Abstract
All viruses that carry a positive-sense RNA genome (+RNA), such as picornaviruses, hepatitis C virus, dengue virus, and SARS- and MERS-coronavirus, confiscate intracellular membranes of the host cell to generate new compartments (i.e., replication organelles) for amplification of their genome. Replication organelles (ROs) are membranous structures that not only harbor viral proteins but also contain a specific array of hijacked host factors that create a unique lipid microenvironment optimal for genome replication. While some lipids may be locally synthesized de novo, other lipids are shuttled towards ROs. In picornavirus-infected cells, lipids are exchanged at membrane contact sites between ROs and other organelles. In this paper, we review recent advances in our understanding of how picornaviruses exploit host membrane contact site machinery to generate ROs, a mechanism that is used by some other +RNA viruses as well.
Keywords: picornavirus, replication organelle, membrane contact site, lipid, phosphatidylinositol 4-phosphate, cholesterol
Trends
Picornaviruses create replication organelles with a unique protein and lipid composition to amplify their genome.
Picornaviruses hijack membrane contact site machinery to shuttle lipids to their replication organelles.
Picornaviruses from different genera employ a cholesterol/PI4P counterflux mechanism to accumulate cholesterol at replication organelles.
Picornaviruses and Their Life Cycle
The Picornaviridae constitute a large family of positive-sense, single-stranded RNA (+RNA) viruses comprising many human and animal pathogens associated with a wide spectrum of diseases (Box 1 ) (reviewed in [1]). The family currently consists of 29 genera (http://www.ictvonline.org). Most studies have focused on the Enterovirus genus, which includes poliovirus, coxsackie A and B viruses, and human rhinoviruses; these viruses have a great clinical and economic impact (e.g., 2, 3, 4, 5, 6).
Box 1. Picornavirus Pathogenesis.
The family Picornaviridae currently consists of 29 genera (http://www.ictvonline.org), which include many clinically and economically important human and animal pathogens. The genus Enterovirus is the largest genus. Examples of human enteroviruses include polioviruses (causative agents of poliomyelitis), coxsackie- and echoviruses (main causes of viral meningitis, but which are also associated with several other conditions such as herpangina, conjunctivitis, and myocarditis), rhinoviruses (causes of common cold, but also exacerbations of asthma and chronic obstructive pulmonary disease), enterovirus-D68 (cause of respiratory illness), and enterovirus-A71 (important cause of hand-foot-and-mouth disease, but which can also cause flaccid paralysis). The genus Cardiovirus includes encephalomyocarditis virus, which can infect many hosts, including rodents and pigs, and cause encephalomyelitis and reproductive failure, as well as saffold virus, a human virus that has not been convincingly associated with a disease. The genus Aphthovirus includes the foot-and-mouth disease virus, which infects a range of cloven-hooved mammals and which causes lesions, but sometimes also myocarditis and death. Other well-known human picornaviruses are the hepatitis A virus (Hepatovirus genus), which causes hepatitis, human parechovirus (Parechovirus genus), which causes respiratory illness, gastroenteritis, myocarditis and encephalitis, and aichivirus (Kobuvirus genus), which causes gastroenteritis.
Enteroviruses are small, nonenveloped viruses with a +RNA genome of ∼7.5 kb. The genome encodes a single open reading frame divided into the regions P1, P2, and P3. The P1 region encodes the four capsid proteins (VP1–VP4), while P2 and P3 encode the nonstructural proteins, which mediate genome replication. P2 encodes the viral proteinase 2Apro, the viroporin 2B, and the RNA helicase 2C, while P3 encodes 3A, which functions in viral RNA replication and in membrane remodeling, the primer for replication 3B, the viral proteinase 3Cpro, and the RNA-dependent RNA polymerase 3Dpol.
The life cycle of enteroviruses starts with receptor-mediated endocytosis of the virion and release of the genome into the cytoplasm of the host cell (reviewed in [7]). Here, the genome is translated into a single large polyprotein, which is subsequently proteolytically processed by the viral proteinases 2Apro and 3Cpro to yield the individual capsid proteins, nonstructural proteins, and some stable and functional precursors such as 2BC, 3AB, and 3CD. Replication of the viral genome by the nonstructural proteins starts with synthesis of complementary negative-strand RNA molecules, which then serve as a template for the production of multiple new +RNA molecules. Newly synthesized +RNA genomes either enter another round of translation and replication or are assembled with capsid proteins into progeny virions. The new virions are released into the extracellular environment upon host cell lysis, although growing evidence suggests that virions can also be secreted via nonlytic release prior to cell lysis and transmitted in vesicles containing multiple virions 8, 9, 10, 11, 12, similar to the mode observed previously for the picornavirus hepatitis A virus (genus Hepatovirus) [13].
Enteroviruses Create ROs for Genome Replication
All +RNA viruses rearrange intracellular membranes into new membranous structures that serve as a scaffold for genome replication. These so-called ROs have been suggested to serve multiple purposes. They may concentrate viral and host proteins and/or mediate proper topology, thereby gathering all necessary components for efficient replication. In addition, the ROs are believed to protect viral RNAs against degradation by cellular RNases and/or detection by RNA sensors that trigger antiviral responses (reviewed in 14, 15, 16).
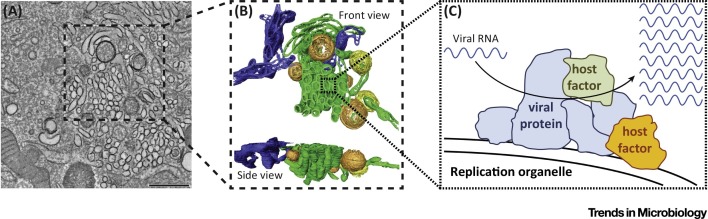
Enterovirus ROs have long been visualized as either single- or double-membrane ‘vesicles’ in two-dimensional electron microscopy studies 17, 18. However, two recent electron tomography studies on poliovirus and coxsackievirus B3 (CVB3) have revealed the three-dimensional structure of ROs (Figure 1 ) 19, 20. Interestingly, the morphology of the ROs transforms during the course of infection, indicating that ROs are dynamic structures. The first structures detected upon infection are single-membrane tubules. As infection progresses, the tubules transform into double-membrane vesicles (DMVs), which are subsequently enwrapped by more tubules to yield multilamellar structures. It remains to be established whether all these different structures support viral RNA replication. At least the tubular structures seem to do so, as the appearance of tubular ROs is concomitant with the exponential increase in viral RNA [19].
Figure 1.
The Ultrastructure of Enterovirus Replication Organelles (ROs). (A) Tomographic slice (thickness 5 nm) through a serial tomogram of a CVB3-infected cell at 5 hours post-infection. Scale bar is 500 nm. (B) Top and side views of a surface-rendered model of the region boxed in panel A showing single-membrane tubules (green), open (orange) and closed (yellow) double-membrane vesicles (DMVs), and endoplasmic reticulum (ER) (blue). (C) Schematic representation of an RO displaying viral RNA genome replication by viral and host proteins. (A, B) Adapted from [19]; the original images have been published under the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported license.
RO formation requires the viral membrane-anchored proteins 2B, 2C, and 3A, but one of the major challenges in the field is to establish how enteroviruses form ROs from host membranes. The emergence of tubular ROs coincides with the disappearance of Golgi membranes, implying that Golgi components might be involved in RO formation [19]. Additionally, the formation of DMVs and their subsequent enwrapping are reminiscent of autophagic mechanisms, suggesting that autophagy plays a role in RO transformation 19, 20. In accordance with this, a number of Golgi proteins 21, 22, 23 and the autophagy marker LC3 9, 24, 25 were found on enterovirus ROs. Importantly, LC3 has been implicated in DMV formation both in an autophagy-dependent and -independent manner [26]. Yet, ROs are not mere remnants of the Golgi or constituents of the autophagy pathway, but instead are new virus-induced organelles with a unique protein and lipid composition (reviewed in [27]).
Enteroviruses Subvert Membrane Contact Site Machinery to Create ROs
Lipids are principal components of biomembranes and critical determinants of membrane properties such as intrinsic curvature, fluidity, and charge, and they play crucial roles in recruiting a wide variety of proteins to membranes (see e.g., 28, 29, 30, 31 for review). Increasing evidence shows that, in addition to host proteins, enteroviruses hijack lipid homeostasis pathways for RO formation. Membrane contact sites (MCSs; Box 2 ) play central roles in the homeostasis of a variety of lipids. In this review, we focus on the emerging concept that enteroviruses subvert host MCS machinery to create ROs with a lipid composition optimally suited to support viral genome replication.
Box 2. Membrane Contact Sites (MCSs).
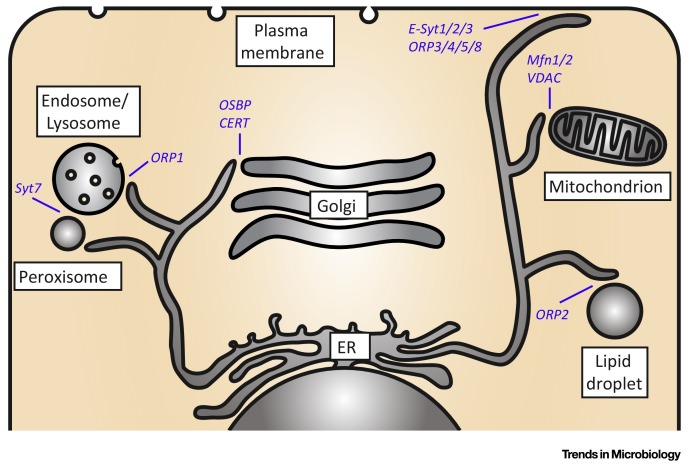
MCSs are sites where two organelles are closely juxtaposed (membranes are typically 15–60 nm apart). MCSs have important functions in signaling and in the transfer of lipids and ions between organelles [89]. Many different types of MCS have been described between a variety of organelles, including ER–Golgi, ER–lipid droplet, ER–mitochondrion, and ER–plasma membrane (PM) (Figure I ). Specialized proteins (dynamically) bridge and/or regulate the distance between opposing membranes in response to triggers (e.g., Ca2+), thus allowing spatiotemporal control of MCSs. MCS proteins include OSBP-related proteins (ORPs; these proteins regulate MCSs and shuttle lipids, including cholesterol) 71, 72 and extended synaptotagmins (E-syts; regulate ER-PM MCSs in response to Ca2+) [90], but our understanding of MCSs is currently limited, and many MCS proteins remain to be identified and characterized.
Figure I.
Examples of Membrane Contact Sites MCSs) and Their Lipid Transfer Proteins. Schematic representation of a cell and MCSs between a variety of cellular organelles. A selection of the currently limited known set of MCS proteins and the MCSs at which they operate is depicted. Abbreviations: CERT, ceramide transfer protein; E-Syt, extended synaptotagmin; Mfn, mitofusin; ORP, OSBP-related protein; OSBP, oxysterol-binding protein; Syt7, synaptotagmin VII; VDAC, voltage-dependent anion channel.
The Essential Role of PI4KB and PI4P in Genome Replication
Phosphoinositides are major determinants of organelle identity and functioning, despite accounting for only a minor fraction of the total cellular phospholipids. One of the most abundant phosphoinositides in the cell is phosphatidylinositol 4-phosphate (PI4P). Recently, PI4P emerged from simply being a precursor of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] to being a key regulator of membrane trafficking and metabolism, particularly by mediating the recruitment to membranes of effector proteins with lipid-transfer functions [32]. PI4P can be synthesized in mammalian cells by four distinct phosphatidylinositol 4-kinases (PI4Ks) [32], type II and type III that both consist of an α- and a β-isoform [33], which reside at different subcellular locations and locally synthesize PI4P. The major pool of PI4P is generated in Golgi membranes, where PI4K type IIIβ (PI4KB) is the predominant isoform 34, 35. All enteroviruses depend on PI4KB activity for genome replication 23, 36. PI4KB is accumulated at ROs by viral nonstructural proteins to locally produce high amounts of PI4P [23]. The small viral protein 3A mediates PI4KB accumulation 23, 37, 38, 39, although 2BC was recently proposed to also play a role [40]. Of note, host factor accumulation is generally denoted ‘recruitment’, although it may also result from retention, or a combination of both.
The Role of Host Proteins in PI4KB Recruitment
Despite intense investigation, the mechanism underlying PI4KB recruitment by 3A remains unclear. Although PI4KB coimmunoprecipitates with 3A, a direct interaction could until now not be shown 37, 39. Hence, 3A may recruit PI4KB indirectly via a cellular interactor. Candidate host proteins for PI4KB recruitment and their role in enterovirus genome replication are discussed below.
GBF1
Enterovirus 3A interacts with the N terminus of GBF1 (Golgi brefeldin A resistant guanine nucleotide exchange factor 1), an essential host factor of enterovirus replication 41, 42, 43. GBF1 is a guanine nucleotide exchange factor and activator of the small GTPase Arf1 (ADP-ribosylation factor 1), which regulates traffic in the secretory pathway by mediating the recruitment of downstream effectors to membranes 44, 45. Since PI4KB is an effector of Arf1 in noninfected cells [35], it was initially proposed that 3A recruits PI4KB via GBF1/Arf1 [23]. However, the 3A proteins of CVB3 and rhinovirus were shown to recruit the kinase independently of GBF1/Arf1 37, 38.
With the finding that PI4KB recruitment is not dependent on GBF1, the role of GBF1 in enterovirus replication remains elusive. Based on the sensitivity of enteroviruses to brefeldin A (BFA), a small molecule inhibitor targeting the enzymatic function of GBF1, it was long believed that enterovirus replication requires GBF1 activity 21, 46. In uninfected cells, activation of Arf1 by GBF1 among others serves to recruit the COPI vesicle coat complex [47]. However, enterovirus ROs are gradually depleted of COPI 23, 38, which exemplifies the selective recruitment of host factors to ROs. Furthermore, this finding implies that the function of GBF1 in enterovirus replication may not be synonymous to its normal cellular function. In agreement with this, other studies showed that enterovirus replication is not sensitive to Arf1 depletion and that overexpression of different Arf proteins cannot relieve virus replication from BFA inhibition 42, 43. Additionally, overexpression of a truncated variant of GBF1 containing an intact N terminus, but lacking the catalytic Sec7 domain, could partially rescue poliovirus replication from the inhibitory effect of BFA [48]. Whatever function GBF1 plays in enterovirus replication, it might be related to the functioning rather than the formation of ROs, because poliovirus proteins expressed in the presence of BFA induced membrane rearrangements indistinguishable from those found in infected cells [43]. A deeper understanding of the role of GBF1 in enterovirus replication might also prove insightful for the as yet unclear role of GBF1 in the life cycle of other distantly related (+)RNA viruses, such as hepatitis C virus and coronaviruses 49, 50.
ACBD3
The Golgi-resident protein ACBD3 (acyl-CoA-binding domain-containing protein 3) was recently identified as another interaction partner of PI4KB [51]. Affinity purification studies demonstrated that multiple enterovirus 3A proteins, including those of poliovirus, coxsackievirus, and rhinovirus, interact with ACBD3, raising the possibility that ACBD3 is the cofactor required for PI4KB recruitment to the ROs [39]. However, PI4KB was still recruited to ROs in CVB3-infected cells depleted of ACBD3, implying that enteroviruses recruit PI4KB independently of ACBD3 37, 38. To date, the importance of ACBD3 for enterovirus infection remains elusive, since two different studies demonstrated that ACBD3 depletion did not inhibit, but instead mildly augmented, replication of CVB3 and poliovirus 37, 52, while in another report poliovirus replication was inhibited, albeit modestly, upon ACBD3 knockdown [39].
C10orf76
C10orf76 is a protein with an unknown function that has been identified as a PI4KB interactor by two separate affinity-purification mass spectrometry (AP-MS) studies 53, 54. A very recent study discovered C10orf76 as a novel enterovirus host factor [55]. In noninfected cells, C10orf76 associated with PI4KB and mediated PI4KB-dependent PI4P synthesis at Golgi membranes. However, C10orf76 was only required for the genome replication of coxsackievirus A10, but not coxsackievirus B1, suggesting a species-specific dependence on this host factor. Future studies should address to which extent different enteroviruses require C10orf76, and whether C10orf76 acts as cofactor in PI4KB recruitment also in infected cells.
In conclusion, a direct interaction between viral 3A and PI4KB could not be shown and – although only a limited number of PI4KB interaction partners have been investigated for their involvement in kinase recruitment to RO – a pathway for indirect PI4KB recruitment has also not been identified yet. Thus, the detailed molecular events underlying the mechanism of PI4KB recruitment remain to be elucidated.
PI4P-Dependent Cholesterol Recruitment to ROs via MCSs
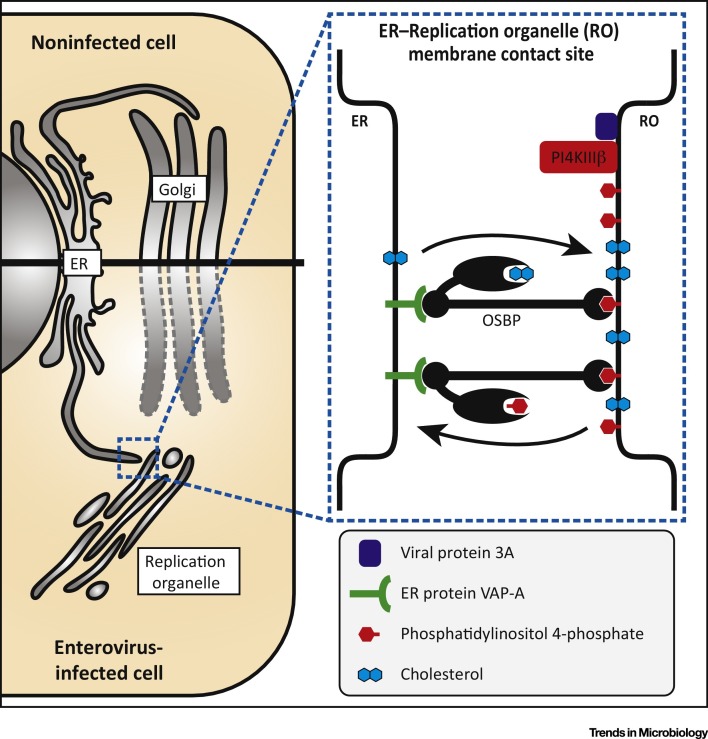
What is the purpose of PI4P in enterovirus replication? A first indication that PI4P might serve as a protein-docking site on RO membranes was the finding that CVB3 3Dpol specifically bound to PI4P in an in vitro assay [23], but this observation awaits validation in infected cells. By recruiting proteins, PI4P could assist in concentrating components required for genome replication and/or RO formation. Notably, PI4P can recruit cellular proteins that have a PI4P-binding pleckstrin homology (PH) domain, including various lipid-transfer proteins 56, 57. One of these is the oxysterol-binding protein (OSBP). In uninfected cells, OSBP simultaneously binds the endoplasmic reticulum (ER) and trans-Golgi membranes, thus bringing them in close proximity and creating MCSs. OSBP interacts with the integral ER membrane proteins VAP-A and VAP-B through its FFAT-motif, while it docks to PI4P and Arf1 in the trans-Golgi via its PH-domain. OSBP binds cholesterol in a pocket in its sterol-binding domain and transports it to Golgi membranes. This cholesterol transfer occurs against the concentration gradient and hence requires energy. This energy is provided by a counterflux of PI4P, which binds in the same pocket of OSBP as cholesterol, from Golgi to ER, where it is hydrolyzed by the phosphatase Sac1 [58].
The PI4P-enriched environment of enterovirus ROs triggers the recruitment of OSBP to tether ER and RO membranes, and to create a novel type of MCS for a cholesterol/PI4P counterflux (Figure 2 ). Similar to the physiological role of OSBP, enteroviruses exploit the PI4KIIIβ–PI4P–OSBP pathway to import cholesterol into their ROs 59, 60, 61. Pharmacological inhibition of this pathway by targeting either PI4P production or OSBP-mediated lipid shuttling efficiently impaired enterovirus replication 23, 36, 59, 60, 61, 62, 63, 64. The observation that replication is hampered when PI4P transported to the ER is not hydrolyzed upon knockdown of Sac1 23, 60 underscores the importance of the cholesterol/PI4P-counterflux for enterovirus replication.
Figure 2.
Enteroviruses Trigger Formation of Membrane Contact Sites (MCSs) between Replication Organelles (ROs) and the Endoplasmic Reticulum (ER) for Lipid Exchange. In enterovirus-infected cells, the viral protein 3A mediates PI4KB recruitment to ROs to locally produce high levels of PI4P. Oxysterol-binding protein (OSBP) binds PI4P at the RO and VAP-A at the ER, and thereby stimulates formation of a novel type of MCS between ER and ROs at which it drives cholesterol/PI4P exchange to accumulate cholesterol at ROs. Adapted from [61]; the original image has been published under the Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported (CC BY-NC-ND 3.0) license.
Cholesterol is crucial for optimal enterovirus genome replication, as treatments that disrupt cholesterol homeostasis inhibited enterovirus replication 65, 66. However, enteroviruses do not depend on ongoing cholesterol synthesis in the ER 60, 66. This raises the question: where does RO cholesterol come from? So far, two cholesterol pools have been linked to replication: cholesterol stored in lipid droplets [60], and cholesterol internalized from the medium or the plasma membrane pool [65]. Lipid droplets are consumed during enterovirus replication to mobilize the stored cholesterylester as a source of free cholesterol [60]. As of yet, it is not known how cholesterol from lipid droplets gets to the ER, and whether MCSs play a role in this. Interestingly, GBF1 and the Arf1/COPI machinery act directly on lipid droplets and are important for lipid droplet-targeting of key enzymes involved in lipid droplet homeostasis [67]. Modulation of the GBF1 and Arf1/COPI machinery by the viral protein 3A may therefore deter lipid storage and promote lipolysis to increase the availability of free cholesterol (and other stored lipids) as building blocks for membrane biogenesis. In addition, enteroviruses were shown to upregulate clathrin-mediated endocytosis [68] to enhance cholesterol (re)uptake from medium/plasma membrane [65]. Internalized cholesterol is delivered to ROs via recycling endosomes, possibly involving an interaction between PI4KIIIβ and the recycling endosome protein Rab11. To date, it has not been established how exactly cholesterol is delivered from recycling endosomes to ROs. Possibly, cholesterol is shuttled at MCSs between recycling endosomes and ROs by cholesterol-transfer proteins, although it cannot be excluded that recycling endosomes fuse with the ROs.
The question remains: what is the function of cholesterol in enterovirus replication? Cholesterol is an essential lipid for cellular membranes and is important for membrane properties such as fluidity, permeability, and the formation of membrane microdomains (rafts). Hence, cholesterol may be required to generate and maintain essential membrane properties that drive RO shape and function. For example, it has been proposed that cholesterol is required for efficient viral polyprotein processing [65]. Acute depletion of cholesterol with methyl-β-cyclodextrins or disruption of cholesterol organization with filipin enhanced the proteolytic processing of the viral precursor protein 3CDpro to 3Cpro and 3Dpol, thereby reducing the amount of 3CDpro, which is required for priming viral RNA synthesis and processing of the capsid proteins. Treatment with the PI4KB inhibitor GW5074, which reduces PI4P and hence OSBP recruitment and cholesterol accumulation at ROs, also affected in vitro polyprotein processing and resulted in an accumulation of the large precursor P2–P3 [69]. Similar to cholesterol depletion, GW5074 treatment decreased the level of 3CDpro and concomitantly increased the amount of 3Dpol.
The Importance of MCSs for Controlling the Metabolism of Other Lipids in Infected Cells
Besides mediating cholesterol/PI4P exchange, OSBP regulates the homeostasis of other lipids via MCSs. Notably, OSBP regulates the ceramide transfer protein (CERT)-dependent shuttling of ceramide (a precursor of sphingomyelin, a lipid that associates with cholesterol in membrane microdomains) from ER to trans-Golgi [70]. It remains to be determined whether CERT and other machinery derived from the ER-Golgi MCS, and the lipids that depend on those lipid-transfer proteins, play a role in enterovirus replication. Additionally, OSBP is part of a family of OSBP-related proteins (ORPs) that have functions in lipid homeostasis at various cellular MCSs (see e.g., 71, 72). A recent study suggested a possible role for other ORPs, besides OSBP, in the replication of rhinoviruses [60], but firm evidence for their involvement remains to be obtained.
Picornaviruses have historically been considered nonenveloped viruses that rely on cell lysis to exit the cell, but this classic view has recently been challenged by a number of findings. First, hepatitis A virus particles were found to be released enwrapped in host-derived membranes [13]. Second, CVB3 particles were also identified within vesicles [12], and third, poliovirus was reported to be able to spread nonlytically [8]. A recent study reported that newly formed poliovirus virions are captured in autophagosome-like vesicular structures closely juxtaposed to ROs, and that both the ROs and the capsid-containing vesicular structures are enriched in the lipid phosphatidylserine [11]. Although it is unknown how exactly the virus-containing vesicular structures are formed, our ultrastructural analysis of CVB3-infected cells suggests that tubular ROs progressively transform into DMVs, which accumulate at later stages of infection [19]. After fusion of such DMVs with the plasma membrane, single-membrane, virus-containing extracellular vesicles would be released [11]. These resulting extracellular vesicles expose phosphatidylserine, a hallmark of apoptotic cells, which triggers uptake of the vesicles in neighboring cells. Transmission of virions in vesicles supports en bloc virus transmission and coinfection of cells by multiple virions [11], but – at least for the picornavirus hepatitis A virus – does not abrogate the need for the virus receptor [13]. How phosphatidylserine becomes enriched in these membranes is unknown. Recently, the OSBP-related proteins ORP5 and ORP8 were shown to mediate phosphatidylserine/PI4P countertransport at MCSs between the ER and the plasma membrane to deliver phosphatidylserine to the plasma membrane [73]. Since RO membranes are enriched in PI4P, ORP5 and/or ORP8 may also be recruited to ER–RO MCSs and transport phosphatidylserine to ROs, from where it may transition to the autophagosome-like DMVs.
Modulation of Lipid Homeostasis Independent of MCSs
Enterovirus replication has been associated with alterations of other lipid metabolic pathways as well. Enteroviruses were shown to activate a long-chain acyl-CoA-synthase 3 to reroute imported fatty acids from storage in lipid droplets to massive production of the major membrane lipid phosphatidylcholine [74]. Importantly, phosphatidylcholine produced during infection is distinct from that synthesized in uninfected cells, as there is a shift towards the generation of phosphatidylcholine with longer acyl chains (C16 and C18, i.e., fatty acyl chains with 16 and 18 carbon atoms, respectively) in infected cells, indicating that RO membranes are significantly different from the pre-existing cellular membranes. The viral proteinase 2Apro was required, but was not sufficient, to activate the import of fatty acids, but this function was independent of the protease activity. It remains to be determined how exactly 2A enhances fatty acid uptake during infection.
Hijacking of MCS by Other +RNA Viruses
Other Picornaviruses
Accumulating evidence suggests that exploiting MCS machineries might be a more widespread phenomenon within the Picornaviridae. Apart from enteroviruses, Aichivirus, a member of the genus Kobuvirus, was also shown to manipulate PI4P metabolism by hijacking PI4KB 39, 51, 75. Similar to enteroviruses, kobuviruses require a nonenzymatic function of GBF1 for efficient replication that is apparently unrelated to PI4KB recruitment 39, 51. Aichivirus recruits PI4KB to its ROs via interaction of several non-structural proteins with ACBD3 [51]. It is tempting to speculate that kobuviruses, like enteroviruses, rely on PI4P to generate MCSs with the ER via OSBP, but this idea awaits experimental validation.
The Cardiovirus genus of the Picornaviridae, which includes encephalomyocarditis virus, Theiler's virus, and the human Saffold virus, was long believed to employ replication strategies substantially distinct from those of enteroviruses, as members of this genus did not show sensitivity to treatment with inhibitors of GBF1 or PI4KB 21, 36, 42, 76, 77. While this implied that cardioviruses do not rely on GBF1/PI4KB-controlled pathways, cardiovirus replication in fact also depends on PI4P lipids, although they are produced by the ER-derived enzyme PI4KA [78]. PI4KA is actively recruited by the viral 3A protein to cardiovirus ROs, which may be derived from the ER. PI4P plays a similar role in cardiovirus replication as in enterovirus replication, that is, to create MCSs between ROs and the ER via OSBP to funnel cholesterol to ROs [78].
Little is known about the host protein and lipid requirements of picornaviruses from other genera. Studies with inhibitors of PI4Ks and OSBP suggest that members of the genera Aphthovirus (e.g., equine rhinitis A virus, a close relative of foot-and-mouth disease virus) and Parechovirus do not rely on PI4K/OSBP functions 61, 62, 78. Whether these or other picornaviruses co-opt cholesterol/PI4P, either via MCSs or via alternative mechanisms, or have evolved to depend on other machineries to establish their ROs remains to be determined.
+RNA Viruses from Other Families
The use of MCS machinery to build replication platforms extends beyond the Picornaviridae family. Similar to cardioviruses, the distantly-related flavivirus hepatitis C virus exploits PI4KA to develop a PI4P-rich network of replication membranes, designated the membranous web [79]. Likewise, OSBP participates in hepatitis C virus infection by forming MCSs between the membranous web and the ER, and by shuttling cholesterol to the membranous web [63]. Unlike hepatitis C virus, the closely related flaviviruses dengue virus or West Nile virus do not require the PI4P–OSBP machinery of MCSs, although these viruses are sensitive to disruptions of cholesterol homeostasis and accumulate cholesterol at their ROs 63, 80, 81, 82.
Tombusviruses, a family of plant viruses, were recently shown to also hijack MCS machinery and shuttle sterols to their ROs. The tombusvirus tomato bushy stunt virus (TBSV) generates invaginations in peroxisomes in which the viral genome is replicated. Studies in both plant cells and in yeast, which can be conveniently used as a surrogate host model system, have revealed that TBSV accumulates ergosterol (the main sterol in plants and yeast) at their ROs dependent on several homologs of OSBP and VAP [83]. It is not known whether TBSV also hijacks PI4P kinases (similar to picornaviruses), but the finding that the nonstructural viral protein p33 binds both ORPs and VAP suggests that TBSV directly recruits MCS machinery. Importantly, electron microscopy studies revealed the presence of p33-containing MCSs between ER and peroxisomes in close proximity to ROs [83]. Similar observations were made for carnation Italian ringspot virus (CIRV), which replicates its genome in invaginations in the mitochondrial outer membrane [83]. Together, these data show that tombusviruses, which, in contrast to picornaviruses and hepatitis C virus, generate ROs of negative curvature, also hijack MCS machinery to shuttle sterols to their ROs.
In vivo Relevance of in vitro Studies
Most studies on virus replication are performed in vitro in nonpolarized immortalized cell lines. To date, it has been poorly explored whether findings in cell culture reflect the in vivo situation. Different cell types can vary greatly in their protein and lipid composition, which may influence virus tropism and replication efficiency. Indeed, the machinery involved in virus entry differs considerably between polarized and nonpolarized cells (e.g., [84]). However, several studies suggest that enterovirus-induced membrane rearrangements observed in polarized cells in vitro and in vivo 25, 85, 86 resembled those in nonpolarized cells 19, 20, suggesting that the machinery involved in RO formation is largely conserved between different cell types. Consistently, studies using an inhibitor revealed that, of the host factors identified in cell culture, at least PI4KB is essential for enterovirus infection in vivo 36, 87.
Notwithstanding, the observation that CVB3 can exploit LC3 in both autophagy-dependent and -independent manners, and can rearrange membranes and replicate in the absence of functional LC3 [26], indicates that enteroviruses – like TBSV [88] and perhaps many other +RNA viruses – can utilize disparate host membranes and machinery to generate ROs, depending on availability in different cell types. Hence, findings in whichever experimental system may only represent part of the story in multicellular organisms with many different cell types.
Concluding Remarks
A number of vastly different +RNA viruses, both those generating positive-membrane-curvature and negative-curvature ROs, have now been shown to require sterols at their ROs and hijack host MCS machinery to shuttle sterols to the ROs. The strategies employed by those viruses to confiscate MCS machinery differ markedly, from direct recruitment of MCS proteins to the generation of physiological-like, PI4P-dependent MCSs. What the apparently universally important role of sterols at ROs is remains to be determined. Furthermore, physiological MCSs support the homeostasis of many lipids other than cholesterol, but it has not yet been investigated whether +RNA viruses also hijack MCS machinery to tune the RO lipid composition. Many aspects regarding biogenesis, composition, architecture, and function of ROs remain to be elucidated (see Outstanding Questions). We expect an important role for MCSs and viral hijacking of MCS machinery in infection to be uncovered in the near future.
Outstanding Questions.
Does the shape of ROs determine their function and, if so, how?
How is the structure of ROs established; what is the role of lipids, and what is the contribution of cellular scaffolding/membrane-shaping proteins in determining RO structure and functionality?
What is the lipid composition of ROs, and what are the roles of the individual components?
Do picornaviruses exchange lipids other than PI4P and cholesterol via the ER–RO MCS?
From where does the cholesterol that is funneled to ROs via MCSs originate?
Do picornaviruses create MCSs with other organelles to exchange lipids?
Author Contributions
H.M. van der Schaar and C.M. Dorobantu contributed equally, as did J.R.P.M Strating and F.J.M. van Kuppeveld.
Acknowledgments
Research in the authors’ laboratory is supported by grants from the Netherlands Organisation for Scientific Research (NWO) (VENI-863.12.005 to HMvdS, VENI-722.012.066 to JRPMS, VICI-91812628 and ALW-820.02.018 to FJMvK) and from the European Union (FP7 Marie Curie ITN “EUVIRNA”, grant agreement number 264286, and Horizon2020 Marie Sklodowska Curie ETN ‘ANTIVIRALS’, grant agreement number 642434) to FJMvK. The funders had no role in preparation of the manuscript or the decision to publish.
References
- 1.Tapparel C. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 2013;14:282–293. doi: 10.1016/j.meegid.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Ooi M.H. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 3.Gern J.E. The ABCs of rhinoviruses, wheezing, and asthma. J. Virol. 2010;84:7418–7426. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Midgley C.M. Severe respiratory illness associated with a nationwide outbreak of enterovirus D68 in the USA (2014): A descriptive epidemiological investigation. Lancet Respir. Med. 2015;3:879–887. doi: 10.1016/S2213-2600(15)00335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamborsky J. Poliomyelitis. In: Hamborsky J., editor. Centers for Disease Controle and Prevention. Epidemiology and Prevention of Vaccine-Preventable Diseases. 13th edn. Public Health Foundation; 2015. pp. 297–310. [Google Scholar]
- 6.Sin J. Recent progress in understanding coxsackievirus replication, dissemination, and pathogenesis. Virology. 2015;484:288–304. doi: 10.1016/j.virol.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuthill T.J. Picornaviruses. Curr. Top. Microbiol. Immunol. 2010;343:43–89. doi: 10.1007/82_2010_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bird S.W. Nonlytic viral spread enhanced by autophagy components. Proc. Natl. Acad. Sci. U.S.A. 2014;111:13081–13086. doi: 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson W.T. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkegaard K., Jackson W.T. Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm. Autophagy. 2005;1:182–184. doi: 10.4161/auto.1.3.2065. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y.H. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson S.M. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014;10:e1004045. doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Z. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero-Brey I. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 16.Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bienz K. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology. 1980;100:390–399. doi: 10.1016/0042-6822(80)90530-9. [DOI] [PubMed] [Google Scholar]
- 18.Kallman F. Fine structure of changes produced in cultured cells sampled at specified intervals during a single growth cycle of polio virus. J. Biophys. Biochem. Cytol. 1958;4:301–308. doi: 10.1083/jcb.4.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limpens R.W.A.L. The transformation of enterovirus replication structures: A three-dimensional study of single- and double-membrane compartments. MBio. 2011;2 doi: 10.1128/mBio.00166-11. e00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belov G.A. Complex dynamic development of poliovirus membranous replication complexes. J. Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazina E.V. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. J. Virol. 2002;76:11113–11122. doi: 10.1128/JVI.76.21.11113-11122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belov G.A. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu N-Y. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong J. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemball C.C. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J. Virol. 2010;84:12110–12124. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alirezaei M. Coxsackievirus can exploit LC3 in both autophagy-dependent and -independent manners in vivo. Autophagy. 2015;11:1389–1407. doi: 10.1080/15548627.2015.1063769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belov G.A., van Kuppeveld F.J. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr. Opin. Virol. 2012;2:740–747. doi: 10.1016/j.coviro.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janmey P.A., Kinnunen P.K. Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 2006;16:538–546. doi: 10.1016/j.tcb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 29.van Meer G. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bigay J., Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: Defining cellular territories in determining specificity. Dev. Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Holthuis J.C., Menon A.K. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 32.De Matteis M.A., Rega L.R. Endoplasmic reticulum-Golgi complex membrane contact sites. Curr. Opin. Cell Biol. 2015;35:43–50. doi: 10.1016/j.ceb.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Balla T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong K. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 1997;272:13236–13241. doi: 10.1074/jbc.272.20.13236. [DOI] [PubMed] [Google Scholar]
- 35.Godi A. ARF mediates recruitment of PtdIns-4-OH kinase-b and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 36.van der Schaar H.M. A novel, broad-spectrum inhibitor of enterovirus replication that targets host cell factor phosphatidylinositol 4-kinase IIIbeta. Antimicrob. Agents Chemother. 2013;57:4971–4981. doi: 10.1128/AAC.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorobantu C.M. Recruitment of PI4KIIIbeta to coxsackievirus B3 replication organelles is independent of ACBD3, GBF1, and Arf1. J. Virol. 2014;88:2725–2736. doi: 10.1128/JVI.03650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorobantu C.M. GBF1- and ACBD3-independent recruitment of PI4KIIIbeta to replication sites by rhinovirus 3A proteins. J. Virol. 2015;89:1913–1918. doi: 10.1128/JVI.02830-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greninger A.L. The 3A protein from multiple picornaviruses utilizes the Golgi adaptor protein ACBD3 to recruit PI4KIIIβ. J. Virol. 2012;86:3605–3616. doi: 10.1128/JVI.06778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arita M. Phosphatidylinositol-4 kinase III beta and oxysterol-binding protein accumulate unesterified cholesterol on poliovirus-induced membrane structure. Microbiol. Immunol. 2014;58:239–256. doi: 10.1111/1348-0421.12144. [DOI] [PubMed] [Google Scholar]
- 41.Wessels E. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell. 2006;11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Lanke K.H. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J. Virol. 2009;83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belov G.A. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 2008;4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D'Souza-Schorey C., Chavrier P. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 45.Donaldson J.G. Multiple activities for Arf1 at the Golgi complex. Biochim. Biophys. Acta. 2005;1744:364–373. doi: 10.1016/j.bbamcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Cuconati A. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 1998;72:6456–6464. doi: 10.1128/jvi.72.8.6456-6464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beck R. The COPI system: Molecular mechanisms and function. FEBS Lett. 2009;583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 48.Belov G.A. Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cell. Microbiol. 2010;12:1463–1479. doi: 10.1111/j.1462-5822.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verheije M.H. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L. ARF1 and GBF1 generate a PI4P-enriched environment supportive of Hepatitis C virus replication. PLoS ONE. 2012;7:e32135. doi: 10.1371/journal.pone.0032135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki J. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 2012;31:754–766. doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teoule F. The Golgi protein ACBD3, an interactor for poliovirus protein 3A, modulates poliovirus replication. J. Virol. 2013;87:11031–11046. doi: 10.1128/JVI.00304-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jovic M. Two phosphatidylinositol 4-kinases control lysosomal delivery of the Gaucher disease enzyme, beta-glucocerebrosidase. Mol. Biol. Cell. 2012;23:1533–1545. doi: 10.1091/mbc.E11-06-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greninger A.L. ACBD3 interaction with TBC1 domain 22 protein is differentially affected by enteroviral and kobuviral 3A protein binding. MBio. 2013;4 doi: 10.1128/mBio.00098-13. e00098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blomen V.A. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 56.Clayton E.L. Mammalian phosphatidylinositol 4-kinases as modulators of membrane trafficking and lipid signaling networks. Prog. Lipid Res. 2013;52:294–304. doi: 10.1016/j.plipres.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Matteis M.A. Phosphatidylinositol-4-phosphate: The Golgi and beyond. Bioessays. 2013;35:612–622. doi: 10.1002/bies.201200180. [DOI] [PubMed] [Google Scholar]
- 58.Mesmin B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 59.Arita M. Oxysterol-binding protein family I is the target of minor enviroxime-like compounds. J. Virol. 2013;87:4252–4260. doi: 10.1128/JVI.03546-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roulin P.S. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Strating J.R. Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 2015;10:600–615. doi: 10.1016/j.celrep.2014.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albulescu L. Broad-range inhibition of enterovirus replication by OSW-1, a natural compound targeting OSBP. Antivir. Res. 2015;117:110–114. doi: 10.1016/j.antiviral.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 63.Wang H. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373–1385. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arita M. Phosphatidylinositol 4-kinase III beta is a target of enviroxime-like compounds for antipoliovirus activity. J. Virol. 2011;85:2364–2372. doi: 10.1128/JVI.02249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ilnytska O. Enteroviruses harness the cellular endocytic machinery to remodel the host cell cholesterol landscape for effective viral replication. Cell Host Microbe. 2013;14:281–293. doi: 10.1016/j.chom.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Albulescu L. Cholesterol shuttling is important for RNA replication of coxsackievirus B3 and encephalomyocarditis virus. Cell. Microbiol. 2015;17:1144–1156. doi: 10.1111/cmi.12425. [DOI] [PubMed] [Google Scholar]
- 67.Wilfling F. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife. 2014;3:e01607. doi: 10.7554/eLife.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cornell C.T. Coxsackievirus B3 proteins directionally complement each other to downregulate surface Major Histocompatibility Complex class I. J. Virol. 2007;81:6785–6797. doi: 10.1128/JVI.00198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ford Siltz L.A. New small molecule inhibitors effectively blocking picornavirus replication. J. Virol. 2014;88:11091–11107. doi: 10.1128/JVI.01877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perry R.J., Ridgway N.D. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell. 2006;17:2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raychaudhuri S., Prinz W.A. The diverse functions of oxysterol-binding proteins. Annu. Rev. Cell Dev. Biol. 2010;26:157–177. doi: 10.1146/annurev.cellbio.042308.113334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olkkonen V.M., Li S. Oxysterol-binding proteins: Sterol and phosphoinositide sensors coordinating transport, signaling and metabolism. Prog. Lipid Res. 2013;52:529–538. doi: 10.1016/j.plipres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Chung J. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 2015;349:428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nchoutmboube J.A. Increased long chain acyl-CoA synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PLoS Pathog. 2013;9:e1003401. doi: 10.1371/journal.ppat.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishikawa-Sasaki K. A complex comprising phosphatidylinositol 4-kinase IIIbeta, ACBD3, and Aichi virus proteins enhances phosphatidylinositol 4-phosphate synthesis and is critical for formation of the viral replication complex. J. Virol. 2014;88:6586–6598. doi: 10.1128/JVI.00208-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van der Linden L. Differential effects of the putative GBF1 inhibitors Golgicide A and AG1478 on enterovirus replication. J. Virol. 2010;84:7535–7542. doi: 10.1128/JVI.02684-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Irurzun A. Involvement of membrane traffic in the replication of poliovirus genomes: Effects of brefeldin A. Virology. 1992;191:166–175. doi: 10.1016/0042-6822(92)90178-r. [DOI] [PubMed] [Google Scholar]
- 78.Dorobantu C.M. Modulation of the host lipid landscape to promote RNA virus replication: The picornavirus encephalomyocarditis virus converges on the pathway used by hepatitis C virus. PLoS Pathog. 2015;11:e1005185. doi: 10.1371/journal.ppat.1005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reiss S. Recruitment and activation of a lipid kinase by Hepatitis C Virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rothwell C. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology. 2009;389:8–19. doi: 10.1016/j.virol.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 81.Mackenzie J.M. Cholesterol manipulation by West Nile Virus perturbs the cellular immune response. Cell Host Microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 82.Martín-Acebes M.A. West Nile Virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS ONE. 2011;6:e24970. doi: 10.1371/journal.pone.0024970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barajas D. Co-opted oxysterol-binding ORP and VAP proteins channel sterols to RNA virus replication sites via membrane contact sites. PLoS Pathog. 2014;10:e1004388. doi: 10.1371/journal.ppat.1004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coyne C.B. Comparative RNAi screening reveals host factors involved in enterovirus infection of polarized endothelial monolayers. Cell Host Microbe. 2011;9:70–82. doi: 10.1016/j.chom.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Volle R. Differential permissivity of human cerebrovascular endothelial cells to enterovirus infection and specificities of serotype EV-A71 in crossing an in vitro model of the human blood-brain barrier. J. Gen. Virol. 2015;96:1682–1695. doi: 10.1099/vir.0.000103. [DOI] [PubMed] [Google Scholar]
- 86.Alirezaei M. Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe. 2012;11:298–305. doi: 10.1016/j.chom.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thibaut H.J. Fitness and virulence of a coxsackievirus mutant that can circumnavigate the need for phosphatidylinositol 4-kinase class III beta. J. Virol. 2014;88:3048–3051. doi: 10.1128/JVI.03177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chuang C. Inactivation of the host lipin gene accelerates RNA virus replication through viral exploitation of the expanded endoplasmic reticulum membrane. PLoS Pathog. 2014;10:e1003944. doi: 10.1371/journal.ppat.1003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Helle S.C. Organization and function of membrane contact sites. Biochim. Biophys. Acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 90.Fernandez-Busnadiego R. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E2004–E2013. doi: 10.1073/pnas.1503191112. [DOI] [PMC free article] [PubMed] [Google Scholar]