Abstract
Three elements are crucial for the programmed frameshifting in translation of dnaX mRNA: a Shine-Dalgarno (SD)-like sequence, a double-shift site, and a 3′ structure. The conformation of the mRNA containing these three elements was investigated using chemical and enzymatic probes. The probing data show that the structure is a specific stem-loop. The bottom half of the stem is more stable than the top half of the stem. The function of the stem-loop was further investigated by mutagenic analysis. Reducing the stability of the bottom half of the stem strongly effects frameshifting levels, whereas similar changes in the top half are not as effective. Stabilizing the top half of the stem gives increased frameshifting beyond the WT efficiency. The identity of the primary RNA sequence in the stem-loop is unimportant, provided that the overall structure is maintained. The calculated stabilities of the variant stem-loop structures correlate with frameshifting efficiency. The SD-interaction and the stem-loop element act independently to increase frameshifting in dnaX.
Keywords: dnaX, frameshifting, recoding, probing, stem-loop structure
Abbreviations: SD, Shine-Dalgarno; DMS, dimethylsulfate; CMCT, 1-cyclohexyl-3-(morpholino-ethyl) carbodiimide metho-p-toluenesulfonate; WT, wild-type; IBV, infectious bronchitis virus; MMTV, mouse mammary tumor virus; HIV, human immunodeficiency virus; PCR, polymerase chain reaction; 2D, two-dimensional; FIV, feline immunodeficiency virus; HTLV-II, human T-cell leukemia virus type II; SRV-1, simian retrovirus type 1
Introduction
A very efficient programmed ribosomal frameshift is used in decoding the Escherichia coli dnaX gene. Two dnaX encoded products, γ and τ, are synthesized in a 1:1 molar ratio and both are subunits of DNA polymerase III. Decoding of the dnaX gene is unusual in that programmed frameshifting yields a product (γ), that is shorter than the non-shift product (τ; Flower and McHenry 1990, Tsuchihashi and Kornberg 1990, Blinkova and Walker 1990). Frameshifting occurs at a specific site two-thirds of the way through the coding region. The second codon in the new frame is a stop codon, and consequently the frameshift product, has only its carboxy-terminal amino acid decoded from the new frame (Tsuchihashi & Kornberg, 1990). As commonly found in programmed frameshifting, the change in reading frame at a particular site is stimulated by recoding signals present in the mRNA (Gesteland & Atkins, 1996). For dnaX frameshifting, three crucial elements have been identified: a 5′ Shine-Dalgarno sequence (SD; Larsen et al., 1994), a very slippery (Weiss et al., 1989) double-shift site and a 3′ structural element Flower and McHenry 1990, Tsuchihashi and Kornberg 1990, Blinkova and Walker 1990. Similar to retroviral frameshifting (Jacks et al., 1988), mRNA dissociates from both A and P site bound tRNAs at the shift site, slips “forwards” by one base, and re-pairs to permit a continuation of triplet reading, but now in the −1 frame. The SD sequence preceding the shift site is not involved in ribosomal initiation. 16 S rRNA in the translocating ribosomes transiently pairs with the SD sequence in the mRNA and stimulates −1 frameshifting 10 nucleotides downstream (Larsen et al., 1994). The 3′ structure, which also stimulates the shift, was proposed to be either a stem-loop Tsuchihashi and Kornberg 1990, Flower and McHenry 1990, Blinkova and Walker 1990 or possibly a pseudoknot (Blinkova & Walker, 1990). Tsuchihashi & Kornberg (1990) replaced blocks of nucleotides in the 3′ region and showed that the putative pseudoknot was not involved. Two different models for the top portion of the stem-loop have been proposed Tsuchihashi and Kornberg 1990, Flower and McHenry 1990, Blinkova and Walker 1990, and the experiments presented here address this issue.
Dual stimulatory signals 5′ and 3′ of programmed frameshift sites are known in three other cases: The −1 frameshift in decoding Shigella dysenteriae IS911 is similar to dnaX in that it has an SD sequence 11 nucleotides 5′ of a double-shift site which is followed by a complex RNA structure (M. F. Prère, J. F. Atkins, O. Fayet, unpublished results; Polard et al., 1991). Mammalian antizyme has a 5′ stimulator element of unknown nature and a pseudoknot 3′ of the +1 shift site Matsufuji et al 1995, Matsufuji et al 1996. In contrast, for eubacterial Release Factor 2 (RF2) +1 frameshifting, while the 5′ element is an SD sequence located 3 nucleotides upstream of the shift site (Weiss et al., 1988), the 3′ element is not a structure but a stop codon overlapping the shift site Weiss et al 1987, Curran and Yarus 1989, Sipley and Goldman 1993.
Mutational analyses have addressed the stimulatory effect of several 3′ structures involved in frameshifting (for reviews; see Miller et al 1995, Brierley 1995, Gesteland and Atkins 1996). Despite the functional importance of these RNA structures in programmed frameshifting, detailed structural information is available only for a few structural elements. Mutational analysis was used to identify pseudoknots involved in frameshifting in avian coronavirus infectious bronchitis virus (IBV; Brierley et al 1989, Brierley et al 1991) and in mouse mammary tumor virus (MMTV; Chamorro et al., 1992). In the latter case structural analyses as well as functional studies were performed on variant pseudoknots resulting in a model based on NMR analysis Chen et al 1995, Chen et al 1996, Shen and Tinoco 1995, Kang et al 1996. A pseudoknot was also shown to promote efficient −1 frameshifting in simian retrovirus type 1 (SRV-1) by mutagenesis and probing analysis (ten Dam et al., 1994). More recently, a general model was proposed for the conformation of pseudoknots involved in retroviral frameshifting (Du et al., 1996). However, some animal viruses, e.g. human astrovirus (Marczinke et al., 1994) and perhaps human immunodeficiency virus (HIV; Parkin et al., 1992) appear to use a stem-loop rather than a pseudoknot, for the stimulation of frameshifting. The 3′ structural elements involved in bacterial recoding are stem-loops with the exception of a pseudoknot in the insertion sequence IS3 (Sekine et al., 1994).
Here, the structure of the dnaX frameshift cassette RNA in solution was characterized using chemical and enzymatic probes.The predicted thermal stabilities of several variants of the 3′ stimulator were correlated with their ability to promote frameshifting in vivo.
Results
To analyze the structure-function relationship of the stem-loop involved in stimulation of frameshifting in the dnaX gene of E. coli: (1) the conformation of the frameshift region was investigated using enzymatic and chemical probes and (2) mutations, predicted to disrupt or alter the proposed structure, were engineered, and tested in vivo in a functional assay using β-galactosidase as a reporter. A region of the dnaX gene between nucleotide 1406 and nucleotide 1468 (Yin et al., 1986) was used because this sequence contains all the elements required for dnaX frameshifting (Figure 1).
Figure 1.
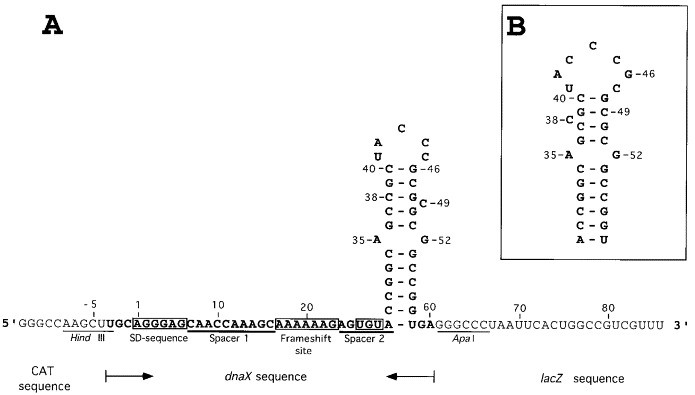
A. Sequence of RNA-2 including the dnaX frameshift cassette. The HindIII and ApaI sites are used for cloning of synthetic oligonucleotides between vector-borne CAT and lacZ. Nucleotides shown in boldface type are dnaX sequence (except for the A to U change in the stop codon). The UGA stop codon in the −1 frame (nucleotides 26 to 28) is changed to a cysteine codon (UGU) (boxed). The 99 nucleotide long RNA-2 is used for enzymatic and chemical probing. Spacer 1 is the nucleotides between the SD sequence and the shift site. Spacer 2 is the nucleotides between the frameshift site and the 3′ stem-loop structure proposed by Tsuchihashi and Kornberg (1990) and by Flower & McHenry (1990). Our numbering begins at the 1st nucleotide of the SD region (Larsen et al., 1994). B, The stem-loop structure proposed by Blinkowa & Walker (1990).
Structural probing
Probing with nucleases, imidazole, and lead was done on 5′-labeled RNA-2, the short transcript. Chemical modification with DMS and CMCT was done on RNA-1, the long transcript, since it was not possible to do primer extension on RNA-2 (see Materials and Methods). Nuclease S1cleaves single-stranded RNA with no known sequence specificity. RNase T1cleaves after unpaired guanosine residues and to a lesser extent, after guanosine residues involved in non-canonical base-pairing (Felden et al., 1997). The V1nuclease from cobra venom preferentially hydrolyzes double-stranded RNA and within regions containing stacked nucleotides. However, Lowman & Draper (1986) suggest that V1nuclease recognizes any 4 to 6 nucleotide segment of polynucleotide backbone with an approximately helical conformation, and does not require that the bases are paired. Detailed structural requirements for Pb(II)-induced hydrolysis are not known, although, site-specific cleavages in a tRNA molecule have been identified Werner et al 1976, Krzyzosiak et al 1988. A particular tertiary folding and an increased flexibility of the phosphate backbone have been suggested as specific structural features for Pb(II)-induced cleavage (Brown et al., 1983). The use of Pb(II) to probe the conformation of 16 S rRNA shows its predominant affinity for loops and inter-helical regions (Gornicki et al., 1989). Imidazole induces cleavages preferentially in single-stranded regions (Vlassov et al., 1995). Probing of Watson-Crick base-pairing positions with DMS (N-3 of C, N-1 of A) and CMCT (N-3 of U, N-1 of G) was done under both native (in the presence of magnesium) and semi-denaturing (in the presence of 1 mM EDTA) conditions.
Representative electrophoretic patterns are presented for several of the probes, and a complete summary of cleavage or modification sites were mapped onto the RNA sequence. The probe-induced cuts or modifications were categorized as weak, intermediate, or strong, according to the intensity of the corresponding band. Figure 2 shows enzymatic probing and the lead-hydrolysis experiments. These results are summarized in Figure 3. Figure 4 shows an autoradiogram of a gel obtained after CMCT modification of dnaX RNA, and the results of both DMS and CMCT modification on RNA-1 are summarized in Figure 5 along with the RNA-2 imidazole data. The very 3′ part of the RNA-1 molecule (nt 71 to 86, downstream of the ApaI cloning site, see Figure 1), and the nucleotides 5′ of the HindIII cloning site (G-13 to A-7), cannot be assessed due to the location of the primer used for primer extension. However, these regions are outside the dnaX sequence. Two degradation sites are present in the control lanes of RNA-1 (between C55 and G56, and between C66 and U67). The reactivities at Watson-Crick pairing positions on the 5′ side of these degradation sites could not be revealed, since during gel electrophoresis those primer extension products co-migrate with the degradation products. RNA-1 was also probed with nuclease S1. The S1pattern was the same for RNA-1 (data not shown) as for RNA-2, indicating that the conformations of the two RNAs are identical in the region of interest.
Figure 2.
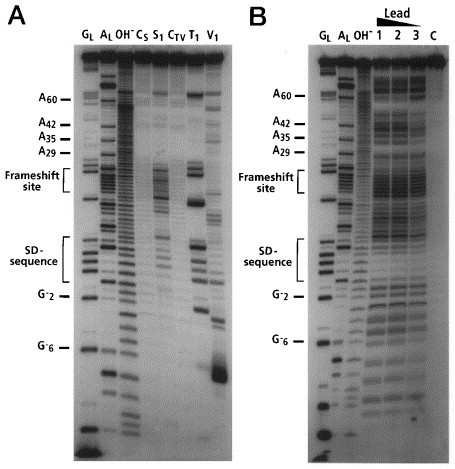
A, Nuclease mapping and B, lead cleavages of 5′-labeled dnaX RNA. Lanes CSand CTVare incubation controls for S1and T1or V1respectively. Lanes GLare RNase T1hydrolysis ladders. Lanes ALare RNase U2hydrolysis ladders. Lanes OH−are alkaline hydrolysis ladders. Lane S1, T1, and V1are nuclease S1, RNase T1, and nuclease V1mapping lanes respectively. Lane C is an incubation control for Pb(II) hydrolysis. Lanes 1, 2, and 3 are Pb(II) hydrolysis by incubation in 2.0 mM, 1.5 mM, and 0.7 mM (final concentration) lead acetate.
Figure 3.
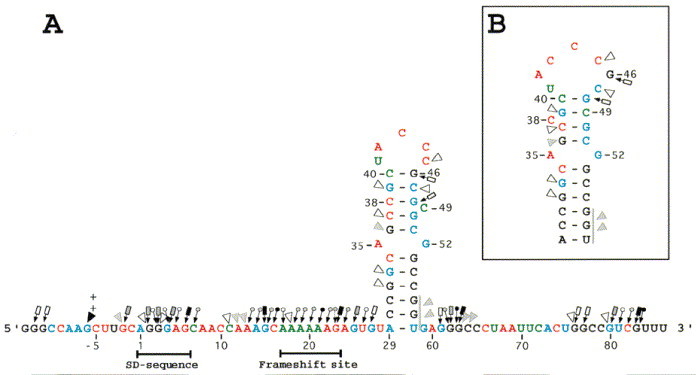


Figure 4.
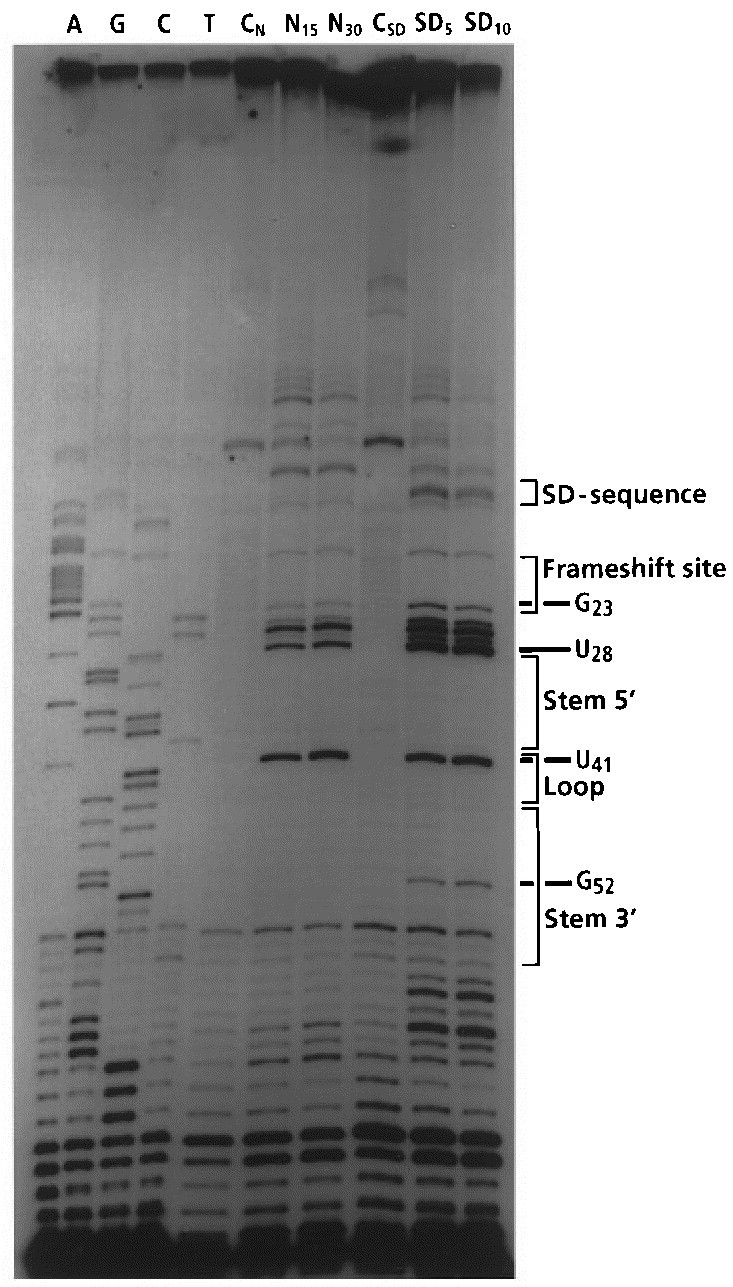
Mapping of N-3 of uracil and N-1 of guanine residues by CMCT using the primer extension method. Lanes A, G, C, and T are sequencing ladders generated in the presence of ddATP, ddGTP, ddCTP, and ddTTP. Lanes CNand CSDare incubation controls in the native and the semi-denaturing buffer respectively. Lanes N15and N30are probings under native conditions (incubation time 15 or 30 minutes). Lanes SD5and SD10are probings under semi-denaturing conditions (five or ten minutes incubation time).
Figure 5.
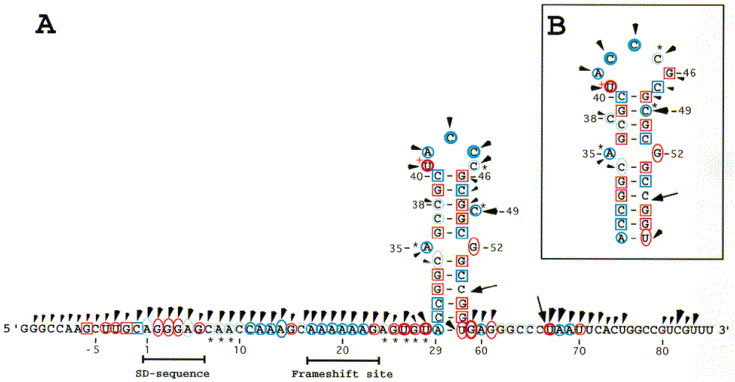
Imidazole induced cleavages and mapping data of Watson-Crick positions of the dnaX frameshift region RNA. Imidazole cleavages are shown on the RNA sequence with arrowheads. The size of the arrowheads refers to the intensity of cleavages (weak, intermediate, and strong). Chemical reactivity of the Watson-Crick positions are indicated on the RNA sequence. DMS reactivities are indicated in blue and CMCT reactivities in red. Reactivities under native conditions (○), under semi-denaturing conditions but not in native conditions (0), or no reactivities under both native and semi-denaturing conditions (□) of adenine residues (atom N-1), cytosine residues (atom N-3), guanine residues (atom N-1), and uracil residues (atom N-3). Weak, intermediate, and strong reactivities are denoted by light, thin, and thick symbols. (∗) indicates that the Watson-Crick position is more strongly modified under semi-denaturing conditions than under native conditions. The two arrows indicate degradation seen in the control lanes. A very strong CMCT modification is denoted by a+next to the nucleotide (U41).
Most of the probing data suggest that the SD sequence (A1 to G6) is single-stranded. Numerous nucleotides in this region are cut by S1, T1, lead (Figure 3) and imidazole (Figure 5). However, some ambiguities are seen: G2 and G3 are not cleaved by lead and they are weakly cut by V1. The N-1 atoms of the guanine and the adenine residues in the SD sequence are only modified by DMS or CMCT in the absence of magnesium.
Spacer 1 (C7 to C16) appears to be single-stranded. All nucleotides in this region are cut by lead and imidazole, and they are all modified by CMCT or DMS. Several S1cuts and a T1cut are also observed in this region. However C11 to A13 are cut by V1(see below).
The frameshift site (A17 to G23) and spacer 2 (A24 to A29) appear to be single-stranded regions. These sequences give intermediate or strong cuts with S1, T1, imidazole and lead, and all Watson-Crick positions are modified. DMS and CMCT modifications of the nucleotides in spacer 2 (A24 to U28) are all stronger under semi-denaturing conditions than under native conditions, indicating that magnesium stabilizes this portion of the RNA sequence. Nucleotides U28 and A29 in spacer 2 are not cut by S1, possibly due to proximity to the strong stem beginning at C30.
The bottom half of the predicted stem appears to be a very stable double-stranded region. The Watson-Crick positions of the nucleotides in the bottom half of the predicted stem are protected from modification by DMS and CMCT. Further, this part of the stem is neither cut by S1nor T1, and several nucleotides are cut by V1. Two intermediate V1cuts are seen between G56 and U58, but it is not possible to localize the exact position of the cleavages because of band compression in the sequencing gels, a further indication of a strong stem. G33 and C34 are partially cleaved by lead and only C34 is cleaved by imidazole. C30·G57 appears to be the very bottom base-pair in the stem. A29 and U58 are both cleaved by imidazole and lead and both are modified at their Watson-Crick positions.
In the proposed structures A35 and G52 are drawn as bulges, but these nucleotides are not cleaved by S1or T1. A35 is cleaved by lead and by imidazole, and G52 is weakly cleaved by lead. The N-1 atom of A35 is modified under native conditions, but N-1 of G52 is only modified in semi-denaturing buffer. These probing data are not sufficient to determine if the two nucleotides are bulged or if an A·G base-pair is present. Four different forms of A·G base-pairs have been described (Tinoco, 1993). Since N-1 of A35 is modified under native conditions two of the four can be excluded (G·A N1-N1, carbonyl-amino, and G·A N-3-amino, amino-N-1). Probing of N-7 of adenine residues is required to determine if one of the other A·G pairs (N-7-N-1, amino-carbonyl, and N-7-amino, amino-N-3) is present in the dnaX stem-loop. Probing of N-7 of adenine residues and NMR spectroscopy are in progress to better define the base arrangement (S. Alam, unpublished results).
The Watson-Crick positions of most of the nucleotides in the top half of the stem are protected from CMCT and DMS modification, and numerous V1cuts are observed. These data imply that the nucleotides are located in a helical domain. However, the top half of the stem appears less stable than the bottom half, as indicated by several minor imidazole and lead cleavages, and by the weak modification of C37 and C38 by DMS. The C37 and C38 modifications can be accounted for by an effect of the bulged nucleotide C49, as depicted in Figure 1A, or it could argue for the presence of a small amount of the structure shown in Figure 1B.
The proposed structure in Figure 1A has a five nucleotide loop (U41-C45) and a bulged nucleotide (C49). The five nucleotides in the predicted loop are all modified by CMCT or DMS and they all give strong or intermediate cuts with imidazole and lead. C49 is modified by DMS and it is strongly cut by both imidazole and lead. These probing data suggest that U41 to C45 and C49 are not involved in base-pairing. The structure shown in Figure 1B shows G46 and C47 as part of the loop sequence, and C38 a bulged nucleotide. G46 is not cleaved by lead and C47 is only weakly cleaved by lead. Furthermore, neither G46 nor C47 are modified in solution by CMCT or DMS, indicating that the two nucleotides are in a stem. These data strongly suggest that the structure in Figure 1A is correct, except that the probing results argue against the formation of the A29 to U58 base-pair.
It is interesting to note that no S1cuts were found in the proposed stem-loop, perhaps due to inaccessibility of the bulky enzyme related to tertiary folding of the loop sequences.
Other structural elements can be proposed in the probed region. For example, a four base-pair stem-loop structure 3′ of the primary stem-loop can be predicted from the lead and imidazole data Figure 3, Figure 5. Base-pairing between G62 and C65 and between G76 and C79 would form the stem. This structure is supported by V1cleavages, but is not consistent with the S1, T1, DMS, and CMCT data. Furthermore, this structure is not part of dnaX; it is in the downstream lacZ sequence. Another small stem-loop structure 5′ of the frameshift site can be predicted. The stem would involve nucleotides G-6 to C-2 and C11 to C16 including a bulged A. This structure is supported by V1cleavages (strong cut at G-6 and cuts at G-2 and C11 to A13), weakly supported by S1, T1, DMS and CMCT data and is inconsistent with both the lead and the imidazole pattern. This structure does include some dnaX sequence (the SD sequence and spacer 1 are part of the structure), but the HindIII cloning site is part of the stem. By adding more 5′ and 3′ dnaX sequence to our constructs, we found that these small structures, if they occur in solution, have insignificant effects on frameshifting.
Structure-function relationship of the 3′ stem-loop
The probing data support the structure proposed by Tsuchihashi and Kornberg 1990, Flower and McHenry 1990 except that the A29·U58 base-pair does not seem to form. The stimulatory effect of this 3′ structure was examined by deletion (SL del) and specific base changes (SLM 1-22). In Figure 6, the functional effects of in vivo mutations are summarized, using β-galactosidase as a reporter. Synthetic oligonucleotides, containing the frameshift cassette, were cloned into a lacZ-containing plasmid. The different constructs were designed on the basis of the two-dimensional (2D) structure predicted from the probing results. The FoldRNA program (the Wisconsin Sequence Analysis Package, GCG Inc.) was used to analyze each mutant construct for alternate secondary structures. In all constructs, the −1 frame stop codon (UGA) was changed to UGU (cysteine) to permit synthesis of β-galactosidase. The change did not affect the efficiency of frameshifting Tsuchihashi and Kornberg 1990, Larsen et al 1994. The efficiency of frameshifting of all the mutants has been normalized to the WT, defined as 100%. In the WT the proportion of ribosomes that shift frame at the dnaX shift site is 50% based on comparison to an in-frame control. As ribosome drop-off is expected to be the same in both the frameshift construct and the in-frame control, we expect the estimated frameshift level to be unbiased by this issue.
Figure 6.
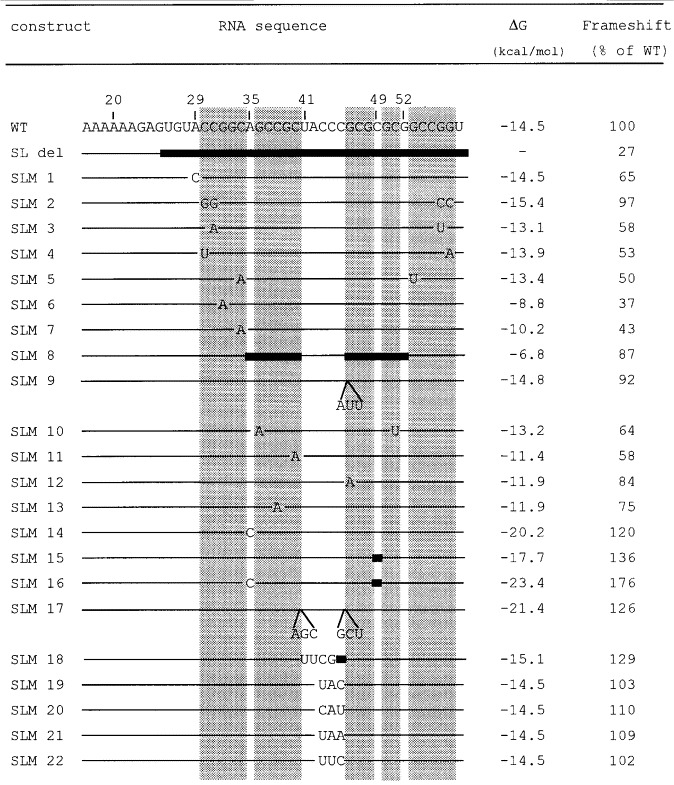
Mutational analysis of the stem-loop. The relevant nucleotide sequence of the wild-type (WT) construct is shown with the stem regions boxed. Our numbering begins at the first base of the SD sequence. Regions identical to the wild-type sequences are denoted by lines, and filld bars indicate deleted regions. Nucleotide substitutions and additions are indicated. Frameshift efficiencies and ΔG values for the mutated stem-loop structures are also listed. Calculations of ΔG (37°C) values (Turner et al., 1988) are based only on the stem-loop sequences. The amount of frameshifting measured using the WT construct is defined as 100%.
Deletion of the 3′ structure (SL del) reduced frameshifting to 27% of WT. The location of the beginning of the stem was tested by substitution of A29 with C (SLM 1). Frameshifting was reduced to 65%. Substitution of the potential partner (U58) was not attempted as the FoldRNA program predicted an alternate structure. The probing data argue against the formation of a canonical A·U base-pair at the base of the stem. However, it is possible that the A29·U58 base-pair does form, and contributes to frameshifting, but it may breath in solution and therefore show up on the structure mapping as single-stranded.
The bottom part of the stem will be considered first. Reversing the two bases at the base of the potential stem had no effect on frameshifting (SLM 2). Substituting a C·G base-pair with an A·U base-pair at the three positions tested lowered frameshifting to 50 to 58% (SLM 3-5). Single substitutions, which disrupted Watson-Crick base-pairs at two positions in the bottom part of the stem, reduced frameshifting to approximately 40% (SLM 6-7).
A small decrease in frameshifting was seen when the top part of the stem was deleted, leaving the loop intact (SLM 8), and when the size of the loop was increased by three nucleotides (SLM 9). Substituting a C·G base-pair with an A·U base-pair in the top part of the stem reduced frameshifting to 64% (SLM 10). Single substitutions which disrupted Watson-Crick base-pairs at three positions in the top part of the stem reduced frameshifting to 58 to 84% (SLM 11 to 13). In contrast, strengthening the stem by substitution of A35 with C, thereby creating a potential C·G base-pair, increased frameshifting to 120% (SLM 14). Deletion of the bulged C (C49) showed a similar effect (SLM 15). Combination of the two mutants (SLM 16) increased frameshifting to 176% of WT (corresponding to 88% frameshifting), giving a shift to non-shift ratio of approximately 9:1. Lengthening the top part of the stem by three base-pairs (SLM 17) increased frameshifting to 126%. Replacing the loop sequence by a tetraloop (Tuerk et al., 1988; UUCG, SLM 18) increased frameshifting to 129%. In general, strengthening the stem-loop gives increased frameshifting. The potential for pairing between the SD sequence and the three Cs in the loop was tested by substitution of the three C residuess (SLM 19 to 22). There was essentially no effect on frameshifting. This result argues against an interaction and is in agreement with the probing data.
Also shown in Figure 6, are the predicted free energy values (ΔG) for stem-loop folding of the various mutants (Turner et al., 1988). In Figure 7A, these values are plotted against frameshifting level. The frameshifting values were corrected for the basal level of activity without the stem-loop. An identical set of mutants were made that lacked the SD sequence in the frameshift window. The data obtained from these mutants are plotted in Figure 7B. Given the uncertainty in the theoretical calculations, a surprisingly good linear correlation exists between predicted stabilities and frameshift levels for both sets of mutants. More stable stem-loop structures produce higher levels of frameshifting. Furthermore, the slopes of the two lines are nearly identical. This strongly suggests that the SD-like interaction, involving mRNA and 16 S rRNA, does not influence the way the stem-loop works to stimulate frameshifting. The two stimulators work independently. Furthermore, there is a threshold stability below which the stem-loop does not stimulate frameshifting (non-zero x intercept). As expected, this threshold is lower for the set of mutants in which the SD sequence is present (line shifted to the right in Figure 7B compared to 7A), since the SD interaction increases frameshifting (Larsen et al., 1994). The two independent stimulatory elements can act together.
Figure 7.
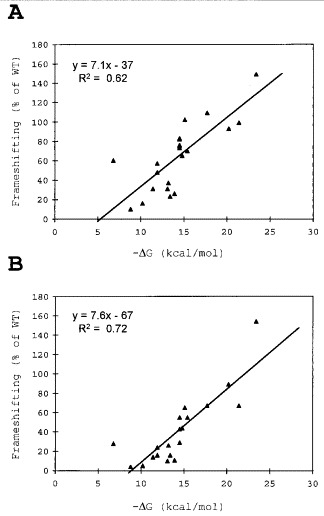
Correlation between the calculated free energy (ΔG) values (Turner et al., 1988) and frameshifting efficiency for all the stem-loop constructs. Values are adjusted by subtracting the level obtained from a stem-loop deletion construct and are expressed as a percentage of the WT level. A linear regression line is shown along with the equation for the line and the regression statistic, R2. Data are shown for the mutants in which the wild-type SD sequence is present (A) and absent (B).
How does the dnaX stem-loop compare to other stem-loops involved in prokaryotic frameshifting? In a construct with the dnaX shift cassette, replacement of the stem-loop with its IS911 (Polard et al., 1991) counterpart reduces frameshifting to 32%. A similar substitution with the IS150 (Vögele et al., 1991) counterpart showed 90% of dnaX WT level frameshifting. However, a hypothetical start codon (GUG) in the −1 frame is present in the loop of IS150 that could contribute to expression of lacZ. When that GUG codon was replaced by a GUC (valine) codon, 69% of dnaX WT frameshifting was measured (data not shown). The stem-loop in dnaX is designed for higher frameshifting efficiency than its IS911 and IS150 counterparts.
Spacer 2: the nucleotides between the frameshift site and the stem-loop structure
Two features of the dnaX frameshift window are similar to those responsible for recoding in several classes of viruses including retroviruses (Jacks et al., 1988) and cornoaviruses (Brierley et al., 1992), a heptanucleotide slippery sequence followed by a 3′ stem-loop or by a pseudoknot. The distance between the slippery sequence and the 3′ stimulator appears to be constrained to 5 to 8 nucleotides in viruses (with two possible exceptions, Brierley, 1995). That precise spacing is critical for high-level frameshifting has been demonstrated in avian coronavirus Infectious Bronchitis Virus (IBV, Brierley et al., 1989), in the Feline Immunodeficiency Virus (FIV, Morikawa & Bishop, 1992), in Human T-cell Leukemia Virus type II (HTLV-II, Kollmus et al., 1994), and in Simian Retrovirus type 1 (SRV-1, ten Dam et al., 1994). Alteration of the relative position of the shift site in IBV by three nucleotides with respect to the pseudoknot, causes a dramatic reduction in frameshifting efficiency. A similar result was observed in FIV. Deletion or insertion of a codon reduced frameshifting to between 2 and 5% compared to about 30% frameshifting in a wild-type construct. In HTLV-II and SRV-1, the optimal spacing was found to be seven nucleotides as is present in the wild-type sequences.
In the dnaX gene, the spacing between the shift site and the 3′ stem-loop is six nucleotides. Previously a three-nucleotide insertion in spacer 2 showed a slight reduction in frameshifting (Tsuchihashi & Brown, 1992). To determine if this spacing is critical several constructs with deletions or additions of nucleotides in spacer 2 were made (Table 1 and Figure 8).) The constructs containing the 5′ SD sequence will be presented first (Figure 8A). Deleting 1 nucleotide in spacer 2 reduced frameshifting to about 50%. The effect of deleting three nucleotides was tested in two different constructs. In one construct, the serine codon (AGU) was deleted and in the other the valine (GUA) codon was deleted. In both, frameshifting dropped to about 17%. This level is lower than that of the construct where the stem-loop was deleted (SL del, 27%). The effect of increasing spacer 2, by one, two, three, or four codons, was tested using two different sets of codons; both yielded similar results. Frameshifting dropped respectively to 64%, 53%, 58%, and 41% when adding one, two, three, and four codons to spacer 2 (Figure 8A).
Table 1.
Deletions and additions in spacer 2
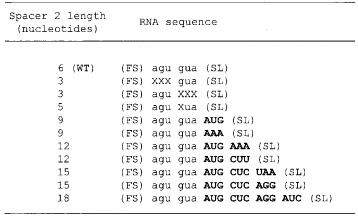
(FS) and (SL) denote the frameshift site (A AAA AAG) and the stem-loop, respectively. Lower case letters are WT spacer 2 sequence. Deleted nucleotides are indicated with Xs. Nucleotides shown in bold are added.
Figure 8.
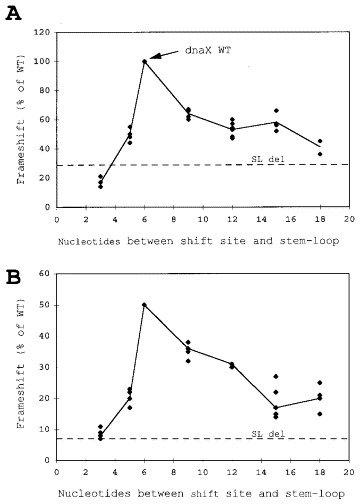
Effects of deletions and additions between the frameshift site and the stem-loop structure on the level of frameshifting. (A) Effect of deletions and additions in spacer 2 of dnaX. The SD sequence is present in the constructs. For reference, the dnaX WT mRNA spacing are indicated with an arrow. The horizontal dashed lines labeled SL del indicate the amount of frameshifting in constructs with the 3′ structure deleted. (B) Same as panel A, except that the SD sequence was deleted from the constructs.
Parallel constructs with the 5′ SD sequence deletion showed similar results (Figure 8B). Deleting one nucleotide or one codon in spacer 2 showed severe effects on the levels of frameshifting. Frameshifting dropped to 36%, 31%, 17% and 20% respectively when one, two, three, and four codons were added to the spacer. The length of spacer 2 is critical for dnaX frameshifting, altering the spacer region reduced frameshifting significantly.
Discussion
Two different structures have previously been proposed for the stem-loop involved in frameshifting in the dnaX gene of E. coli Tsuchihashi and Kornberg 1990, Flower and McHenry 1990, Blinkova and Walker 1990. Our probing data distinguish between the two, and strongly suggest that the model illustrated in Figure 1A is correct, except that the probing results argue against the formation of the A29·U58 base-pair. The data also rule out possible pairing between the SD sequence and nucleotides in the loop. The five nucleotides in the loop are all accessible to cleavage by imidazole and lead, and their Watson-Crick positions are all modified by DMS and CMCT. Theoretical calculations of ΔG values (Turner et al., 1988) show that the stability of the stem-loop structure correlates with the frameshifting efficiency Figure 6, Figure 7. The more stable stem-loop structures cause increased frameshifting even above WT levels. The strength of the stem-loop is not the only factor influencing frameshifting. The SD sequence 10 bases 5′ of the shift site is also a significant contributor to frameshifting, both in dnaX mRNA and mRNAs from other genes (Larsen et al., 1994). Our data strongly suggest that the two stimulators work independently. The SD-like mRNA:rRNA interaction does not influence the way the stem-loop works to stimulate frameshifting and vice versa. To a first approximation, the identity of any single nucleotide in the stem-loop is unimportant provided that the overall structure is maintained. A similar result was observed for the pseudoknot involved in IBV frameshifting (Brierley et al., 1991).
It is unlikely that the stem-loop binds a protein to influence the oncoming ribosome. Single base substitutions in the stem-loop did not influence frameshifting levels significantly, and deleting the top half of the dnaX stem only reduced frameshifting to 87% of WT. Presumably the RNA structure directly influences the oncoming ribosome. A similar deduction was made by ten Dam et al. (1994) for simian retrovirus −1 frameshifting; adding SRV pseudoknot RNA to reticulocyte lysate programmed with SRV mRNA caused no decrease in the frameshifting level. If a specific pseudoknot-binding protein was involved, it might have been titrated out in such an experiment.
A construct containing only the shift site and a stem-loop with uninterrupted Watson-Crick base-pairing (SLM 16 with the SD sequence deleted) gave 160% of WT levels, or 80% frameshifting (see Figure 7B). Notably, this frameshift efficiency is achieved without a second stem and formation of a pseudoknot; in contrast efficient eukaryotic −1 frameshifting events utilize pseudoknots. The most efficient eukaryotic frameshifts known are in IBV and MMTV gag-pro where the efficiencies are 23 to 30% Brierley et al 1987, Moore et al 1987, Jacks et al 1987. In addition to MMTV, other retroviruses that require two shifts to make the Gag-Pro-Pol precursor have similar high efficiency for the gag-pro shift (Nam et al., 1993). A high efficiency shift (26 to 29%) has also been reported for a plant virus, cocksfoot mottle sobemovirus, but the nature of the stimulatory element(s) has not yet been experimentally investigated (Makinen et al., 1995). In distinction to eukaryotes, the only reported case of a pseudoknot stimulator in prokaryotic frameshifting is in decoding the insertion sequence IS3 (Sekine et al., 1994). Further work is needed to compare the relative contributions of stem-loops and pseudoknots in prokaryotic frameshifting.
The mutational analysis presented shows that the bottom half of the dnaX stem is sufficient to provide a strong stimulatory effect on frameshifting. A stem-loop, of five base-pairs, made by deleting the top part of the WT stem gave 17% frameshifting (SLM 8 with the SD sequence deleted). This is likely due to the fact that the bottom half of the stem is more stable than the top, as substantiated by the probing data. In another report, a construct with a five-base-pair stem and six-nucleotide loop from beet western yellow virus, situated six nucleotides 3′ of an A AAA AAG shift site, gave only 4% frameshifting in E. coli (Garcia et al., 1993). However, when we substituted the beet western yellow virus stem-loop for its dnaX counterpart in a construct lacking the SD sequence, we detected 14% frameshifting, consistent with the 17% value obtained from SLM 8 with the SD sequence deleted.
In addition to the strength of the stem-loop structure, its spacing from the shift site is important. Several groups have moved a 3′ stimulator for different cases of programmed frameshifting, one codon in either direction, and a substantial decrease in frameshifting was observed. Moving the dnaX stem-loop structure upstream (decreasing spacer 2) has a dramatic effect on frameshifting. Moving the structure downstream also reduced −1 frameshifting, but not as severely as in the other studies.
A stabilized stem-loop without the SD sequence can give WT levels of frameshifting (SLM 14 with the SD sequence deleted). Why is an SD sequence used in addition to a stem-loop? We don’t know. SD sequences are utilized in other cases of −1 frameshifting, but at least with IS911 decoding there is a dual initiation role (M. F. Prère, J. F. Atkins & O. Fayet unpublished results). An SD sequence is also involved in stimulating the obligatory +1 frameshifting required for expression of eubacterial RF2. In this case, it is clear why the SD sequence is present; the 3′ stimulator for the +1 frameshift is inadequate on its own to promote frameshifting at the required level (Weiss et al., 1987).
In addition to the dilemma of the reason for the involvement of the SD sequence, another unknown is why the top part of the stem is present. Presumably a small addition to the bottom half of the stem in the absence of the top half would give WT level frameshifting. It could be that the complex nature of the shift signals is an evolutionary relic which reflect an unknown origin. Perhaps they once served some other function and got trapped by the requirement for optimum efficiency. One could imagine that retention of the top part of the stem-loop may reflect selection at the amino acid level, but this is difficult to envisage for the SD sequence. It is not obvious that regulation is the reason for the complexity. The dnaX frameshifting ratio appears to be set; there is no evidence for autoregulation as in RF2 frameshifting Craigen and Caskey 1986, Kawakami and Nakamura 1990. Adding purified τ and γ subunits to an in vitro translation system had no effect on frameshifting (Tsuchihashi, 1991). However, there could be some reason for the complexity that is not yet apparent. Evolution experiments starting with engineered variants of the chromosomal gene may address this point. The chromosomal dnaX gene could be replaced by a derivative in which frameshifting is required for expression of the essential τ (Blinkova et al., 1993) but not for γ. Having a simple weak frameshifting stimulator could create conditions for selection of potentially informative stronger frameshifting signals or perhaps even mutants in other DNA polymerase III components.
During purification of DNA polymerase III proteolytic cleavage of τ by the outer membrane protein T protease, OmpT, to a γ-like derivative is detectable (Pritchard et al., 1996). Why is γ produced by frameshifting rather than by some proteolytic mechanism? Maybe the ability of the frameshifting mechanism to generate a 1:1 ratio of the two subunits is the reason for its utilization.
The overall DNA sequence identity of the dnaX genes in E. coli and Salmonella typhimurium (Blinkova et al., 1996) is 83%, close to the current estimate, 84.4%, of divergence of average genes between these species (Sharp, 1991). The 58 nucleotides frameshift cassettes are identical, apart from two base substitutions in spacer 1 and two base substitutions in the loop. This high degree of conservation in important parts of the cassettes means that the shift cassette will have to be analyzed, in more distant bacteria, to detect variability. Despite considerable horizontal transfer between bacterial “species”, it has been estimated that the divergence of E. coli and Salmonella typhirurium happened 100 million years ago Doolittle et al 1996, Lawrence and Ochman 1997.
In RF2 frameshifting, an SD-like interaction between rRNA and mRNA is crucial. Strengthening the pairing potential above that found in WT reduces the frameshift (Weiss et al., 1987). Stabilizing the IS150 stem-loop did not increase the rate of frameshifting above the WT level (Vögele et al., 1991). In contrast, strengthening the dnaX stem-loop beyond the WT level increased frameshifting. The shift: non-shift product ratios can be set at a wide range of levels. However, in the WT dnaX mRNA the stimulators are tuned to give a 1:1 ratio of τ and γ. It is remarkable that levels as high as 88% frameshifting can be achieved (SLM 16). It remains to be seen if 100% can be attained.
Materials and methods
Enzymes and chemicals
Nucleases S1, T1, V1, U2, nucleotides (deoxynucleotides and dideoxynucleotides), and calf intestinal alkaline phosphatase were from Pharmacia (Piscataway, New Jersey). Phage T4 polynucleotide kinase was obtained from New England BioLabs (Beverly, MA). Nuclease-free water and T7 RNA polymerase were from Promega (Madison, Wisconsin). Total yeast tRNA, used as carrier RNA to supplement labeled RNA, CMCT (1-cyclohexyl-3-(morpholino-ethyl) carbodiimide metho-p-toluenesulfonate) and lead acetate were from Sigma (St. Louis, Missouri). DMS (dimethylsulfate) and imidazole were purchased from Aldrich (Milwaukee, Wisconsin). [γ-32P]ATP was from DuPont NEN (Wilmington, Delaware).
Structural probing
Two different RNAs were used for the probing experiments. Both contain dnaX sequences cloned downstream from a phage T7 RNA polymerase promoter. A 2000 nucleotide long RNA (RNA-1) was prepared by in vitro T7 transcription of the wild-type (WT) plasmid previously used for in vivoβ-galactosidase measurements (See Figure 2 in Larsen et al., 1994). This plasmid was linearized by EcoRV prior to transcription. A plasmid used for expressing a shorter RNA (RNA-2) was constructed using polymerase chain reaction (PCR), and the WT plasmid was used as template. From its 5′ end, one primer (62 nucleotides total) had a tail consisting of an AatII site, the T7 phage RNA polymerase promoter, and 28 nucleotides of the WT sequence. The other primer had a tail consisting of an EagI site, a PmeI site and 18 nucleotides of the WT sequence. The PCR product was purified on a centricon 30 (Amicon, Beverly, MA) and digested with AatII and EagI. The cut fragment was ligated into AatII-EagI cut pBR322 and the DNA sequence was confirmed by plasmid sequencing. Before transcription, the plasmid was linearized by Pme I. Both plasmids were transcribed with T7 RNA polymerase following a protocol from Promega. After transcription, RNA-1 was washed with RNase free H2O on a microcon 10 spin-column (Amicon). RNA-2 was gel-purified (12%, w/v PAGE), electroeluted, and ethanol precipitated. RNA-2 was used for 5′ end-labeling, and RNA-1 was used for the primer extension experiments, since it was not possible to do primer extension on RNA-2.
Before 5′ labeling, RNA-2 was dephosphorylated with alkaline phosphatase (Silberklang et al., 1977). RNA-2 and the oligonucleotide used for primer extension were labeled with [γ-32P]ATP using phage T4 polynucleotide kinase. After labeling, both the RNA and the oligonucleotide were gel purified, eluted, and ethanol-precipitated.
Both RNA-1 and RNA-2 were renatured by heating to 80°C for 2.5 minutes in the digestion buffers, and allowed to cool slowly to room temperature for 15 minutes before enzymatic digestion or chemical modification. All probing experiments were done at 37°C.
Probing of RNA-2 was performed as follows: Reaction mixtures (total volume 20 μl) contained 5′-end-labeled RNA (50,000 to 100,000 cpm/reaction) supplemented with 1 μg of carrier tRNA. Digestions with ribonucleases V1and T1were done in a buffer containing 50 mM sodium cacodylate (pH 7.5), and 20 mM magnesium acetate. The same buffer was used for nuclease S1digestion except that 1 mM zinc acetate was added. A titration of each enzyme was done to define the optimal conditions for probing (data not shown). The following amounts of enzyme were added (units were defined by Pharmacia): 5 units of nuclease S1, 0.06 units of RNaseT1, and 0.05 units of nuclease V1. Incubation time was seven minutes. Reactions were stopped by phenol extraction followed by ethanol-precipitation of the RNA. Imidazole-induced cleavage of RNA was performed as described (Vlassov et al., 1995), using an imidazole buffer at 2.0 M for a 12 hour incubation time. Probing with lead acetate was done as described (Krzyzosiak et al., 1988) for five minutes in a buffer containing Pb(II) acetate ranging from 0.7 to 2.0 mM. The products of limited cleavage of RNA-2 were assessed by 12% denaturing PAGE, and the cut positions were identified by comparison with parallel ladders created by alkaline hydrolysis, RNase T1, and RNase U2digestion.The buffers used for generating the adenine and guanine ladder were those recommended by the manufacturer.
Mapping of Watson-Crick positions with DMS and CMCT were done on RNA-1 as described Peattie and Gilbert 1980, Felden et al 1994. The reaction mixtures contained the appropriate buffer (in a total volume of 200 μl), supplemented with 4 μg of RNA, and 1 μl of a 50% (v/v) DMS diluted in 100% ethanol or 50 μl of 42 mg/ml CMCT in H2O. DMS modification was done for six minutes under both native (with 20 mM magnesium acetate) and semi-denaturing (without magnesium and supplemented with 1 mM EDTA) conditions. CMCT modification was done for 15 and 30 minutes under native conditions and five and ten minutes under semi-denaturing conditions. The reactions were stopped and modification sites were assigned as described previously (Romby et al., 1987). Modification sites were detected by analyzing DNA sequencing patterns generated by primer extension with reverse transcriptase of the modified RNAs.
The amount of label in each sample was determined before loading, and the same number of counts were loaded in each lane. The band intensities were assessed manually.
In vivo β-galactosidase activity measurements
Site-directed mutagenesis of the dnaX frameshift window was performed by cloning two complementary synthetic oligonucleotides into a lacZ-containing plasmid RW201 (a gift from R. Weiss). As described previously, the wild-type construct (WT) contains 63 bp of the dnaX sequence (Larsen et al., 1994). In all constructs, translation from the −1 frame of dnaX after the shift site, can continue unimpeded into the zero frame of lacZ. The UGA stop codon in the −1 frame, 3′ of the frameshift site, was modified to UGU (cysteine) to measure β-galactosidase activity for the frameshift product (Figure 1). The DNA sequence of all constructs was verified by sequencing.Whole-cell β-galactosiase assays (100 μl of culture grown in Luria-Bertani medium) were performed under a modification (Weiss et al., 1987) of the Miller (1972) conditions. Four separate cultures were made from each of two independent primary isolates of each mutant and assayed. The β-galactosidase levels measured were within ±10%.
Spacer 2 mutants
The constructs used to assay the effect of different spacer 2 lengths are shown in Table 1. The constructs were made both with and without the SD sequence present. When necessary, correction of the reading frame was accomplished downstream of the dnaX stem-loop.
Acknowledgements
We thank Brice Felden for invaluable advise on the structural probing including interpretation of the data and for constructive comments on the manuscript. The critical reading by Norma Wills and especially Harold Swerdlow is much appreciated. We also thank Dr J. Walker for informing us of the Salmonella typhimurium dnaX sequence prior to its deposition in the database. R. F. G. is an investigator of the Howard Hughes Medical Institute. This work was also supported by a grant (to J. F. A.) from NIH (RO1-GM48152-05).
Footnotes
Edited By D. E. Draper
References
- Blinkova A.L, Walker J.R. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucl. Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkova A, Hervas C, Stukenberg P.D, Onrust R, O’Donnell M.E, Walker J.R. The Escherichia coli DNA polymerase III Holoenzyme contains both products of the dnaX gene, τ and γ, but only τ is essential. J. Bacteriol. 1993;175:6018–6027. doi: 10.1128/jb.175.18.6018-6027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkova A, Burkart M.F, Owens T.D, Walker J.R. Salmonella typhimurium dnaX. Database Accession U66040 Genbank; USA: 1996. [Google Scholar]
- Brierley I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Brierley I, Boursnell M.E.G, Binns M.M, Bilimoria B, Blok V.C, Brown T.D.K, Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Digard P, Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Rolley N.J, Jenner A.J, Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Jenner A.J, Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.S, Hingerty B.E, Dewan J.C, Klug A. Pb(II)-catalysed cleavage of the sugar-phosphate backbone of yeast tRNAPheimplications for lead toxicity and self-splicing RNA. Nature. 1983;303:543–546. doi: 10.1038/303543a0. [DOI] [PubMed] [Google Scholar]
- Chamorro M, Parkin N, Varmus H.E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl Acad. Sci. USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chamorro M, Lee S.I, Shen L.X, Hines J.V, Tinoco I, Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kang H, Shen L.X, Chamorro M, Varmus H.E, Tinoco I. A characteristic bent conformation of RNA pseudoknots promotes −1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- Craigen W.J, Caskey C.T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Doolittle R.F, Feng D-F, Tsang S, Cho G, Little E. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science. 1996;271:470–476. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- Curran J.F, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J. Mol. Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- Du Z, Giedroc D.P, Hoffman D.W. Structure of the autoregulatory pseudoknot within the gene 32 messenger RNA of bacteriophages T2 and T6: A model for a possible family of structurally related RNA pseudoknots. Biochemistry. 1996;35:4187–4198. doi: 10.1021/bi9527350. [DOI] [PubMed] [Google Scholar]
- Felden B, Florentz C, Giege R, Westhof E. Solution structure of the 3′-end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J. Mol. Biol. 1994;235:508–531. doi: 10.1006/jmbi.1994.1010. [DOI] [PubMed] [Google Scholar]
- Felden B, Himeno H, Muto A, McCutcheon J.P, Atkins J.F, Gesteland R.F. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA) RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- Flower A.M, McHenry C.S. The γ subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 1990;87:3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A, van Duin J, Pleij C.W. Differential response to frameshift signals in eukaryotic and prokaryotic translational systems. Nucl. Acids Res. 1993;21:401–406. doi: 10.1093/nar/21.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R.F, Atkins J.F. Recoding: Dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- Gornicki P, Baudin F, Romby P, Wiewiorowski M, Kryzosiak W, Ebel J.P, Ehresmann C, Ehresmann B. Use of lead(II) to probe the structure of large RNAs. Conformation of the 3′ terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J. Biomol. Struct. Dyn. 1989;6:971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- Jacks T, Townsley K, Varmus H.E, Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc. Natl Acad. Sci. USA. 1987;84:4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Madhani H.D, Masiarz F.R, Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Hines J.V, Tinoco I. Conformation of a non-frameshifting RNA pseudoknot from Mouse Mammary Tumor Virus. J. Mol. Biol. 1996;259:135–147. doi: 10.1006/jmbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Nakamura Y. Autogenous suppression of an opal mutation in the gene encoding peptide chain release factor 2. Proc. Natl Acad. Sci. USA. 1990;87:8432–8436. doi: 10.1073/pnas.87.21.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus H, Honigman A, Panet A, Hauser H. The sequences of and distance between two cis-acting signals determine the efficiency of ribosomal frameshifting in human immunodeficiency virus type 1 and human T-cell leukemia virus type II in vivo. J. Virol. 1994;68:6087–6091. doi: 10.1128/jvi.68.9.6087-6091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzosiak W.J, Marciniec T, Wiewiorowski M, Romby P, Ebel J.P, Giege R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988;27:5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- Larsen B, Wills N.M, Gesteland R.F, Atkins J.F. rRNA-mRNA base-pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.G, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Lowman H.B, Draper D.E. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
- Mäkinen K, Næss V, Tamm T, Truve E, Aaspollu A, Saarma M. The putative replicase of the cocksfoot mottle sobemovirus is translated as a part of the polyprotein by -1 ribosomal frameshift. Virology. 1995;207:566–571. doi: 10.1006/viro.1995.1118. [DOI] [PubMed] [Google Scholar]
- Marczinke B, Bloys A.J, Brown T.D, Willcocks M.M, Carter M.J, Brierley I. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J. Virol. 1994;68:5588–5595. doi: 10.1128/jvi.68.9.5588-5595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins J.F, Gesteland R.F, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S, Matsufuji T, Wills N.M, Gesteland R.F, Atkins J.F. Reading two bases twice: Mammalian antizyme frameshifting in yeast. EMBO J. 1996;15:1360–1370. [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Miller W.A, Dinesh-Kumar S.P, Paul C.P. Luteovirus gene expression. Crit. Rev. Plant Sci. 1995;14:179–211. [Google Scholar]
- Moore R, Dixon M, Smith R, Peters G, Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: Two frameshift suppression events are required for translation of gag and pol. J.Virol. 1987;61:480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S, Bishop D.H. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S.H, Copeland T.D, Hatanaka M, Oroszlan S. Characterization of ribosomal frameshifting for expression of pol gene products of human T-cell leukemia virus type I. J. Virol. 1993;67:196–203. doi: 10.1128/jvi.67.1.196-203.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N.T, Chamorro M, Varmus H.E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D.A, Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P, Prère M.F, Chandler M, Fayet O. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol. 1991;222:465–477. doi: 10.1016/0022-2836(91)90490-w. [DOI] [PubMed] [Google Scholar]
- Pritchard A.E, Dallmann H.G, McHenry C.S. In vivo assembly of the τ-complex of the DNA polymerase III holoenzyme expressed from a five-gene artificial operon. J. Biol. Chem. 1996;271:10291–10298. doi: 10.1074/jbc.271.17.10291. [DOI] [PubMed] [Google Scholar]
- Romby P, Moras D, Dumas P, Ebel J.P, Giege R. Comparison of the tertiary structure of yeast tRNAAsp and tRNAPhe in solution. Chemical modification study of the bases. J. Mol. Biol. 1987;195:193–204. doi: 10.1016/0022-2836(87)90336-6. [DOI] [PubMed] [Google Scholar]
- Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- Sharp P.M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J. Mol. Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- Shen L.X, Tinoco I. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Silberklang M, Prochiantz A, Haenni A.L, RajBhandary U.L. Studies on the sequence of the 3′ terminal region of turnip yellow mosaic virus. Eur. J. Biochem. 1977;72:465–478. doi: 10.1111/j.1432-1033.1977.tb11270.x. [DOI] [PubMed] [Google Scholar]
- Sipley J, Goldman E. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc. Natl Acad. Sci. USA. 1993;90:2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E, Brierley I, Inglis S, Pleij C. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucl. Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I. Structure of base-pairs involving at least two hydrogen bonds. In: Gesteland R.F, Atkins J.F, editors. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 603–607. [Google Scholar]
- Tsuchihashi Z. Translational frameshifting in the Escherichia coli dnaX gene in vitro. Nucl. Acids Res. 1991;19:2457–2462. doi: 10.1093/nar/19.9.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z, Brown P.O. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unsatable interaction between tRNALysand an AAG lysine codon. Genes Dev. 1992;6:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Kornberg A. Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc. Natl Acad. Sci. USA. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gauss P, Thermes C, Groebe D.R, Gayle M, Guild N, Stormo G, d’Aubenton-Carafa Y, Uhlenbeck O.C, Tinoco I, Brody E.N, Gold L. CUUCGG hairpins: Extraordinary stable RNA secondary structures associated with various biochemical processes. Proc. Natl Acad. Sci. USA. 1988;85:1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.H, Sugimoto N, Freier S.M. RNA structure prediction. Annu. Rev. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Vlassov V.V, Zuber G, Felden B, Behr J.-P, Giege R. Cleavage of tRNA with imidazole and spermine imidazole constructs: a new approach for probing RNA structure. Nucl. Acids Res. 1995;23:3161–3167. doi: 10.1093/nar/23.16.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögele K, Schwartz E, Welz C, Schiltz E, Rak B. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucl. Acids Res. 1991;19:4377–4385. doi: 10.1093/nar/19.16.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.B, Dunn D.M, Atkins J.F, Gesteland R.F. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R.B, Dunn D.M, Dahlberg A.E, Atkins J.F, Gesteland R.F. Reading frame switch caused by base-pair formation between the 3′ end of 16 S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.B, Dunn D.M, Shuh M, Atkins J.F, Gesteland R.F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New. Biol. 1989;1:159–169. [PubMed] [Google Scholar]
- Werner C, Krebs B, Keith G, Dirheimer G. Specific cleavages of pure tRNAs by plumbous ions. Biochim. Biophys. Acta. 1976;432:161–175. doi: 10.1016/0005-2787(76)90158-1. [DOI] [PubMed] [Google Scholar]
- Yin K.C, Blinkowa A, Walker J.R. Nucleotide sequence of the Escherichia coli replication gene dnaZX. Nucl. Acids Res. 1986;14:6541–6549. doi: 10.1093/nar/14.16.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]


