Abstract
The Nipah Virus (NiV) was first isolated during a 1998–9 outbreak in Malaysia. The outbreak initially infected farm pigs and then moved to humans from pigs with a case-fatality rate (CFR) of about 40%. After 2001, regular outbreaks occurred with higher CFRs (~71%, 2001–5, ~93%, 2008–12). The spread arose from drinking virus-laden palm date sap and human-to-human transmission. Intrinsic disorder analysis revealed strong correlation between the percentage of disorder in the N protein and CFR (Regression: r2 = 0.93, p < 0.01, ANOVA: p < 0.01). Distinct disorder and, therefore, genetic differences can be found in all three group of strains. The fact that the transmission modes of the Malaysia strain are different from those of the Bangladesh strains suggests that the correlations may also be linked to the modes of viral transmission. Analysis of the NiV and related viruses suggests links between modes of transmission and disorder of not just the N protein but, also, of M shell protein. The links among shell disorder, transmission modes, and virulence suggest mechanisms by which viruses are attenuated as they passed through different cell hosts from different animal species. These have implications for development of vaccines and epidemiological molecular analytical tools to contain outbreaks.
Keywords: Intrinsically disordered protein, Nucleocapsid, Nipah, Virulence, Viral protein, Protein structure, Protein function, Shell
1. Introduction
The Nipah virus (NiV) was first discovered, when an outbreak happened in Malaysia and Singapore in 1998–99. About 270 people were infected with a case-fatality ratio (CFR) of 40% [[1], [2], [3]]. All of the cases involved close contacts with infected farm pigs. Many of the pigs were initially infected by eating fruits that were partially eaten by fruits bats, which had left their virus-laden saliva behind (4–6). Upon isolation and sequencing of the virus, it was revealed that the culprit was a novel RNA virus from Paramyxoviridae family, which includes the Hendra (HeV), mumps (MuV), and measles (MeV) viruses [[4], [5], [6], [7]]. The symptoms for Nipah infections often include an initial flu-like fever that often follows by irritations, coma, and then death.
In 2001, a different NiV strain was observed in Bangladesh [4]. There were outbreaks in India or Bangladesh each year thereafter. While human infections in the Malaysian outbreak arose mainly from close contacts with infected pigs, the patients in Bangladesh were often infected by drinking date palm sap that was previously consumed by bats. The virulence was also different. For example, the Bangladesh outbreaks in 2004 involved a CFR of 75%, while the 2008–10 outbreaks had a CFR of 89% (Kalkarni et al. [8]. The reason for these remarkable differences in the virulence of different NiV strains remains largely unknown [9]. This paper will put forth a set of evidence demonstrating that the variability in the intrinsic disorder propensities of some of the viral proteins from different strains NiV may act as an underlying reason for the differences in their virulence with the potential links to the modes of transmission. It is worth mentioning here the same underlying reason for high virulence and differences in the transmission modes had been previously found for other viruses, such as MERS-CoV, SARS-CoV, Ebola virus, flaviviruses, and HIV [[10], [11], [12], [13], [67]].
An important concept that will be used in this paper involves intrinsically disordered proteins, which are biologically active proteins that have no unique structures. While it had been initially observed that unique structures dictate the functions of the protein, it was also found that many proteins lack structure, and the lack of structure itself provides protein with new functional means [[14], [15], [16], [17]]. As a result of the recognition of the biological and pathological importance of intrinsically disordered proteins and proteins containing intrinsically disordered regions, multiple disorder predictors have been elaborated to recognize disordered regions [[18], [19], [20]]. One of the earliest disorder predictors is the PONDR® VLXT [[21], [22], [23]] which has been shown to be useful in the analysis of proteins from various viruses, such as HIV, influenza A 1918H1N1 and H5N1 viruses, poliovirus, SARS-CoV, MERS-CoV, smallpox virus, Ebola virus, and HCV [[10], [11], [12], [13],[24], [25], [26], [27], [28], [29], [30], [31], [67]].
While NiV is of the Paramyxoviridae, this virus, together with HeV, belongs to the henipavirus genus characterized by larger genomes, when compared to the other paramyxoviruses. In fact, 18.6 kb-long negative-sense single-stranded RNA (ssRNA) genome of NiV contains 6 genes (Fig. 1 ) that encode 9 proteins: nucleoprotein (N), phosphoprotein (P), the interferon antagonists W and V, the viral C protein, a matrix protein (M), viral fusion and glycoproteins (F and G, respectively), and a large polymerase (L), with the proteins P, V, W, and C about the genomic organization), which are used by the virus to overcome the innate immune response [32,33], being all encoded by the P gene that undergoes specific mRNA editing that results in the appearance of the specific reading frame shifts [[34], [35]]. A homopolymer of the major nucleocapsid protein N condenses and encases the genomic RNA within the long helical nucleocapsids, which serve as biologically active templates for the viral RNA synthesis by the viral RNA-dependent RNA polymerase [[25], [35], [36]]. Structurally, N protein contains two globular domains, the N-terminal (NNTD, residues 32–258) and C-terminal (NCTD, residues 259–371) domains decorated with two projections, N-terminal (NTARM, residues 1–31) and C-terminal subdomains (CTARM, residues 372–383), and a long and highly disordered C-terminal tail (NTAIL, residues 384–532) that protrudes outside the nucleocapsid. These different structural parts of N have divergent biological functions, where NNTD and NCTD enwrap the genomic RNA to protect it against nucleases [[37], [38], [39], [40], [41]], NTARM and CTARM from adjacent protomers are exchanged to ensure stable lateral contacts needed for stabilization of the N homopolymer 2005) [[37], [38], [39], [40], [41]], and a highly disordered Ntail is utilized in binding to the C-terminal domain of the phosphoprotein P (PXD) [[42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54]]. Therefore, the viral shell contains N as a major nucleocapsid protein alongside with the phosphoprotein, P, and a large protein L that serves as an RNA polymerase during viral replication [6,8,52,55]. and which is also found in close proximity. The matrix protein, M, that can be found underlying the viral membrane, is in contact with the fusion protein, F, and the glycoprotein, G, bound to the lipid membrane [7].
Fig. 1.
An illustrated organization of the NiV genome (Acheson 2007).
2. Material and methods
PONDR® VLXT (www.pondr.com) is the main tool used in our investigation. It is a neural network that reads inputs from the sequence of a protein and then predicts regions that are expected to be disordered as output [21,23]. The PONDR® VLXT has been found to be the most accurate predictor in situations that involve protein-protein interactions, which are essential in viral shell proteins [56, 57]. This is likely the reason for it to be rather successful in the various studies of viral shell proteins as aforementioned.
The sequences of the various NiV strains were selected and downloaded from UniProt (http://www.uniprot.org). The programming language JAVA was then used to retrieve and record into MYSQL. This process was done through a JAVA-JDBC driver. The program also calculates the percentage of intrinsic disorder (PID), which is defined as the number of predicted disordered residues divided by the total number of residues in the protein. In order to obtain a clearer visualization, a search for Nipah nucleocapsid was made using NCBI-Protein Databank (NCBI-PDB, http://www.ncbi.nlm.nih.gov/Structure/index.shtml). The accession codes were recorded and the respective PDB and FASTA files were downloaded and placed into the MySQL database. The information stored in the database can later be retrieved using a JAVA program that would generate codes to be used in molecular viewing software, Jmol (www.jmol.org) [29]. Statistical analyses were done using one-way analysis of variance (ANOVA) via R statistical package. The case-fatality ratios were calculated based on published data from the World Health Organization (WHO) (http://www.searo.who.int/entity/emerging_diseases/links/nipah_virus_outbreaks_sear/en/).
A phylogenetic tree was constructed using Clustal Omega (EMBL-EBI, https://www.ebi.ac.uk/Tools/msa/clustalo/) and the mentioned sequences of NiV N proteins from UniProt. The NiV strains and the respective calculated N PIDs (disorder levels) were carefully denoted on the tree.
3. Results
3.1. Nipah virulence and CFR analysis by year of infection
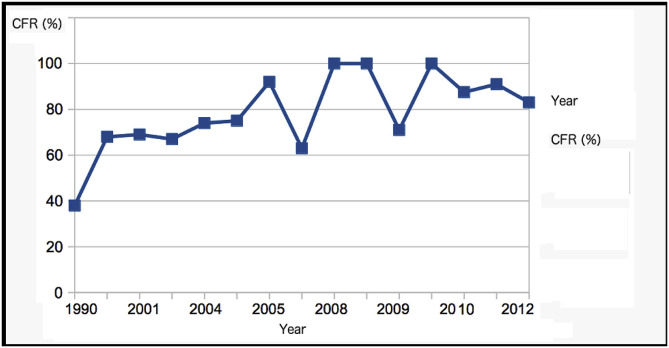
It has already been known that the virulence of the Malaysia-Singapore 1998-9 NiV strain was lower than those of strains that appeared in Bangladesh and India in later years. This can be reaffirmed by looking at Fig. 2 showing a CFR plot of the historical records from the WHO. It should be noted that the plot is adjusted for the “noise”. The CFR of the 1998–9 outbreak, which involved 270 people, was about 40%, while subsequent 2004 outbreaks in Bangladesh and India showed CFR of ~75%, and the outbreaks caused by the NiV strains after 2007 showed even higher fatality rates, which average at about 85%. Statistical analyses using One-Way ANOVA (F-9.93, p < 0.01) and regression reaffirm the statistical significance of the differences in the virulence strength among the strains (r2 = 0.83, F = 42.43, p < 0.01). These results therefore justify the grouping of NiV into three main strains with the following calculated averages: 1) Malaysia-Singapore 1998–9 (CFR, 39.5 ± 1%), 2) Bangladesh-India 2001–5 (CFR, 71 ± 3.5%), and 3) Bangladesh-India 2007/8–12 (92.8 ± 6%). As we shall see from the analyses below, there is also noticeable difference in the viral shell disorder and, therefore, genetic difference, among these three groups of the NiV strains.
Fig. 2.
A CFR (Case-Fatality Ratio) as shown by outbreaks by year. The calculations and data shown have been adjusted for “noise” especially involving samples with the number of cases in the outbreaks that are too small to be reliable. Regression analysis (p < 0.01, F = 44.1, r2 = 0.81) shows correlation between the CFR and year of infection. One-Way ANOVA analysis based on dividing three groups, 1998–9, 2001–6, and 2008–12, has yielded a statistically significant result (One-Way ANOVA: F = 49, p < 0.01). Average CFRs (~40% in 1998–9, ~75% in 2001–5, and ~85% in 2007–12).
3.2. Positive correlation between the disorder predisposition of Nipah N protein and virulence
A search in UniProt for sequences of Nipah N protein provided us with the access to sufficient variety of NiV strains, as seen in Table 1 . Table 1 also shows that based on the year of the strain, there is a clear pattern in the levels of disorder, PID. Analyses of the correlation between disorder levels and CFR via regression (r2 = 0.92, F = 39.4, p < 0.01) and ANOVA (r2 = 0.92, F = 39.4, p < 0.01) indicated that the differences observed in the disorder predispositions of the N proteins from various NiV strains positively correlated with the virulence of the corresponding strains. This is further illustrated by both Fig. 2 and Table 1, where it is clearly seen that in a similar way as three categories of NiV strains are characterized by the increasing virulence, the disorder levels in their N proteins are also changing as we move from one category of strains to another or from year to year. Table 1 shows that N proteins from all strains analyzed in this study are characterized by high level of predicted intrinsic disorder, which defines this protein as highly disordered following the accepted classification of proteins based on their PID, where proteins are considered as highly ordered, moderately disordered, or highly disordered if their PID <10%, 10% ≤ PID <30%, or PID >30%, respectively [[57], [58]].
Table 1.
A comparison of disorder levels (PID) of the nucleocapsid (N) proteins across NiV types with CFR.
| Outbreak Strains | Number of Cases (Deaths) | Fatality Rate (CFR, %)a | PID (%) N | UniProt accession numbers |
|---|---|---|---|---|
| Malaysia-Singapore 1998-99 | 276 (106) | 38 | 41.73 ± 0.01b | Q9IK92, A0A1LK92 |
| Bangladesh-India 2001–2005 | 170 (123) | 72 | 42.67 ± 0.27b | Q4VCQ2, H6V858 |
| Bangladesh-India 2007-10 | 103 (88) | 85 | 43.32 ± 0.24b | D2DEB7, A0A141HUS6c, H6V855, H6V857, E4Z9F9d |
Statistical Analyses: ANOVA, p < 0.01, Regression Analysis, p < 0.01, r2 = 0.93.
a CFR are calculated from the total number of death divided by the total number of cases.
b The numeric value after “+” is the standard error. It should be noted that the mean PID values are misleadingly close to each other. It does not need to be the case since the N protein is huge and most of the mutations arise from or from areas close to the Ntail.
b Samples from two anamalous strains are not included: H6V868, H6V854. In general, a few anamalous strains should be expected in cases where a large number of outbreaks/strains is involved. The data used involve 9 isolates and 6 strains.
c This Thailand sample was collected in 2010 from bats. This sample makes the study interesting as the PID and genetic makeup is closer to those of the Bangladesh-India 2007-10 group despite the fact that Thailand is geographically closer to Malaysia-Singapore. However, because the PID and genetic makeup is closer to that of the 2008-10 strains and because 2007 is close to 2008, we suspect that this strain falls into the 2008-10 strains.
d Sample was collect from Malaysian bats in 2008. It is of great interest since its PID and genetic makeup is closer to the Bangladesh-India 2007-10 samples than to those of Malaysia-Singapore 1998–99.
3.3. Disorder differences between the N proteins from different strains necessarily mean the presence of genetic differences
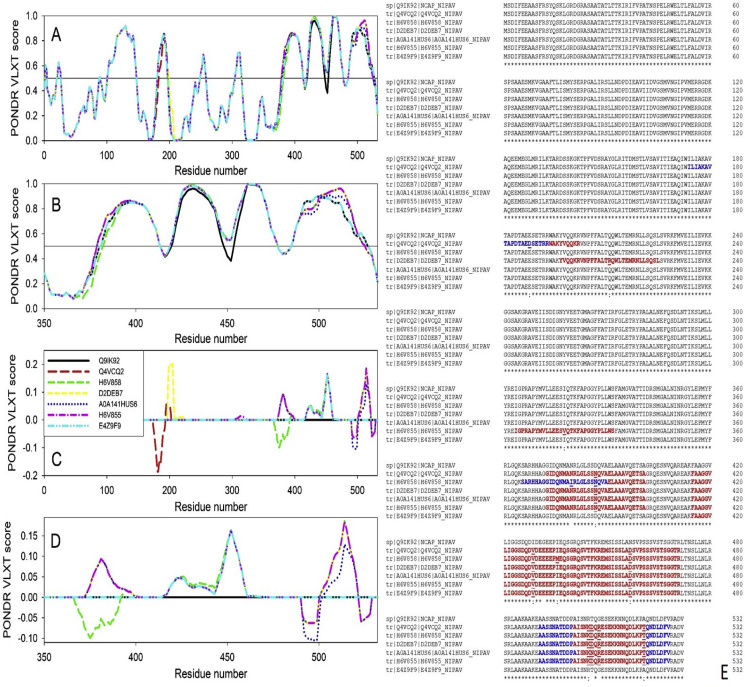
While Table 1 summarizes the predicted disorder levels in N proteins from different NiV strains in terms of their PID, Fig. 3 provides insights into the reasons for the observed PID differences by showing the PONDR® VLXT disorder profiles and corresponding protein sequences. Fig. 3A represents the PONDR® VLXT profiles for all N proteins listed in Table 1 and shows that although profiles of the N-terminal halves of these proteins are almost indistinguishable, some noticeable variability can be found within their C-terminal regions. To further illustrate these differences, Fig. 3B represents the zoomed-in view of the C-terminal regions, whereas Fig. 3C and D correspondingly show a set of differential profiles, where the PONDR® VLXT profile of the N protein of the NiV strain responsible for the Malaysia-Singapore 1998–9 outbreak is subtracted from the PONDR® VLXT profiles and the zoomed-in view of the parts of these differential profiles corresponding to the C-terminal regions of N proteins. Finally, Fig. 3E shows the results of the multiple sequence alignment of all the NiV N proteins analyzed in this study. Again, one can see that the C-terminal regions of these proteins are characterized by the largest sequence variability. In order to better visualize the correlation between the sequence variability of the N proteins and their intrinsic disorder predispositions, positions with higher disorder levels in the Bangladesh-India strains as compared with the original Malaysia-Singapore 1998-9 strain are indicated by bold red font, whereas positions with higher levels of predicted disorder in the Malaysia-Singapore 1998-9 strains than in the strains associated with the subsequence outbreaks are shown by bold blue font. Exact positions of mutations in N proteins from various strains relative to the sequence of N protein from the original 1998-9 Malaysia-Singapore strain are underlined. Visual inspection of this graph provides useful information on the location of mutations causing changes in the local intrinsic disorder predisposition of the corresponding N proteins. Fig. 3 shows that mutations are causing noticeable distortions in the local intrinsic disorder predisposition and that these distortions typically propagate well beyond the mutation site. Furthermore, Fig. 3 illustrates that although N proteins generally contain high levels of intrinsic disorder, their C-terminal domains (residues 370–532) are almost completely disordered. This is a peculiar observation indicating that strain-specific mutations are predominantly located within these highly disordered C-terminal domains suggesting their importance for the strain evolution, probably via modulation of the biological activities of the Nipah N protein.
Fig. 3.
Differences in nucleocapid disorder of various Nipah virus strains. A. PONDR® VLXT plots of the nucleocapsid proteins of various NiV strains listed in Table 1. B. Zoomed-in PONDR® VLXT plots of the C-terminal regions of the nucleocapsid proteins of various NiV strains. C. Disorder “difference spectra” representing differences between the various NiV strains in relation to the Malaysia-Singapore 1998-9 strain. D. Zoomed-in disorder “difference spectra” representing differences between the various NiV strains in relation to the Malaysia-Singapore 1998-9 strain with the focus on the C-terminal regions of the nucleocapsid proteins. In these plots, the corresponding disorder “difference spectra” were calculated by subtracting the PONDR® VLXT profile of the Malaysia-Singapore 1998-9 strain from the PONDR® VLXT profiles of other NiV strain. Positive peaks here correspond to the region of newer strains that are more disordered than the corresponding regions in the Malaysia-Singapore 1998-9 strain, whereas negative peaks show regions with higher disorder in the Malaysia-Singapore 1998-9 strain relative to the newer NiV strains. E. Multiple sequence alignments of N proteins from different NiV strains listed in Table 1. Alignment was conducted by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) using the default parameters. Residues, which are predicted to be more disordered in the Malaysia-Singapore 1998-9 strain relative to the newer NiV strains are shown by bold blue font, whereas bold red font is used to show residues predicted to be more disordered in the more recent NiV strains relative to the Malaysia-Singapore 1998-9 strain.
3.4. Strain-specific differences tend to be located near the C-terminus of the N protein used for interactions with the P protein
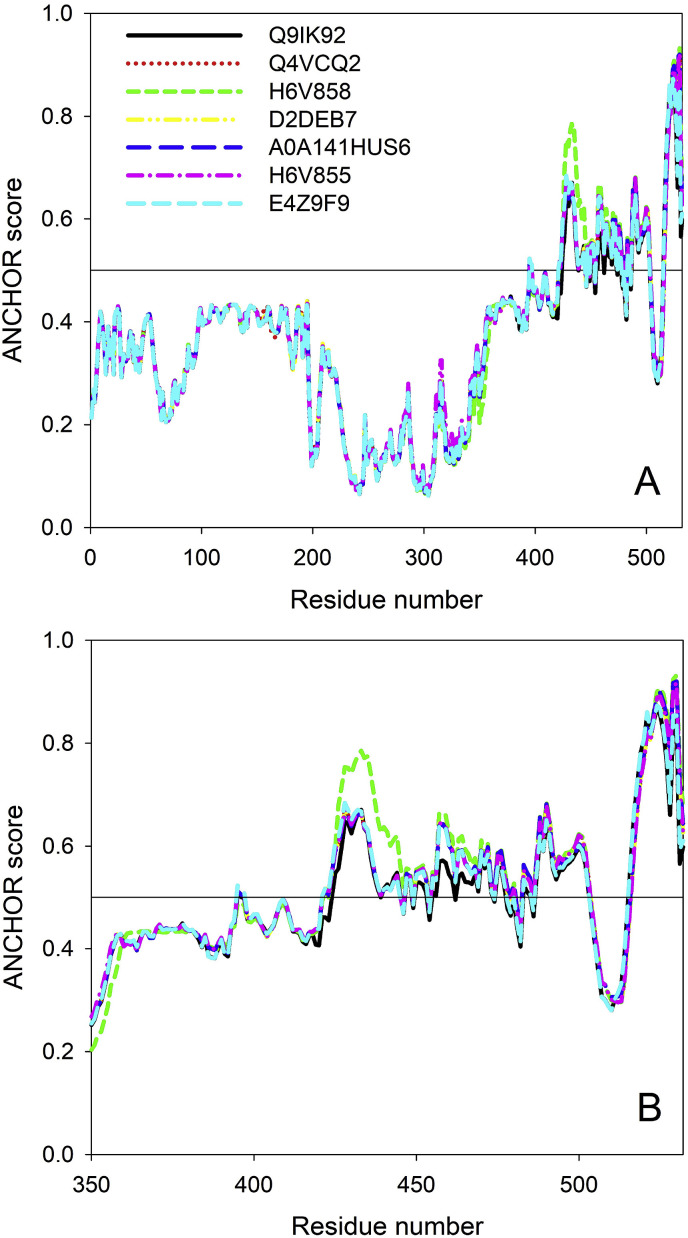
As it follows from Fig. 3, most strain-specific differences are found within the highly disordered C-terminal regions of the N proteins. This is a very important observation, since previous structural analysis revealed that this region represents an interaction site of N nucleoprotein for the P protein, and the binding of N to P is crucial for the appropriate co-localization of the complex during viral replication [52,59]. Fig. 4 shows that the C-tail of the N protein contains several disorder-based binding sites, molecular recognition features (MoRFs), which can undergo disorder-to-order transitions at interaction with their biological partners, and which can be identified by the ANCHOR algorithm [60,61]. Fig. 4 also illustrates that the strain-specific mutations are expected to affect interactability of N-protein, since all of them increase ANCHOR scores of the C-terminal region.
Fig. 4.
ANCHOR-based analysis of the potential intrinsic disorder-based interactivity of N proteins from different NiV strains. A. ANCHOR plot for the full-length proteins. B. Zoomed-in ANCHOR plot showing C-terminal regions of the N-proteins that are most affected by the strain-specific variability.
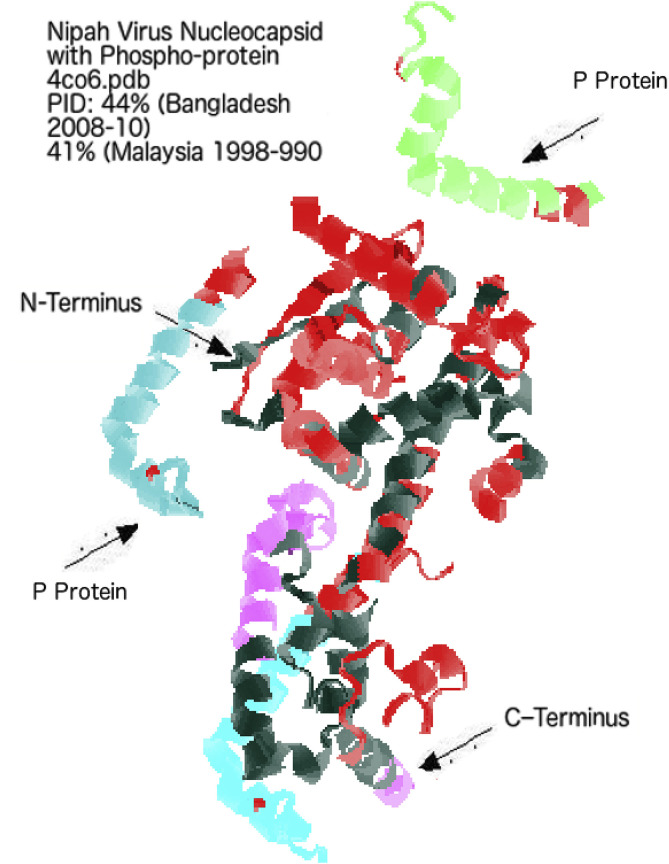
The crystal structure of the N–P protein complex with disorder annotations in N protein shown in Fig. 5 re-affirms the mostly C-terminal location of the regions with strain-specific differences in disorder (pink) among strains. The figure is somewhat misleading by showing only one pink region (around residue 315). This is due to the fact that the other regions with strain-specific variability in disorder content shown in Fig. 3 are missing because of the fact that the construct used in the crystallization experiments (residues 28–383) exclusively contained the globular NNTD and NCTD domains and CTARM, whereas NTARM and NTAIL were removed to avoid complications associated with an attempt to crystalize long regions with high disorder content and to avoid unwanted self-assembly of N [62]. Interestingly, Fig. 5 shows the pink area virtually touching the protein, P, which is shaded in dark blue. Although the most ordered part of N protein was used in the crystallization experiments and although N protein was co-crystallized with the P nucleocapsid-binding domain PXD (residues 655–709), the target complex formed a trimer of heterodimers, where all involved components contained significant amount of intrinsic disorder, as can be judged by the presence of noticeable regions of missing electron density. In fact, all three chains of N found in this trimer of heterodimers contained variable levels of structural disorder, with residues 28–31, 116–121, 186–189, and 372–383 in chain A, residues 28–31, 118, 182–187, 241–247, 346–347, and 370–383 in chain B, and residues 28–42, 61–64, 79–153, 169–199, 239–248, and 369–383 in chain C being located within regions with missing electron density. Similarly, 12 to 14 of the C-terminal residues of the three chains of the PXD were missing in the trimer of heterodimers structure (PDB ID: 4CO6) [62].
Fig. 5.
3D Crystal Structure Representation of Nipah Nucleocapsid with Disorder Annotated in Red. The region in pink (around location 315) represents the area that have larger predicted disorder residues in Bangladesh-India 2007-12 strains when compared to that of Malaysia-Singapore 1998–9.
3.5. Nipah phylogenetic and disorder analyses
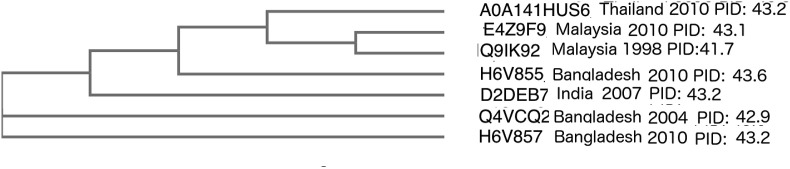
Phylogenetic analyses of NiV have already been performed in the past (1,4). For instance, Lo Presti et al. used the NiV N protein in the construction of a phylogenetic tree that suggests that NiV first entered Southeast Asia around 1947. We have made a phylogenetic analysis to compliment our disorder study as seen in Fig. 6 . Fig. 6 is able to reveal results that would have been impossible to obtain using disorder or phylogenetic method alone. We are able to see that the Malaysia strains cluster together with strains from nearby regions such as Thailand regardless of differences in both N disorder (PID) and year. This reasserts past observations of co-evolutions among strains from different regions ie. Bangladesh-India and Malaysia [1].
Fig. 6.
Phylogenetic tree based on N protein of selected NiV strains with disorder. accession code, location, year and PID (disorder level) denoted.
4. Discussion
4.1. Phylogenetic and disorder evidence of co-evolutions and possible roles of migratory bats
We have demonstrated the presence of a correlation between the Nipah virulence and its N protein disorder. The virulence of the Nipah virus can be traced to the year that the virus sample was obtained even more so than the geographic localization from which the sample was retrieved. For example, samples taken from Malaysia and Thailand in 2008 and 2010 have PID values closer to those of samples from Bangladesh-India 2007/8–12 than to those of Bangladesh-India 2001–5 and Malaysia-Singapore 1998–9, despite the fact Malaysia is in close proximity to Thailand. Without phylogenetic analysis, it would be easy to misinterpret the results by assuming that migratory bats may have brought the various strains of different N PID from elsewhere. Our combined disorder and phylogenetic analysis says, however, that this is not necessarily the case. As seen in Fig. 6, the Thailand 2010, Malaysia 2010 and 1998 strains are all clustered together despite huge differences in year and PID. Thailand and Malaysia are in close geographic proximity together. This basically implies that the strains co-evolved at distinct locations (i.e. Southeast Asia and Bangladesh) as already suggested by others [1,4], even if the various strains in separate locations assume different N disorder as seen in Fig. 6.
Strangely, though, the phylogenetic-disorder analysis also reveals that there is room for the role of migratory bats as Fig. 6 shows that a specific strain from Bangladesh-2008 (H6V855) is closer to the strains from Southeast Asia than many of its counterparts from Bangladesh-India, regardless of year and PID. This last observation suggests that there is some circulation of strains over the long distances, presumably with the help of migratory bats. A previous study, using GPS, supports this hypothesis by showing that bats can travel a round trip of over 1000 km in just one month [63]. While our study presents some cases of crossover strains from migratory bats, it also conversely shows also the strong presence of co-evolution as explained above. This study therefore paints a picture of the virus strains facing various but often similar evolutionary pressures at distinct geographic regions. The pressures could have arisen from factors which are the results of varying preferential modes of transmission dependent on the species of bat hosts and other host types such as pigs and humans. The pressures are therefore evolutionarily manifested as the differences in N PID as we shall re-visit later.
4.2. C-terminus of N protein touches P protein: the focal point
The N, P, and L proteins play important roles in the replication of the viral RNA [64,65]. The N protein when bound to the P protein, encapsidates the viral RNA during the replication of the latter. When P binds to N, the complex stabilizes in order to have greater affinity for the viral RNA. Also, when P protein binds to N protein, it induces the binding of the L protein close to the binding region of the N protein, and together the N–P-L complex functions as an RNA polymerase during the viral replication [52,64].
The fact that most of the areas affected by strain-specific mutations lie near the C-terminus of N protein (Fig. 3, Fig. 4, Fig. 5), which represent or are located in close proximity to the regions that bind to the P protein (Fig. 5), suggests that the greater disorder of the more virulent strains may be involved in modulating the aforementioned functions of the viral proteins N, P, and L. It is likely that the regions of greater disorder found in the more virulent strains, such as that shown in pink in Fig. 5, help to recognize and assist in the binding of N to P and perhaps, also to L. A more efficient replication process will mean greater viral load that makes the strain more virulent.
4.3. Virulence arises from “Trojan horse” replication: reproducibility of past research
We need to be reminded that correlations between the inner shell and the virulence of the virus have been determined not just in Nipah virus as we have shown in the current study, but also in Ebola virus and flaviviruses, such as dengue and yellow fever viruses [[10], [11], [30], [67]]. This is a reflection of the reliability of the applications of PONDR® VLXT for the analysis of viral shell proteins and is based on the fact that viral shells often have similar functions across the different viral species. While high disorder at the outer shell represents a way of immune evasion that involves the inability to neutralize the virus easily, greater disorder at the inner shell represents a different means of the immune evasion. The former has been found to be a “true viral shapeshifters” that includes the HIV, whereas the latter class has been describe as “Trojan horses”, which are the viral shapeshifters of a different kind. It is named a “Trojan horse” as its strategy to evade the host immune system involves rapid reproduction before the immune system can recognize the foreign invader.
4.4. Empirical evidence of a correlation between the disorder status of N protein and NiV virulence in the wild
Interestingly, past experiments using mutations of N proteins of NiV cousins, canine distemper virus (CDV) and MeV, have shown that greater disorder at the Ntail cause the viruses to be more virulent to ferrets in a laboratory [64]. While this was done experimentally using MeV and CDV in a laboratory setting, our results show for the first time that this also can be the case for NiV, but using an empirical analysis of NiV in the wild. An additional interesting note is the fact that mutations that affect the pathogenesis of the virus is found further upstream to location 315 as illustrated by Fig. 3, Fig. 4, Fig. 5, not noticed by any previous literature. Apparently, greater disorder around 315 facilitates easier binding of the P protein to the C-terminus of N protein.
The findings above are a reflection of the way the virus evolves. While they obviously do not provide all the answers, these observations do provide hints of how we could go about seeking answers to some of the questions involving Nipah evolution. We know that the 1998-9 human outbreak in Malaysia-Singapore involved mainly transmission by close contacts with infected pigs. The outbreaks in Bangladesh-India, however, involved patients consuming virus-laden date palm sap or human-to-human transmission [1,2]. Little is currently known about bat-to-bat transmission in the wild. It is likely that consideration of the variability in PIDs of N proteins could also provide an evolutionary clue on Nipah transmission and its intricate link to virulence.
4.5. Evidence of a link between NiV transmission modes, virulence, and shell disorder
The fact that Nipah strain from 1998 to 9 has a PID value for N protein of 41.73 ± 0.01%, whereas those of N proteins from the 2001–5 and 2007-12 strains are increasing to 42.67 ± 0.27% and 43.32 ± 0.24%, suggests that the differences in the disorder levels of N protein of the Malaysia-Singapore and Bangladesh-India strains can be associated with the differences in the preferred modes of transmission of NiV. Such findings are in line with the results of previous studies where it has been shown that changes in the disorder of the shells can be used to predict the mode of transmission of coronaviruses, including SARS-CoV and MERS-CoV [25,26]. In those studies, we have found that the more disordered the shells of coronaviruses are, the greater the contribution of the respiratory transmission and the lesser the fecal-oral transmission component. The results of the current research seem to extend this finding from coronaviruses to NiV (and, probably, to other RNA viruses from Paramyxoviridae family). In fact, we can see that the lower PID level of N protein from the 1998-9 strain allows greater fecal-oral component, such that contacts with fecal and bodily fluid of pigs provide for easier spread of the virus to the farmers. The higher PIDs of N protein in the 2001-12 strains, on the other hand, facilitate greater chances of human-to-human transmission by respiratory contacts. Further evidence of links between the shell disorder and modes of transmission can also be found in other viruses, including HIV and other retroviruses.
It should be noted that we had previously been able to establish a more comprehensive relationship between coronavirus shell disorder and modes of transmission because much was already known about the behaviors of various porcine coronaviruses. On the contrary, little is currently known about modes of NiV transmission among bats in the wild. The results of the analysis reported in this paper suggest the ways we can further study NiV in both the wild and laboratories using disorder levels of N protein as a useful proxy. Related to this, the results reported in this paper highlight one reason for the differences among the virulence of various NiV strains is likely to be arising from the evolutionary pressures pertaining to the fitness of a particular mode of transmission. For example, a bat-to-bat transmission may favor respiratory transmission, whereas bat-to-pig and pig-to-human transmissions could favor the fecal-oral route. Fig. 7 re-affirms that this link is not confined to NiV but extends to some of the NiV's cousins with respect to the disorder levels of not just N protein but also that of the M protein.
Fig. 7.
Relationship Between Shell Disorder and Modes of Transmission. The level of disorder (PID) rises as the fecal-oral component decreases. It should be noted that HeV is known to spread by contact with bodily fluid and therefore has a higher fecal-oral component. Depending of the strain, NiV can spread via respiratory and fecal-oral routes including close contacts with bodily fluids. MeV and HIV are spread mainly by respiratory and sexual modes respectively. For comparative purposes, only maximal PIDs found for a virus and protein are used.
4.6. Links between shell disorder and modes of viral transmission across the cousins
Fig. 7 compares the PID values for the N and M proteins of NiV and some of its cousins, with HIV-1, which is an unrelated virus, being used as a reference. The various NiV cousins are chosen to make interesting comparison as they have known to have differing modes of transmission. HeV has infected horses from bats via fecal materials and horse-to-horse transmissions usually require close contacts that involve bodily fluids (6). NiV spread is more complicated. While NiV transmission from bats to humans or pigs usually involves consumption of fecal or salivary contaminated fruits or palm date saps, the human transmission of NiV Malaysia-Singapore 1998-9 strain involved only close human contacts with infected pigs via bodily fluids, unlike the evidence of Bangladesh-India human-to-human transmission, which is usually indicative of the presence of the respiratory mode [10,25,26]. In sharp contrast, MeV spreads virtually entirely by respiratory mode [7,66].
A look at Fig. 7 allows us to see the link between the viral shell disorder and the modes of viral transmission, especially across the NiV relatives. The viral shells include both N and M proteins. A noticeable trend is the increase in the disorder of the shell proteins as the components of the respiratory and fecal-oral modes increases and decrease respectively. This is consistent with findings with regard to other viruses in the past. Viruses that rely heavily on fecal-oral transmission tend to have harder shells that are needed to persistent in the environment for a longer period of time. On the other hand, viruses that depends on respiratory modes of transmission have moderate disorder on their outer shells, but are likely to have highly disordered inner shells. Sexually transmitted viruses, such as HIV-1, on the other hand, could have highly disordered outer shell and have inner shells with reasonably high disorder levels. These are the trends we are seeing in Fig. 7.
This analysis can be further extended by considering a correlation of the disorder status N proteins from various NiV and the transmission modes of these strains. The Bangladesh NiV strains that are characterized by higher respiratory transmission component have highest PID values. Furthermore, while the maximal PID value determined among the N proteins of the various NiV strains is higher than that of the HeV N PID, a comparison of Table 1 and Fig. 7 shows that the N protein from the Malaysia-Singapore NiV strain has an PID value (41.73 ± 0.01%) comparable to that of the N protein from HeV (PID: 41%). In agreement with their lower PID values, both the Malaysia-Singapore NiV and HeV have greater fecal-oral components, since the transmissions of both viruses require close contacts in the presence of bodily fluids. This refers to the horse-to-horse and pig-to-human transmissions of the HeV and Malaysia-Singapore 1998-9 NiV strain, respectively. No human-to-human spread was observed in this NiV strain.
The MeV, however, represents an interesting contrast, as, unlike HeV and NiV, it relies mostly on respiratory transmission for its spread. Fig. 7 shows that PIDs of both M and N proteins from MeV are noticeably higher than those of its somewhat distant cousins shown. The maximal PID values of N proteins from HeV and NiV are 41% and 43% respectively, whereas that of the MeV is 46%. Similarly, the maximal PID values for MeV, HeV, and NiV are 20%, 8%, and 14%, respectively. These observations suggest that the evolutionary need of persistence in the environment (which is associated with the disorder levels of the viral N and M proteins) depends on the modes of transmission that the viruses rely on.
HIV-1 is, of course, an unrelated virus, but it makes an interesting reference point in Fig. 7, as it is sexually transmitted. It has been theorized that the highly disordered outer shell of HIV-1 makes it difficult for an effective vaccine to be found. As we can see from Fig. 7, none of the PID values for the M proteins of the paramyxoviruses are higher than that of HIV. Therefore, the successful development effective vaccines is likely to be feasible for NiV, MeV, and HeV. In agreement with this hypothesis, the vaccines for MeV and HeV have been already elaborated [6,7].
5. Conclusion
There have been few studies on the virulence and infectivity of NiV. The results of this paper correlate infectivity and virulence of NiV to the levels of intrinsic disorder in the N protein and hints to the relationship of disorder levels to modes of viral transmission. These findings have multiple implications for the future research into not just NiV virulence and infectivity, but also that of related viruses, as the same protein is likely to be playing the same role among related viruses. Needless to say, the Paramyxoviridae family is an important class of viruses that includes some medically interesting ones, such as MeV, CDV, HeV and, of course, NiV.
It should be understood that the differences in disorder of the N protein and the behavior of the virus in terms of virulence and infectivity can be related to the way the virus evolves. We have seen that virulence in NiV is the result of the virus's “Trojan horse” strategy of producing viral particles rapidly before the host immune system is able to detect and neutralize them. The greater virulence is likely to have arisen from the higher viral load of strains that comes with greater N protein disorder.
Given that the level of N protein disorder is associated with NiV virulence, this research may pave a way for some new strategies for the development of attenuated vaccines, not just against HiV, but also against viruses related and unrelated to NiV. Another possible application is the potential of disorder analysis of N protein disorder as a tool for analyzing the outbreaks and deciding policies and approaches for preventing further spread. The detection of links between modes of transmission and shell disorder will also be particularly helpful in the development of tools to anticipate the nature of an outbreak via molecular analysis of the viral shells. Since little is currently known about NiV virulence and infectivity, any new knowledge pertaining to the potential virulence and infectivity of a virus in an outbreak can prove to be invaluable.
Lastly, the results reported here suggest an answer to an age-old biomedical mystery, where it is not understood why viruses become attenuated, when they are passed to the cells that are from different animal species and of different cell types for many generations as in the case of the preparation of Sabin polio vaccine [66]. Our results seem to hint that when the host cell is changed, there will be a switch of preference for a different mode of transmission as the cell type and animal species have been change, and the in process the virus becomes attenuated. The “preference” is likely the result of the way the virus evolves or has evolved to optimally adjust to the cell types and the animal species.
Funding
JAF is funded in part by NSF DBI0939454, BEACON Center for Evolution in Action.
Declaration of competing interest
GKMG is an independent researcher, and the owner of Goh's BioComputing, Singapore. GKMG has also written a book on a related subject. The authors have no other potential conflict of interests.
Contributor Information
Gerard Kian-Meng Goh, Email: gohsbiocomputing@yahoo.com.
A. Keith Dunker, Email: kedunker@iupui.edu.
James A. Foster, Email: foster@uidaho.edu.
Vladimir N. Uversky, Email: vuversky@health.usf.edu.
References
- 1.Angeletti S., Lo Presti A., Cella E., Ciccozzi M. Molecular epidemiology and phylogeny of nipah virus infection: a mini review. Asian Pac J Trop Med. 2016;9:630–634. doi: 10.1016/j.apjtm.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang B.S.P., Lim T.C.C., Wang L. Nipah virus infection. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01875-17. e1875-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mire C.E., Satterfield B.A., Geisbert J.B., Agans K.N., Borisevich V., Yan L., Chan Y.P., Cross R.W., Fenton K.A., Broder C.C. Pathogenic differences between nipah virus Bangladesh and Malaysia strains in primates: implications for antibody therapy. Sci. Rep. 2016;6:30916. doi: 10.1038/srep30916. Mire et al. 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo Presti A., Cella E., Giovanetti M., Lai A., Angeletti S., Zehender G., Ciccozzi M. Origin and evolution of nipah virus. J. Med. Virol. 2016;88:380–388. doi: 10.1002/jmv.24345. [DOI] [PubMed] [Google Scholar]
- 5.Sun B., Jia L., Liang B., Chen Q., Liu D. Phylogeography, transmission, and viral proteins of Nipah virus. Virol. Sin. 2018;33:385–393. doi: 10.1007/s12250-018-0050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weingartl H.M. Hendra and nipah viruses: pathogenesis, animal models and recent breakthroughs in vaccination. Vaccine Dev. Ther. 2015;2015:59–74. [Google Scholar]
- 7.Acheson Acheson N. Wiley; 2007. Fundamentals of Molecular Virology. [Google Scholar]
- 8.Kulkarni D.D., Tosh C., Venkatesh G., Senthil Kumar D. Nipah virus infection: current scenario. Indian J. Virol. 2013;24:398–408. doi: 10.1007/s13337-013-0171-y. Kalkani 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Witt de Wit E., Munster V.J. Animal models of disease shed light on nipah virus pathogenesis and transmission. J. Pathol. 2015;235:196–205. doi: 10.1002/path.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh G.K. Simplicity Research Institute; Singapore: 2017. Viral Shapeshifters: Strange Behavoirs of Hiv and Other Viruses. [Google Scholar]
- 11.Goh G.K., Dunker A.K., Uversky V.N. Correlating flavivirus virulence and levels of intrinsic disorder in shell proteins: protective roles vs. Immune evasion. Mol. Biosyst. 2016;12:1881–1891. doi: 10.1039/c6mb00228e. [DOI] [PubMed] [Google Scholar]
- 12.Goh G.K., Dunker A.K., Uversky V.N. A comparative analysis of viral matrix proteins using disorder predictors. Virol. J. 2008;5:126. doi: 10.1186/1743-422X-5-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh G.K., Dunker A.K., Uversky V.N. Shell disorder, immune evasion and transmission behaviors among human and animal retroviruses. Mol. Biosyst. 2015;11:2312–2323. doi: 10.1039/c5mb00277j. [DOI] [PubMed] [Google Scholar]
- 14.Dunker A.K., Lawson J.D., Brown C.J., Williams R.M., Romero P., Oh J.S., Oldfield C.J., Campen A.M., Ratliff C.M., Hipps K.W. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 15.Uversky V.N., Gillespie J.R., Fink A.L. Why are "natively unfolded" proteins unstructured under the physiological conditions? Proteins Struct. Funct. Genet. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. Uversky et al. 2000. [DOI] [PubMed] [Google Scholar]
- 16.Wright P.E., Dyson H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 17.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. Tompa, 2002) [DOI] [PubMed] [Google Scholar]
- 18.He B., Wang K., Liu Y., Xue B., Uversky V.N., Dunker A.K. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 19.Meng F., Uversky V.N., Kurgan L. Computational prediction of intrinsic disorder in proteins. Curr. Protein Pept. Sci. 2017;88(2):16 11–1612 16 14. doi: 10.1002/cpps.28. Meng et al. 2017. [DOI] [PubMed] [Google Scholar]
- 20.Peng Z.L., Kurgan L. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci. 2012;13:6–18. doi: 10.2174/138920312799277938. [DOI] [PubMed] [Google Scholar]
- 21.Garner E., Romero P., Dunker A.K., Brown C., Obradovic Z. Predicting binding regions within disordered proteins. Genome Informatics. 1999;10:41–50. [PubMed] [Google Scholar]
- 22.Li X., Romero P., Rani M., Dunker A.K., Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 23.Romero P., Obradovic Z., Li \ X., Garner E.C., Brown C.J., Dunker A.K. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Goh G.K., Dunker A.K., Uversky V.N. Detection of links between Ebola nucleocapsid and virulence using disorder analysis. Mol. Biosyst. 2015;11:2337–2344. doi: 10.1039/c5mb00240k. [DOI] [PubMed] [Google Scholar]
- 25.Goh G.K., Dunker A.K., Uversky V.N. Prediction of intrinsic disorder in MERS-CoV/HCoV-EMC supports a high oral-fecal transmission. PLoS Curr. 2013;5 doi: 10.1371/currents.outbreaks.22254b58675cdebc256dbe3c5aa6498b. Goh et al. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goh G.K., Dunker A.K., Uversky V.N. Understanding viral transmission behavior via protein intrinsic disorder prediction: Coronaviruses. J Pathog. 2012;2012:738590. doi: 10.1155/2012/738590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue B., Williams R.W., Oldfield C.J., Goh G.K., Dunker A.K., Uversky V.N. Viral disorder or disordered viruses: do viral proteins possess unique features? Protein Pept. Lett. 2010;17:932–951. doi: 10.2174/092986610791498984. Xue et al. 2010. [DOI] [PubMed] [Google Scholar]
- 28.Goh G.K., Dunker A.K., Uversky V.N. Protein intrinsic disorder and influenza virulence: the 1918 H1N1 and H5N1 viruses. Virol. J. 2009;6:69. doi: 10.1186/1743-422X-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goh G.K., Dunker A.K., Uversky V.N. Protein intrinsic disorder toolbox for comparative analysis of viral proteins. BMC Genomics. 2008;9(Suppl. 2):S4. doi: 10.1186/1471-2164-9-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goh G.K., Dunker A.K., Foster J.A., Uversky U.N. HIV vaccine mystery and viral shell disorder. Biomolecules. 2019;9:178. doi: 10.3390/biom9050178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue B., Blocquel D., Habchi J., Uversky A.V., Kurgan L., Uversky V.N., Longhi S. Structural disorder in viral proteins. Chem. Rev. 2014;114:6880–6911. doi: 10.1021/cr4005692. Xue 2014. [DOI] [PubMed] [Google Scholar]
- 32.Park M.S., Shaw M.L., Munoz-Jordan J., Cros J.F., Nakaya T., Bouvier N., Palese P., Garcia-Sastre A., Basler C.F. Newcastle disease virus (ndv)-based assay demonstrates interferon-antagonist activity for the ndv v protein and the nipah virus v, w, and c proteins. J. Virol. 2003;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basler C.F. Nipah and hendra virus interactions with the innate immune system. Curr. Top. Microbiol. Immunol. 2012;359:123–152. doi: 10.1007/82_2012_209. Basler et al. 2012) [DOI] [PubMed] [Google Scholar]
- 34.Shaw M.L. Henipaviruses employ a multifaceted approach to evade the antiviral interferon response. Viruses. 2009;1:1190–1203. doi: 10.3390/v1031190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin B., Rahmeh A.A., Whelan S.P. Mechanism of rna synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 2012;31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnheiter H., Davis N.L., Wertz G., Schubert M., Lazzarini R.A. Role of the nucleocapsid protein in regulating vesicular stomatitis virus rna synthesis. Cell. 1985;41:259–267. doi: 10.1016/0092-8674(85)90079-0. Amheiter et al, 2012) [DOI] [PubMed] [Google Scholar]
- 37.Desfosses A., Goret G., Farias Estrozi L., Ruigrok R.W., Gutsche I. Nucleoprotein-rna orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J. Virol. 2011;85:1391–1395. doi: 10.1128/JVI.01459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruigrok R.W., Crepin T. Nucleoproteins of negative strand rna viruses; RNA binding, oligomerisation and binding to polymerase co-factor. Viruses. 2010;2:27–32. doi: 10.3390/v2010027. Ruigrok et al. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tawar R.G., Duquerroy S., Vonrhein C., Varela P.F., Damier-Piolle L., Castagne N., MacLellan K., Bedouelle H., Bricogne G., Bhella D. Crystal structure of a nucleocapsid-like nucleoprotein-rna complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. Tavar et al. 2009. [DOI] [PubMed] [Google Scholar]
- 40.Albertini A.A., Wernimont A.K., Muziol T., Ravelli R.B., Clapier C.R., Schoehn G., Weissenhorn W., Ruigrok R.W. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 41.Green T.J., Zhang X., Wertz G.W., Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 42.Karlin D., Ferron F., Canard B., Longhi S. Structural disorder and modular organization in paramyxovirinae N and P. J. Gen. Virol. 2003;85:3239–3252. doi: 10.1099/vir.0.19451-0. [DOI] [PubMed] [Google Scholar]
- 43.Bourhis J.M., Johansson K., Receveur-Brechot V., Oldfield C.J., Dunker K.A., Canard B., Longhi S. The C-terminal domain of measles virus nucleoprotein belongs to the class of intrinsically disordered proteins that fold upon binding to their physiological partner. Virus Res. 2004;99:157–167. doi: 10.1016/j.virusres.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Longhi S. Nucleocapsid structure and function. Curr. Top. Microbiol. Immunol. 2009;329:103–128. doi: 10.1007/978-3-540-70523-9_6. Longhi, 2009. [DOI] [PubMed] [Google Scholar]
- 45.Longhi S., Oglesbee M. Structural disorder within the measles virus nucleoprotein and phosphoprotein. Protein Pept. Lett. 2010;17:961–978. doi: 10.2174/092986610791498894. [DOI] [PubMed] [Google Scholar]
- 46.Jensen M.R., Communie G., Ribeiro E.A., Jr., Martinez N., Desfosses A., Salmon L., Mollica L., Gabel F., Jamin M., Longhi S. Intrinsic disorder in measles virus nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9839–9844. doi: 10.1073/pnas.1103270108. Jensen et al. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habchi J., Longhi S. Structural disorder within paramyxovirus nucleoproteins and phosphoproteins. Mol. Biosyst. 2012;8:69–81. doi: 10.1039/c1mb05204g. Habachi et al. 2012. [DOI] [PubMed] [Google Scholar]
- 48.Longhi S. The measles virus N(tail)-xd complex: an illustrative example of fuzziness. Adv. Exp. Med. Biol. 2012;725:126–141. doi: 10.1007/978-1-4614-0659-4_8. Longhi et al. 2012. [DOI] [PubMed] [Google Scholar]
- 49.Communie G., Habchi J., Yabukarski F., Blocquel D., Schneider R., Tarbouriech N., Papageorgiou N., Ruigrok R.W., Jamin M., Jensen M.R. Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of Hendra virus. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003631. Communie et al. 2013. e1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baronti L., Erales J., Habchi J., Felli I.C., Pierattelli R., Longhi S. Dynamics of the intrinsically disordered c-terminal domain of the Nipah virus nucleoprotein and interaction with the x domain of the phosphoprotein as unveiled by nmr spectroscopy. Chembiochem. 2015;16:268–276. doi: 10.1002/cbic.201402534. Boronti et al. 2013. [DOI] [PubMed] [Google Scholar]
- 51.Erales J., Blocquel D., Habchi J., Beltrandi M., Gruet A., Dosnon M., Bignon C., Longhi S. Order and disorder in the replicative complex of paramyxoviruses. Adv. Exp. Med. Biol. 2015;870:351–381. doi: 10.1007/978-3-319-20164-1_12. [DOI] [PubMed] [Google Scholar]
- 52.Habchi J., Longhi S. Structural disorder within paramyxoviral nucleoproteins and phosphoproteins in their free and bound forms: from predictions to experimental assessment. Int. J. Mol. Sci. 2015;16:15688–15726. doi: 10.3390/ijms160715688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Longhi S. Structural disorder within paramyxoviral nucleoproteins. FEBS Lett. 2015;589:2649–2659. doi: 10.1016/j.febslet.2015.05.055. [DOI] [PubMed] [Google Scholar]
- 54.Longhi S., Bloyet L.M., Gianni S., Gerlier D. How order and disorder within paramyxoviral nucleoproteins and phosphoproteins orchestrate the molecular interplay of transcription and replication. Cell. Mol. Life Sci. 2017;74:3091–3118. doi: 10.1007/s00018-017-2556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourhis J.M., Canard B., Longhi S. Structural disorder within the replicative complex of measles virus: functional implications. Virology. 2006;344:94–110. doi: 10.1016/j.virol.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 56.Cheng Y., Oldfield C.J., Meng J., Romero P., Uversky V.N., Dunker A.K. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oldfield C.J., Cheng Y., Cortese M.S., Romero P., Uversky V.N., Dunker A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 58.Rajagopalan Rajagopalan K., Mooney S.M., Parekh N., Getzenberg R.H., Kulkarni P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011;112:3256–3267. doi: 10.1002/jcb.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Omi-Furutani M., Yoneda M., Fujita K., Ikeda F., Kai C. Novel phosphoprotein-interacting region in nipah virus nucleocapsid protein and its involvement in viral replication. J. Virol. 2010;84:9793–9799. doi: 10.1128/JVI.00339-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meszaros B., Simon I., Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000376. e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dosztanyi Z., Meszaros B., Simon I. Anchor: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yabukarski F., Lawrence P., Tarbouriech N., Bourhis J.M., Delaforge E., Jensen M.R., Ruigrok R.W., Blackledge M., Volchkov V., Jamin M. Structure of nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat. Struct. Mol. Biol. 2014;21:754–759. doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
- 63.Weller T.J., Castle K.T., Liechti F., Hein C.D., Schirmacher M.R., Cryan P.M. First direct evidence of long-distance seasonal movements and hibernation in a migratory bat. Sci. Rep. 2016;6:34585. doi: 10.1038/srep34585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thakkar V.D., Cox R.M., Sawatsky B., da Fontoura Budaszewski R., Sourimant J., Wabbel K., Makhsous N., Greninger A.L., von Messling V., Plemper R.K. The unstructured paramyxovirus nucleocapsid protein tail domain modulates viral pathogenesis through regulation of transcriptase activity. J. Virol. 2018;92 doi: 10.1128/JVI.02064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ong S.T., Yusoff K., Kho C.L., Abdullah J.O., Tan W.S. Mutagenesis of the nucleocapsid protein of Nipah virus involved in capsid assembly. J. Gen. Virol. 2009;90:392–397. doi: 10.1099/vir.0.005710-0. [DOI] [PubMed] [Google Scholar]
- 66.Sompayrac L. Jones and Barlett Publishers; Sudbury, Massachusetts: 2002. How Pathogenic Viruses Work. [Google Scholar]
- 67.Goh G.K., Dunker A.K., Foster J.A., Uversky V.N. Zika and flavivirus shell disorder: Virulence and fetal morbidity. Biomolecules. 2019;9(11):E710. doi: 10.3390/biom9110710. [DOI] [PMC free article] [PubMed] [Google Scholar]