But on the other hand it may generate siRNAs against those targets. In the present study, we used a cocktail siRNA approach to initially prove the amenability of UL-6 gene (one of the only two gene sequences of the virus available) for silencing by RNAi. CEF adapted Ranipet strain of the virus was readapted to homologous cell culture system, i.e. DEF cell line. The virus readily adapted in DEF cell line and displayed 80–90% CPE within 48 h post-infection. Cocktail siRNAs generated by RNase-III digestion of dsRNA produced from the PCR product was introduced to DEF cells followed by infection with the virus. The effect of siRNA mediated suppression of virus replication was monitored through CPE and virus titre. Though RnaseIII generate siRNA has limitation, unambiguous visual appreciation of reduction in CPE and virus titre were observed confirming that UL-6 indeed is an effective target for RNAi based viral suppression.
Consequently, two unique siRNA targets were also identified in the UL-6 genome. These siRNAs, i.e. AHV-UL6-AsiRNA and AHV-UL6-BsiRNA were synthesized by an alternative method using Silencer™ siRNA construction kit of Ambion®. Both the siRNAs were administered to DEF cells in separate experiments followed by infection with 1 m.o.i. AHV-1. More predominant reduction in CPE and virus titre was observed than the cocktail siRNAs. This could be due to the differences in the effective concentration of intracellular siRNAs as RNase-III used in the Ambion kit need not essentially generate siRNAs of 21 bp. It is also known that RNase-III generates predominantly siRNAs of 13–15 bp (Byrom et al., 2003). However, it is known that siRNAs of 21 bp is most suited for binding to RNA induced silencing complex (RISC) that eventually effects the degradation of specific mRNA. Further, the targeted region for cocktail siRNA contained many overlapping sequences of potential siRNAs. All these factors could have considerably reduced the effective molar concentration of 21 bp long siRNAs within the cocktail digest in comparison to the unique siRNAs used. However, the cocktail siRNA approach still remains one of the most suited, simple exploratory tool for identifying target sequence for gene silencing. Substitution of RNase-III with Dicer enzyme for generating cocktail would have been more effective, but not tested in the present study.
The present experiment was primarily intended to prove the amenability of siRNA as a method of control against AHV-1. Hence, quantitative technique like Real Time-PCR was also used to estimate the virus load following RNAi. Wang et al. (2004) have already used the same technique for monitoring the effect of siRNA in SARS replication. SYBR® Green based Q-PCR was effective in differentiating the viral DNA concentration in different experimental and control groups. The differences in threshold cycles (ΔC t) while using same volume of template material indicates a direct difference in the initial concentrations of template molecule in the different samples tested. One of the problems of this evaluation method is that viral DNA can get into culture supernatants as infectious virions, non-infectious, physically or genetically defective virions, or as free DNA released from lysed cells that may affect the the ΔC t. Hence the real time data may indicate directly the difference in amount of viral genomic DNA rather than the real infectious particle. On the other hand the viral titration directly indicate the difference in infectious particles. But since the same method is used for both experimental and control groups the ΔC t should indicate a difference in the initial viral concentration between the two. Further the same difference was estimated by virus titration as well to confirm the results. The difference in threshold cycle (ΔC t) between control and AHV-UL6-BsiRNA was 4.51 cycles, whereas between control and AHV-UL6-AsiRNA ΔC t was 4.26 cycles, clearly indicating the superiority of AHV-UL6-BsiRNA in eliciting RNAi (Fig. 2A and B). This finding had been supported by viral titration data also. We propose AHV-UL6-BsiRNA based plasmid vector or Adeno virus/Retro virus based vector for developing an effective prophylactics against AHV-1.
Acknowledgements
T.J.R. is thankful to Department of Biotechnology, Ministry of Science and Technology, Govt. of India for funding the project on Duck Plague Virus. S.K.M. and B.S. are thankful to Indian Council of Agricultural Research, Govt. of India, and Council of Scientific and Industrial Research, Govt. of India, respectively, for providing the fellowships.
References
- Adelman Z.N., anchez-Vargas I., Travanty A., Carlson J.O., Beaty B.J., Blair C.D., Olson K.E. RNA silencing of Dengue virus type-2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 2002;76:12925–12933. doi: 10.1128/JVI.76.24.12925-12933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G.M., Medzhitov R. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14943–14945. doi: 10.1073/pnas.242594499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernads R., Agami R.A. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Burgess E.C., Ossa J., Yuill T.M. Duck plague: a carrier state in waterfowl. Avian Dis. 1979;24:940–949. [PubMed] [Google Scholar]
- Byrom M.W., Chang A.M., Ford L.P. Inducing RNAi with siRNA cocktails generated by RNase-III. Ambion Tech. Notes. 2003;10:4–6. [Google Scholar]
- Capodici J., Kariko K., Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interferance. J. Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- Chen W., Yan W., Du Q., Fei L., Liu M., Ni Z., Sheng Z., Zheng Z. RNA interference targeting VP1 inhibits Foot-and-Mouth Disease virus replication in BHK-21 cells and suckling mice. J. Virol. 2004;78:6900–6907. doi: 10.1128/JVI.78.13.6900-6907.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn G.A., Cullen B.R. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interferance. J. Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Horsefield R., Braunstein T.H., Baulcombe D.C. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardiri A.H. Duck viral enteritis (Duck plague) characteristics and immune response of the host. Am. J. Vet. Res. 1975;36:535–538. [PubMed] [Google Scholar]
- Devos A., Viaene N., Staelens H. Duck plague in Belgium. Vlaams Diergeneeskd Tijdschr. 1964;33:260–266. [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Weber K., Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Erickson, G.A., Proctor, S.J., Pearson, J.E., Gustafson, G.A., 1974. Diagnosis of Duck virus enteritis (Duck plague). Proceedings of the 17th Ann. Meet. Am. Ass. Vet. Lab. Diag. Roanoke, pp. 85–90.
- Foulon T. Herpesviridae: classification et structure en 1991. Comp. Immunol. Microbiol. Infect. Dis. 1992;15:13–29. doi: 10.1016/0147-9571(92)90098-c. [DOI] [PubMed] [Google Scholar]
- Fukuchi F., Sudo M., Lee Y.S., Tanaka A., Nonoyama M. Structure of Marek's disease virus DNA: detailed restriction map. J. Virol. 1984;51:102–109. doi: 10.1128/jvi.51.1.102-109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R., Wilkerson J., Johnson J.C. Molecular characterization of the DNA of Anatid herpesvirus 1. Intervirology. 1993;36:99–112. doi: 10.1159/000150328. [DOI] [PubMed] [Google Scholar]
- Gitlin L., Andino R. Nucleic acid based immune system: the anti-viral potential of mammalian RNA silencing. J. Virol. 2003;77:7159–7165. doi: 10.1128/JVI.77.13.7159-7165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L., Karelsky S., Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- Hall S.A., Simmons J.R. Duck plague (Duck virus enteritis) in Britain. Vet. Rec. 1972;90:691. doi: 10.1136/vr.90.24.691. [DOI] [PubMed] [Google Scholar]
- Hu W., Myers C., Kilzer J., Pfaff S., Bushman F. Inhibition of retroviral pathogenesis by RNA interferance. Curr. Biol. 2002;12:1301. doi: 10.1016/s0960-9822(02)00975-2. [DOI] [PubMed] [Google Scholar]
- Jacque J.M., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interferance. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J. Duck plague. JAVMA. 1968;152:1009–1016. [PubMed] [Google Scholar]
- Kaleta E.F. Herpesviruses of birds. A review. Avian Path. 1990;19:193–211. doi: 10.1080/03079459008418673. [DOI] [PubMed] [Google Scholar]
- Kronke J., Kittler R., Buchholz F., Windish M.P., Pietschmann T., Bartenschlager R., Frese M. Alternative approaches for efficient inhibition of Hepatitis C Virus RNA replication by small interfering RNAs. J. Virol. 2004;78:3436–3446. doi: 10.1128/JVI.78.7.3436-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.S., Dohjima T., Bauer G., Li H., Li J.M., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Nakai H., Pandey K., Huang Z., Salazar F.H., Xu H., Wieland S.F., Marion P.L., Kay M.A. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 2003;21(6):639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Miyagishi M., Taira K. U6 promoter-driven siRNA with four uridines 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Montali R.J., Bush M., Greenwell G.A. An epornitic of duck viral enteritis in a zoological park. JAVMA. 1976;169:954–958. [PubMed] [Google Scholar]
- Mourrain P., Beclin C., Elmayan T., Feuerbach F., Godon C., Morel J.B., Jouette D., Lacombe A.M., Nikic S., Picault N., Remoue K., Sanial M., Vo T.A., Vaucheret H. Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Novina C.D., Murray M.F., Dykxhoorn D.M., Beresford P.J., Reiss J., Lee S.K., Collman R.G., Lieberman J., Shankar P., Sharp P.A. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Paddison P.J., Caudy A.A., Hannon G.J. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W.S., Miyano-Kurosaki N., Hayafune M., Nakajima E., Matsuzaki T., Shimada F., Takaku H. Prevention of HIV-1 infection in human peripheral blood mononuclear cells by specific RNA interferance. Nucleic Acids Res. 2002;30:4830–4835. doi: 10.1093/nar/gkf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer P.J., Alefantis T., Kaplan S., O’Connell P., Shawky S., Schat K.A. Detection of Duck enteritis virus by polymerase chain reaction. Avian Dis. 1998;42:554–564. [PubMed] [Google Scholar]
- Pritchard L.I., Morrissy C., Phuc K.V., Daniels P.W., Westbury H.A. Development of polymerase chain reaction to detect Vietnamese isolates of Duck virus enteritis. Vet. Microbiol. 1999;68:149–156. doi: 10.1016/s0378-1135(99)00071-1. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent end points. Am. J. Hygiene. 1938;27:493–497. [Google Scholar]
- Roizman, B., Pellett, P.E., 2001. The family Herpesviridae: a brief introduction. Field's Virology, fourth ed., pp. 2381–2397.
- Shen C., Buck A.K., Liu X., Winkler M., Reske S.N. Gene silencing by adenovirus-delivered siRNA. FEBS Lett. 2003;539:11–114. doi: 10.1016/s0014-5793(03)00209-6. [DOI] [PubMed] [Google Scholar]
- Sui G.C., Soohoo C., Affair E.B., Gay F., Shi Y., Forrester W.C., Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surabhi R.M., Gaynor R.B. RNA interferance directed against viral and cellular targets inhibits human immunodeficiency virus type-1 replication. J. Virol. 2002;76:12963–12973. doi: 10.1128/JVI.76.24.12963-12973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G., Singer O., Ikawa M., Verma I.M. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ren L., Zhao X., Hung T., Meng A., Wang J., Chen Y.-G. Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J. Virol. 2004;78:7523–7527. doi: 10.1128/JVI.78.14.7523-7527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang M.B., Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- Wilson J.A., Jayasena S., Khvorova A., Sabatinos S., Rodrigue-Gervais I.G., Arya S., Sarangi F., Harris-Brandts M., Beauliu S., Richardson C.D. RNA interferance blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.Y., DeRuiter S.L., Turner D.L. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]


