Abstract
Background & Aims: Severe acute respiratory syndrome (SARS) is a recently emerged infection from a novel coronavirus (CoV). Apart from fever and respiratory complications, gastrointestinal symptoms are frequently observed in patients with SARS but the significance remains undetermined. Herein, we describe the clinical, pathologic, and virologic features of the intestinal involvement of this new viral infection. Methods: A retrospective analysis of the gastrointestinal symptoms and other clinical parameters of the first 138 patients with confirmed SARS admitted for a major outbreak in Hong Kong in March 2003 was performed. Intestinal specimens were obtained by colonoscopy or postmortem examination to detect the presence of coronavirus by electron microscopy, virus culture, and reverse-transcription polymerase chain reaction. Results: Among these 138 patients with SARS, 28 (20.3%) presented with watery diarrhea and up to 38.4% of patients had symptoms of diarrhea during the course of illness. Diarrhea was more frequently observed during the first week of illness. The mean number of days with diarrhea was 3.7 ± 2.7, and most diarrhea was self-limiting. Intestinal biopsy specimens obtained by colonoscopy or autopsy showed minimal architectural disruption but the presence of active viral replication within both the small and large intestine. Coronavirus was also isolated by culture from these specimens, and SARS-CoV RNA can be detected in the stool of patients for more than 10 weeks after symptom onset. Conclusions: Diarrhea is a common presenting symptom of SARS. The intestinal tropism of the SARS-CoV has major implications on clinical presentation and viral transmission.
Abbreviations: CoV, coronavirus; ICU, intensive care unit; PCR, polymerase chain reaction; RT, reverse transcription; SARS, severe acute respiratory syndrome
Severe acute respiratory syndrome (SARS) is a newly emerged infection characterized by fever and pneumonia. This disease may progress rapidly to acute respiratory distress syndrome with considerable morbidity and mortality. The disease was first identified in the Guandong Province of China in November 2002, and a major outbreak occurred in Hong Kong in March 2003.1, 2 The disease then quickly spread around the world at an alarming rate in <2 months. As of May 31, 2003, SARS has been described in 32 countries affecting 8360 individuals and causing 764 deaths.3 The overall mortality for this illness is about 16% in Hong Kong1 and can be as high as 43% in elderly patients.4 Shortly after the description of this disease, a novel form of coronavirus, named the SARS-CoV, that does not closely resemble any of the 3 previously known groups of coronaviruses was identified to be the causative agent.5, 6 This virus can be detected by culture and polymerase chain reaction (PCR) from most patients with SARS, and specific antibodies against this new virus can also be shown in the infected host. Furthermore, a monkey inoculation experiment produces similar disease as in humans, fulfilling the Koch’s postulate.7
The coronaviruses are a diverse group of large, enveloped, positive-stranded RNA viruses. Coronavirus is known to cause a wide spectrum of diseases in animals. In particular, respiratory and enteric infections are the most common presentation. However, it is uncertain whether respiratory and enteric coronaviruses in animals are distinctive in their genetic sequences. Emerging data suggest the differences in the spike protein of the virus may underlie the tissue tropism of bovine coronavirus.8 In humans, there are 2 forms of coronavirus, HCoV-229E and HCoV-OC43, that are associated with about 30% of mild upper respiratory tract infections. Moreover, human coronaviruses or coronavirus-like particles have been linked to diarrheal disease in susceptible hosts, including patients with acquired immunodeficiency syndrome9 and children from developing countries.10, 11 Early data also linked the coronavirus to necrotizing enterocolitis in infants, but the real significance remains undetermined.12, 13
Although SARS is characterized by fever and lower respiratory tract infection, data from recent outbreaks suggest that concurrent gastrointestinal symptoms are not uncommon.2, 14, 15 In this study, we characterize the enteric manifestations of the first cohort of patients with SARS in the Hong Kong outbreak2 with histologic and virologic correlations.
Materials and methods
Patients
A major outbreak of SARS occurred in the Prince of Wales Hospital of Hong Kong in March 2003, affecting more than 100 health care workers and hospitalized patients.2 This is a retrospective study that evaluated the enteric manifestations of the first 138 consecutive patients of this cohort. The study was approved by the Clinical Research Ethics Committee of the Chinese University Hong Kong.
The diagnosis of SARS was based on the criteria defined by the Centers for Disease Control and Prevention and the World Health Organization.16, 17 Briefly, SARS is defined by the presence of fever (38°C or higher), cough, or shortness of breath that is associated with new pulmonary infiltrates on chest radiography or high-resolution computed tomography in the absence of an alternative diagnosis to explain the clinical presentation. A history of contact with patients with SARS is also required.
The clinical course of these patients on presentation and their subsequent hospital stay, including supplementary oxygen requirement, admission to the intensive care unit (ICU), and need for mechanical ventilation, were recorded. Enteric symptoms, particularly diarrhea (defined as passing loose stool >3 times per day), were noted. All patients were kept in the hospital for a minimum of 21 days from the onset of symptoms for public health reasons.
Microbiologic and virologic investigations
Diagnosis of SARS-CoV infection was performed by serology, virus isolation, and detection of viral RNA in nasopharyngeal aspirates. The presence of antibody against coronavirus was detected by indirect immunofluorescent technique based on Vero cells infected with SARS-CoV isolated from a patient with SARS. Paired sera taken during acute (within 7 days after the onset of fever) and convalescent (>21 days after the onset of fever) phases were tested at serial dilutions starting from 1:40. The results were crosschecked by 2 experienced technicians. Positive serologic evidence of coronavirus infection was defined as having a seroconversion or ≥4-fold increase in antibody titer.18
Virus isolation was performed by inoculating samples onto multiple cell lines, including rhesus monkey kidney (LLC-MK2), human laryngeal carcinoma (HEp-2), Madin Darby canine kidney (MDCK), human embryonic lung fibroblast, and African green monkey kidney (Vero) monolayers. All cell cultures were incubated at 37°C, except for MDCK and one additional LLC-MK2 cell culture tube that were incubated at 33°C. Cell monolayers were examined daily up to 14 days for cytopathic effect. Hemadsorption was performed for MDCK and LLC-MK2 cells to detect influenza and parainfluenza viruses. When a refractile, diffuse rounding cytopathic effect was observed on Vero cell monolayer, the cell culture supernatant was passaged to another Vero tube to confirm the reproducibility of the cytopathic effect. In addition, the cell culture supernatant was used for detection of coronavirus by reverse-transcription (RT)-PCR. The SARS-CoV specific primers COR-1 (sense) 5′ CAC CGT TTC TAC AGG TTA GCT AAC GA 3′ and COR-2 (antisense) 5′ AAA TGT TTA CGC AGG TAA GCG TAA AA 3′ were used to detect the presence of SARS-CoV RNA.19
In patients with diarrhea, stool samples were tested for common enteric pathogens (e.g., Salmonella, Shigella, and Escherichia coli) by culture and for Clostridium difficile enterotoxin. Moreover, attempts were made in some patients to detect the presence of SARS-CoV in stool.
Endoscopic and histologic examination
Colonoscopy was performed in one patient with persistent diarrhea to exclude the possibility of inflammatory colitis or other cause of diarrhea. The procedure was performed in a negative-pressure room, and the patient was given light sedation. Endoscopists and assistants were equipped with protective gear, including surgical gloves, eye goggles, a face shield, an N95-grade mask, and water-resistant gowns. Endoscopic appearance was noted and random biopsy specimens were taken from the colon and terminal ileum for histologic examination and viral detection.
Postmortem examination was performed in 5 patients who died from SARS. In addition to examination of the respiratory system, macroscopic appearances of the small and large intestine were noted. All intestinal specimens were fixed in 10% buffered formalin and embedded in paraffin for routine histologic examination. For ultrastructural study, fresh tissues were fixed in 2.5% glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.2) and processed for examination by electron microscopy. Fresh tissues were also used for culture of coronavirus as previously described.
Statistics
All statistics were computed using SPSS software (version 11.0; SPSS Inc., Chicago, IL). Data are presented as mean ± SD unless specified. Patients with diarrhea and those without were compared by unpaired Student t test for numerical data and χ2 test for categorical data. A 2-sided P value of <0.05 was considered statistically significant.
Results
Clinical features
The results of the first 138 consecutive patients (65 men and 73 women; 39.3 ± 16.8 years) admitted between March 11 and March 25, 2003, with the diagnosis of SARS were analyzed. All patients fulfilled the Centers for Disease Control and Prevention or World Health Organization definition of SARS and had evidence of SARS-CoV infection either by serology or direct detection of virus by culture or RT-PCR in nasopharyngeal aspirates. Specifically, 124 patients (89.9%) had a positive serology test, 24 (17.4%) had virus isolated by culture, and 12 (8.7%) had viral RNA detected by RT-PCR. There were 69 health care workers (50%), 16 medical students (11.6%), and 53 patients or their relatives (38.4%) who contracted the disease during a hospital visit. Twenty patients (14.5%) had coexisting medical illnesses, including diabetes mellitus in 5, cardiovascular disease in 5, chronic liver disease in 3, chronic lung disease in 3, chronic renal failure in 2, and myelodysplastic syndrome in 2.
All patients had fever on presentation, and chest x-ray abnormalities were noted in all patients either on admission or during the subsequent course of the disease. Apart from fever, other common presenting symptoms included chills and/or rigor (73.2%), myalgia (60.9%), cough (57.3%), and headache (55.8%). Among this cohort, 36 patients (26.1%) required care in the ICU and 21 patients (15.2%) were ventilated. The overall 3-month mortality was 10.9% (95% confidence interval, 6.2%–17.3%).
Gastrointestinal manifestation
Watery diarrhea is the most common gastrointestinal symptom in these patients and was reported in 28 patients (20.3%) on presentation. Eight patients (5.8%) presented with fever and diarrhea in the absence of respiratory symptoms. The diarrhea was mainly watery in nature without blood or mucus. Abdominal pain was minimal in most cases. Of the remaining patients who did not report diarrhea on presentation, 25 more patients developed symptoms of watery diarrhea during hospitalization. In total, 53 patients (38.4%) had symptoms of diarrhea in the first 3 weeks of illness. The distribution of patients with diarrhea over this 3-week period is shown in Figure 1. Most patients had watery diarrhea in the first week of illness. In patients with diarrhea, the average (±SD) duration of diarrhea was 3.7 ± 2.7 days. The severity of diarrhea varied among patients. Most patients had a few loose stools per day but some patients had up to 30 bowel motions per day, leading to dehydration and hypokalemia.
Figure 1.
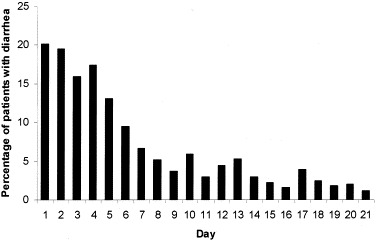
Percentage of SARS patients with diarrhea during the first 21 days of illness.
There was no correlation between diarrhea on presentation with subsequent requirement of supplementary oxygen and mortality. However, as shown in Table 1, patients who had diarrhea during the course of illness had a higher percentage requiring ventilatory support (26.4% vs. 8.2%; P = 0.004) and higher admission rate to the ICU (49.0% vs. 11.8%; P < 0.001). There was no specific identifiable pathogen, including C. difficile toxin, detectable in the stool of patients with diarrhea.
Table 1.
Demographic Data and Clinical Features of SARS Patients With or Without Diarrhea
| Patients with diarrhea (%) | Patients without diarrhea (%) | P | |
|---|---|---|---|
| No. of patients | 53 (38.4) | 85 (61.6) | |
| Male sex | 28 (52.8) | 37 (43.5) | 0.29 |
| Mean age ± SD (yr) | 42.1 ± 16.5 | 37.8 ± 16.6 | 0.13 |
| Concurrent medical illnesses | 11 (20.8) | 9 (10.6) | 0.1 |
| Mean hospital stay ± SD (days) | 24.0 ± 14.6 | 20.0 ± 11.6 | 0.07 |
| Use of supplementary oxygen | 35 (66.0) | 44 (51.8) | 0.21 |
| Ventilatory support | 14 (26.4) | 7 (8.2) | 0.004 |
| ICU care | 26 (49.0) | 10 (11.8) | <0.001 |
| Death | 8 (15.1) | 7 (8.2) | 0.21 |
Histologic and virologic findings
Colonoscopy was performed on day 13 after symptom onset in one patient with diarrhea. The colonic mucosa and terminal ileum looked normal on endoscopy. Microscopically, biopsy specimens taken from the colon and terminal ileum showed normal architecture with no evidence of villous atrophy, inflammatory infiltrates, bacterial invasion, viral inclusion, or granuloma (Figure 2). Postmortem examinations were performed in 5 patients who died from SARS. Again, there was no specific change on gross examination and light microscopy apart from autolytic changes.
Figure 2.
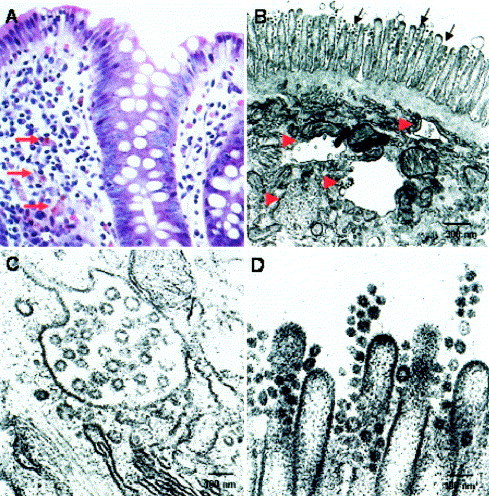
Histologic and ultrastructural appearances of the colon in a patient with SARS-CoV infection. (A) Endoscopic colonic biopsy specimens with scattered lipofusin-ladened macrophages in the lamina propria indicative of melanosis coli. The macrophages are indicated by the red arrows, and there were no significant inflammatory cell infiltrates (H&E; original magnification 200×). (B) Dilated cytoplasmic vesicles, which were consistent with dilated endoplasmic reticulum, were seen toward the apical cytoplasm (indicated by red arrowheads) and some were filled with viral particles. A number of viral particles were also seen on the surface microvilli (indicated by black arrows). (C) Higher magnification of the virus-containing vesicles. The viral particles had mild variation in size and ranged from 60 to 90 nm in dimension, which is consistent with coronavirus morphologically. (D) Viral particles were detected on the luminal surface of the enterocytes. Some viral particles appeared to attach onto the microvilli, whereas some appeared to be detached from the cell.
By electron microscopy, viral particles (60–90 nm in size) that were consistent with coronavirus were detected in all 5 small intestine tissues obtained on autopsy as well as the terminal ileal and colonic biopsy specimens obtained on colonoscopy Figure 2, Figure 3. Viral particles were confined to the epithelial cells, primarily in the apical surface enterocytes and rarely in the glandular epithelial cells. Intracellularly, viral particles were contained within dilated cytoplasmic vesicles consistent with dilated endoplasmic reticulum. The vesicles containing the viral particles were often seen toward the apical cytoplasm. Clusters of coronavirus were also detected on the surface microvilli, which may suggest virus leaving from the luminal surface of enterocytes. There was no evidence of villous atrophy despite viral adhesion and colonization.
Figure 3.
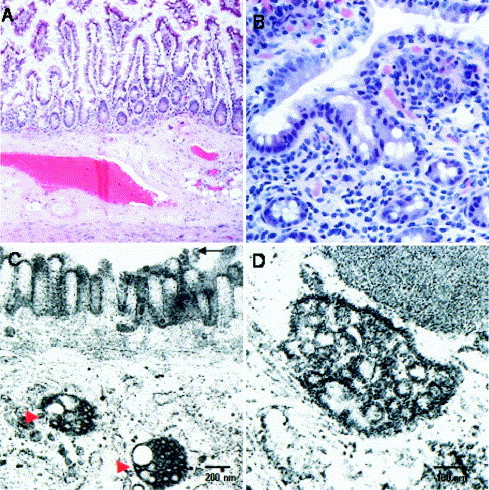
Histologic and ultrastructural appearances of the small intestine in patients with SARS-CoV infection. (A) Section of the small intestine of an autopsy specimen with unremarkable mucosa, submucosa, and muscle layer (H&E; original magnification 40×). (B) Endoscopic ileal biopsy specimen with no inflammatory process (tangentially sectioned, H&E; original magnification 200×). (C) Dilated cytoplasmic vesicles filled with viral particles in the small intestine (indicated by red arrowheads). Scattered viral particles were also detected on the surface microvilli of this surface enterocyte (indicated by black arrow). (D) Higher magnification of a virus-containing vesicle.
SARS-CoV was isolated by culture in the small intestinal tissues of 5 patients: 4 obtained from autopsy and one from colonoscopy. The one with negative viral culture also had virus RNA detected by RT-PCR. On the other hand, coronavirus could only be recovered from the postmortem lung tissues in 2 patients and not in other organs sampled.
An attempt was made to detect SARS-CoV in the stool of some patients. The overall detection rate of SARS-CoV RNA in stool specimens by RT-PCR was 16%, which was comparable to the detection rate in nasopharyngeal aspirates. Moreover, prolonged fecal shedding of viral RNA is common. The longest period in which viral RNA can be detected in a stool sample was up to 73 days after symptom onset in one patient. However, none of the patients had successful virus isolation from stool samples.
Discussion
SARS was first described by Dr. Urbani of the World Health Organization in March 2003. It is generally considered a disease of the lower respiratory tract transmitted by droplet. However, results from this study suggest that enteric involvement is common in SARS, which may carry paramount implications on disease manifestation, mechanism of spread, and infection-control measures.
In this study, we show that about 20% of patients had watery diarrhea on presentation. In fact, some patients presented with fever and diarrhea only before onset of respiratory symptoms. Overall, 38% of patients had diarrhea during the course of illness. The mean duration of diarrhea was 3.7 days, and most cases of diarrhea subsided spontaneously. It is possible that diarrhea in some of our cases is related to the use of antibiotics at the early phase of treatment. However, routine bacteriologic culture and detection of C. difficile toxin failed to identify any other culprit for diarrhea. Also, many patients who presented with fever and diarrhea had no prior exposure to antibiotics.
In line with our findings, diarrhea was also reported in 2 other SARS cohorts. In the Toronto outbreak, 23.6% of the 144 patients had diarrhea on presentation.15 Peiris et al. report that up to 70% of their 75 patients in the community outbreak from Hong Kong developed watery diarrhea, although the frequency of patients with diarrhea on admission was not provided.14 In their study, the mean duration of diarrhea was 4 days, which is similar to our findings. The higher prevalence of diarrhea in their report when compared with our data is postulated to be linked to the different modes of transmission. Our cohort was believed to be infected by droplet transmission, whereas the community outbreak reported by Peiris et al. was linked to the faulty sewage system in an apartment complex in which fecal-oral transmission might be a major route of transmission.20
In addition, this study shows the tissue tropism of SARS-CoV in the intestinal tract. Although coronavirus affects both the respiratory and intestinal tract in animals, this is the first report to show the presence of active viral replication in the small and large intestine of patients with SARS. Despite a relatively normal endoscopic and microscopic appearance, the presence of coronavirus in the large and small bowel can be shown by both electronic microscopy and viral culture. There was also evidence of active viral replication in intestinal cells with accumulation of virus inside endoplasmic reticulum and possibly leaving of virus from the apical membrane of the enterocytes. Moreover, small bowel biopsy specimens obtained from postmortem examination, which is usually performed a few days after the death of patients, still yielded viable SARS-CoV. In contrast, SARS is associated with epithelial cell proliferation and an increase in macrophages in the lung.21 It is suggested that cytokine dysregulation plays an instrumental role in the determination of severity of lung disease. However, the current study shows that there was only minimal disruption of intestinal cells by the virus despite the tropism. Given the minimal destruction of enterocytes, diarrhea associated with SARS-CoV infection may be more related to proteins or toxins produced during viral replication than malabsorption or inflammation.
Notably, the culture yield from the small intestine was even higher than that of specimens obtained from lung tissues, which is generally believed to be the prime target organ of this virus. All other organ tissues obtained during autopsy did not have viable virus recovered. By molecular diagnostic method, the detection rates of viral RNA from stool and nasopharyngeal aspirate samples were comparable in our study. On the other hand, our detection rate of viral RNA in nasopharyngeal aspirate was lower than the 32% reported by Peiris et al.14 The difference may be accounted for by the differences in primer sequences and the timing of obtaining these specimens. Despite this, all of these data favor the intestinal tropism of SARS-CoV. From the virologic point of view, this intestinal tropism is unusual because most diarrhea-associated viruses (e.g., rotavirus, calicivirus, enteric adenovirus, Norwalk virus) are nonenveloped. Viruses without envelope are generally more resistant and have a better chance of survival in the intestinal tract. The intestinal tropism of SARS-CoV raises the question of whether this virus is more resistant to disinfection than other enveloped viruses.
Although diarrhea on presentation was not associated with oxygen requirement and overall mortality of these patients, patients with diarrhea had higher rates of admission to the ICU and intubation. Given the many potential confounding factors complicating diarrhea in these patients with prolonged hospitalization, this association should be interpreted with caution. However, routine bacteriologic culture and detection of C. difficile toxin failed to identify any other culprit for diarrhea. Further studies are necessary to determine whether intestinal viral load has any correlation with enteric manifestation and, more importantly, the clinical outcome of these patients.
Another important issue relating to the intestinal tropism is the shedding of viral RNA in stool. Peiris et al. reported that the virus can be detected by RT-PCR in the stool samples of all 20 patients with positive nasopharyngeal aspirates at day 10 after onset of symptoms.14 At day 21, 67% of these initially positive patients still had viral RNA detected in stool samples. Our data further showed that virus can be detected by PCR in stool for up to 73 days from symptom onset in a patient, which may have substantial implications on infection control. Although viable virus is not recovered from stool samples, the question of whether stool remains a potential source of the virus in recovering or even recovered patients deserves further evaluation. It is also imperative that strict precautions be used in handling the excreta of these patients in addition to routine droplet and formite precautions to minimize the risk of viral transmission. With the high viral load in the intestinal tract, the potential risk to the endoscopist performing colonoscopy in patients with SARS is considerable if barrier precaution is not strictly enforced.
Our data support the intestinal tropism of SARS virus, and diarrhea is a common presenting symptom of this emerging infection. In addition to the call for increasing awareness for this atypical presentation, the findings of this study open up a new area for future research on this important virus. Understanding of the tissue tropism of SARS-CoV on intestinal cells may also help to elucidate the pathogenetic mechanism of this virus and possibly help in the development of novel antiviral therapy.
Footnotes
The authors thank Man-yee Yung, Sara Fung, Dr. Bonnie Kwan, and Dr. Thomas Li for their help in retrieving patient information.
References
- 1.Department of Health. Latest figures on severe acute respiratory syndrome. Available at: http://www.info.gov.hk/dh/diseases/ap/eng/infected.htm/. Accessed June 3, 2003.
- 2.Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt G.M, Ahuja A, Yung M.Y, Leung C.B, To K.F, Lui S.F, Szeto C.C, Chung S, Sung J.J. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.World Heath Organization. Cumulative number of reported probable cases of SARS. Available at: http://www.who.int/csr/sars/country/2003_05_31/en/. Accessed June 2, 2003.
- 4.Donnelly C.A, Ghani A.C, Leung G.M, Hedley A.J, Fraser C, Riley S, Abu-Raddad L.J, Ho L.M, Thach T.Q, Chau P, Chan K.P, Lam T.H, Tse L.Y, Tsang T, Liu S.H, Kong J.H, Lau E.M, Ferguson N.M, Anderson R.M. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ksiazek T.G, Erdman D, Goldsmith C.S, Zaki S.R, Peret T, Emery S, Tong S, Urbani C, Comer J.A, Lim W, Rollin P.E, Dowell S.F, Ling A.E, Humphrey C.D, Shieh W.J, Guarner J, Paddock C.D, Rota P, Fields B, DeRisi J, Yang J.Y, Cox N, Hughes J.M, JeDuc J.W, Bellini W.J, Anderson L.J, SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 6.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt H.R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier R.A, Berger A, Burguiere A.M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J.C, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk H.D, Osterhaus A.D, Schmitz H, Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Fouchier R.A, Kuiken T, Schutten M, Van Amerongen G, Van Doornum G.J, Van Den Hoogen B.G, Peiris M, Lim W, Stohr K, Osterhaus A.D. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez C.M, Izeta A, Sanchez-Morgado J.M, Alonso S, Sola I, Balasch M, Plana-Duran J, Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt W, Schneider T, Heise W, Weinke T, Epple H.J, Stoffler-Meilicke M, Liesenfeld O, Ignatius R, Zeitz M, Riecken E.O, Ullrich R. Stool viruses, coinfections, and diarrhea in HIV-infected patients. Berlin Diarrhea/Wasting Syndrome Study Group. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:33–38. doi: 10.1097/00042560-199609000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Gerna G, Passarani N, Battaglia M, Rondanelli E.G. Human enteric coronaviruses: antigenic relatedness to human coronavirus OC43 and possible etiologic role in viral gastroenteritis. J Infect Dis. 1985;151:796–803. doi: 10.1093/infdis/151.5.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simhon A, Mata L. Fecal rotaviruses, adenoviruses, coronavirus-like particles, and small round viruses in a cohort of rural Costa Rican children. Am J Trop Med Hyg. 1985;34:931–936. doi: 10.4269/ajtmh.1985.34.931. [DOI] [PubMed] [Google Scholar]
- 12.Resta S, Luby J.P, Rosenfeld C.R, Siegel J.D. Isolation and propagation of a human enteric coronavirus. Science. 1985;229:978–981. doi: 10.1126/science.2992091. [DOI] [PubMed] [Google Scholar]
- 13.Holmes K.V. Enteric infections with coronaviruses and toroviruses. Novartis Found Symp. 2001;238:258–269. doi: 10.1002/0470846534.ch16. [DOI] [PubMed] [Google Scholar]
- 14.Peiris J.S.M, Chu C.M, Cheng V.C.C, Chan K.S, Hung I.F.N, Poon L.L.M, Law K.I, Tang B.S.F, Hon T.Y.W, Chan C.S, Chan K.H, Ng J.C.S, Zheng B.J, Ng W.L, Lai R.W.M, Guan Y, Yuen K.Y, members of the HKU/UCH SARS Study Group Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Booth C.M, Matukas L.M, Tomlinson G.A, Rachlis A.R, Rose D.B, Dwosh H.A, Walmsley S.L, Mazzulli T, Avendano M, Derkach P, Ephtimios I.E, Kitai I, Mederski B.D, Shadowitz S.B, Gold W.L, Hawryluck L.A, Rea E, Chenkin J.S, Cescon D.W, Poutanen S.M, Detsky A.S. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 2003;289:2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. Updated interim U.S. case definition of severe acute respiratory syndrome (SARS). Available at: http://www.cdc.gov/ncidod/sars/casedefinition.htm/. Accessed June 10, 2003.
- 17.World Health Organization. Case definitions for surveillance of severe acute respiratory syndrome (SARS). Available at: http://www.who.int/csr/sars/casedefinition/en/. Accessed June 2, 2003.
- 18.World Health Organization. Use of laboratory methods for SARS diagnosis. Available at: http://www.who.int/csr/sars/labmethods/en/. Accessed June 16, 2003.
- 19.World Health Organization. PCR primers for SARS developed by WHO network laboratories. Available at: http://www.who.int/csr/sars/primers/en/. Accessed June 2, 2003.
- 20.WHO environmental health team reports on Amoy gardens. Available at: http://www.info.gov.hk/gia/general/200305/16/0516114.htm. Accessed June 2, 2003.
- 21.Nicholls J.M, Poon L.L, Lee K.C, Ng W.F, Lai S.T, Leung C.Y, Chu C.M, Hui P.K, Mak K.L, Lim W, Yan K.W, Chan K.H, Tsang N.C, Guan Y, Yuen K.Y, Peiris J.S. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


