Abstract
A study was performed to evaluate porcine torovirus (PToV) seroprevalence and infection in three multi-site farms from the North-eastern region of Spain. Serum samples from 120 piglets and faecal samples from 36 piglets were longitudinally collected at 1, 3, 7, 11 and 15 weeks of age. Serum samples from their dams (n = 30) were also taken 1-week post-farrowing. PToV antibodies in serum were monitored by ELISA, while viral infection was assessed by real-time RT-PCR in faeces. A high seroprevalence (about 100%) was observed in animals older than 11 weeks and in adult sows. Moreover, all 1-week-old animals were seropositive, indicating maternal antibody transference through colostrum. The antibody titers declined to close to or below the ELISA cut-off value by the age of weaning (3 weeks of age). Development of a significant antibody response to PToV occurred before 7 weeks of age in about 50% of piglets, and the remaining animals developed the response by weeks 11 or 15. These results indicate that PToV infection occurred soon after weaning. Although the prevalence of infection in suckling piglets varied among the studied farms, PToV prevalences in 7 and 11-week-old pigs were between 50–67% and 58–75%, respectively, in all farms. Sequencing results indicated that more than one PToV strains were circulating in the studied farms. Present data suggest that PToV was endemic on the studied farms, and provide new insights on the epidemiology of PToV.
Abbreviations: ELISA, enzyme-linked immunoassay; CPS, commercial porcine serum; RT-PCR, reverse transcription and polymerase chain reaction
Keywords: Torovirus, Pig, Diagnostic, ELISA, Real-time RT-PCR
1. Introduction
Toroviruses are enveloped viruses with a single stranded RNA genome, which belong to the Torovirinae subfamily from the Coronaviridae family within the Nidovirales order (ICTV web site: www.ictvonline.org). Toroviruses are enteric viruses affecting calves (Woode et al., 1982, Brown et al., 1987), horses (Weiss et al., 1983), pigs (Scott et al., 1987) and humans (Beards et al., 1986), and probably other species of domestic animals (Weiss et al., 1984). The first torovirus identified was the equine torovirus, known as Berne virus or BEV (Weiss et al., 1983). It is the only torovirus that has been adapted to grow in tissue culture, and thus, is the most thoroughly studied at the molecular level, and the prototype member of the torovirus genus. Nonetheless, most of the available information about torovirus epidemiology has been obtained from bovine torovirus (BToV), since successful experimental infections of gnotobiotic calves can be readily performed, and this, in turn, has facilitated the development of diagnostic methods to detect antibodies in serum samples (Brown et al., 1987) and viral particles in faecal specimens (Koopmans et al., 1990). BToV was first isolated in United States (Woode et al., 1982), but it has been later found in other countries such as Canada (Duckmanton et al., 1998), Japan (Ito et al., 2007), South Korea (Park et al., 2007), Austria (Haschek et al., 2006), United Kingdom (Liebler et al., 1992), The Netherlands (Koopmans et al., 1989), Germany (Koopmans et al., 1989), Italy (Lavazza, 1989) and South Africa (Vorster and Gerdes, 1993). Moreover, the infectious cycle of BToV under natural field conditions was established by compiling information from different studies (Hoet and Saif, 2004). In contrast, little is known about the porcine torovirus (PToV) epidemiology, despite it has been detected in Canada (Durham et al., 1989), South Africa (Penrith and Gerdes, 1992), and in different European countries as United Kingdom (Scott et al., 1987), The Netherlands and Belgium (Kroneman et al., 1998), Italy (Lavazza et al., 1996, Smits et al., 2003), Hungary (Matiz et al., 2002) and, recently also in Spain (Pignatelli et al., 2009, Pignatelli et al., 2010). The results of these studies indicated a high prevalence of PToV in porcine herds as well as an extensive geographical distribution of the virus. However, little information about the characteristics of PToV spreading or about the dynamics of PToV infection in piglets is available.
Recently, two new assays for PToV diagnosis have been reported: an enzyme-linked inmunosorbent assay (ELISA) based on a recombinant PToV nucleocapsid protein (Pignatelli et al., 2009), to detect antibodies against PToV, and a real-time RT-PCR method to detect PToV viral RNA in stool samples (Pignatelli et al., 2010). In the present work both diagnostic methods have been used to carry out an epidemiological survey to establish the incidence and the dynamics of PToV infection in piglets from three Spanish farms.
2. Materials and methods
2.1. Cells and viruses
Equine dermal cells (E. Derm) (ATCC® CCL-57™) were kindly provided by R.J. de Groot (Utrecht University, Utrecht, The Netherlands). They were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% foetal calf serum (FCS), 100 units per ml of penicillin, and 100 mg/ml of streptomycin. The equine torovirus, BEV (strain P138/72), was propagated in E. Derm cells as described previously (Weiss and Horzinek, 1986), and BEV particles were purified by sucrose gradient ultracentrifugation.
2.2. Field sample collection
Field samples including serum and stool samples were collected from three multi-site farms with all in-all out flow production system, located in North-eastern Spain. Piglets were weaned at 17–23 days of age and transferred to nursery units, where litters from different sows were mixed. At 9 weeks of age, pigs were moved to the growing-finishing units.
Selected piglets (n = 120, 40 per farm, coming from 10 different sows; 4 piglets per sow, being a total of 54 females and 66 males) were ear-tagged at the 1st week of age and blood samples were collected from each piglet at 1, 3, 7, 11 and 15 weeks of age. Blood samples from the corresponding sows (n = 30) were also collected 1 week post-farrowing. Blood (Vacutainer®, Becton-Dickinson, Meylan Cedex, France) was centrifuged at 1500 × g during 10 min at 4 °C. The obtained sera were aliquoted and stored at −80 °C until their use.
From a subpopulation of the pigs (n = 36, 12 per farm, coming from 3 different sows; 4 piglets per sow), faecal samples were also obtained by means of rectal swabs. Swabs were immersed in 900 μl of sterile PBS and stored at −80 °C until RNA isolation.
No gastroenteritis episodes were observed during the period of sample collection.
2.3. Serological methods
A commercial porcine serum (CPS) (AbD Serotec, Kidlington, UK) previously determined to contain antibodies against PToV, and a mixture of serum samples from caesarean-derived, colostrum-deprived (CD/CD) pigs kept under germ free conditions (spf) were used in all serological assays as positive and negative controls, respectively (Pignatelli et al., 2009).
2.4. Serum neutralization assay
BEV neutralization assay was performed as previously described (Pignatelli et al., 2009). The virus-neutralizing antibody titre from each serum sample was expressed as the reciprocal of the highest serum dilution giving 50% reduction in the number of plaques (neutralization dose 50 or ND50) with respect to that obtained with the virus without serum. Serum samples were tested by duplicated and negative (spf) and positive (CPS) control sera were included in each test. Neutralization titers were calculated using the Spearman–Kärber formula and serum samples with neutralizing antibody titers equal or higher than 4 (or 0.6 in logarithmic scale) were considered positive.
2.5. ELISA
To test for the presence of antibodies against PToV in pig serum samples, a previously described indirect ELISA using a recombinant purified PToV nucleocapsid protein as antigen was used (Pignatelli et al., 2009). All collected sow and piglet serum samples were tested on duplicated wells at 1:100 dilution. The ELISA cut-off value (O.D. 492 nm = 0.270) had been established previously using 69 serum samples from CD/CD piglets that were tested in five independent experiments performed in different days, and represented the mean value obtained plus three times the standard deviation. Nonetheless, a mixture of those serum samples were used as negative control, and were analyzed in parallel in each ELISA plate, as well as a positive serum (CPS). In addition, the same purified protein preparation was used to carry out all the ELISA assays.
2.6. PToV molecular detection methods
All RNA extraction procedures, cDNA synthesis and real-time RT-PCR reactions were performed in independent rooms and in PCR hoods, using exclusive pipettes, filter tips and disposable aliquots of reagents and PCR grade water. Besides, RNA extraction, reverse transcription and real-time RT-PCR negative controls were analyzed in parallel with field samples in each plate to discard cross-contamination of samples.
2.7. RNA isolation and reverse transcription
All collected rectal swabs were homogenized by vortexing in 900 μl of PBS. Then, 250 μl of the homogenate were centrifuged 10 min at 9000 rpm, and 200 μl of the resulting supernatant was used for RNA isolation with a commercial kit (High pure RNA isolation Kit, Roche Diagnostic S.L., Applied Science, Madrid, Spain). RNA was recovered in 60 μl and stored at −80 °C until use. An in vitro transcribed RNA template control, T7-N RNA, containing 109 molecules was used as positive control. Reverse transcription reaction was performed using SuperScript III reverse transcriptase (Invitrogen, Life Technologies Corp., Carlsbad, CA, USA) as previously described (Pignatelli et al., 2010). The cDNA preparations were aliquoted and conserved at −20 °C until use.
2.8. Real-time PCR amplification
Real-time PCR was performed in all collected swabs using SYBR Green detection method and oligonucleotide primers rtNII5′ and rtNII3′ corresponding to the sequence coding for PToV-N protein, as previously described (Pignatelli et al., 2010). For virus quantitation 10-fold dilutions of the T7-N control cDNA were analyzed in each experiment. Field samples, T7-N cDNA standard curve and the negative controls from RNA extraction, reverse transcription and PCR procedures were assayed by duplicated in the same plate. Results were analyzed using SDS v1.2 software (Applied Biosystems, Life Technologies Corp., Carlsbad, CA).
2.9. PCR amplification of the PToV-HE gene
Thirty-two out of the samples giving positive results by real-time PCR were randomly selected for sequencing purposes. To amplify the complete HE gene from rectal swab samples, an amplification reaction was performed from the corresponding cDNA samples and using the primer PToV-HE5′ (5′-GCGGATCCTTAGATGATGTT-3′), that contains a BamHI restriction site, and the previously described PToV-HE3′ primer (Pignatelli et al., 2009). Alternatively, the primer set ToV-M5′-PToV-HE3′ was used to amplify a region comprising the 3′-end of M gene and the complete HE gene under previously established conditions (Pignatelli et al., 2009). Briefly, 2 μl of cDNA were added to a reaction mix containing 1× high fidelity buffer, 200 μM of each dNTP, 200 nM of each primer and 0.71 U/μl of Platinum® Taq DNA polymerase high fidelity (Invitrogen, Life Technologies Corp, Carlsbad, CA, USA). Thermal cycling conditions were as follows: 1 cycle of 94 °C for 10 min; 30 cycles of denaturation at 94 °C for 20 s, annealing at 50 °C for 20 s and elongation at 72 °C for 2 min.
2.10. Sequencing
The amplified products obtained by real-time RT-PCR were isolated from the reaction mix using PCR isolation kit (Qiagen GmbH, Hilden, Germany) and were directly sequenced using the rtNII3′ primer. For HE gene sequencing, PCR amplified fragments were cloned into pGemT easy vector (Promega Corp., Madison, WI, USA), and three independent clones obtained from each fragment were used for sequencing. Sequencing was carried out by Secugen (Secugen S.L., Madrid, Spain) and the obtained chromatograms were analyzed using SeqMan software (DNAstar, Inc., Madison, WI, USA). Phylogenetic analyses of the new PToV sequences were performed using MegAlign Software (DNAstar, Inc., Madison, WI, USA) by comparing the sequences obtained with PToV and BToV sequences available at the GenBank using ClustalW algorithm. For comparison purposes, N and HE gene sequences from PToV strains Markelo, P4, p9, P10, p78 and BRES (accession numbers AJ575358, AJ575359, AJ575361, AJ575362, FJ232068 and AJ575363, AJ575364, AJ575365, AJ575366, AJ575367, FJ232070, respectively), and sequences from the same genes from the European BToV strains B6, B145, B150, B155, B156 and B1314 (accession numbers AJ575389, AJ575388, AJ575387, AJ575386, AJ575385, AJ575384 and AJ575378, AJ575379, AJ575380, AJ575381, AJ575382, AJ575383, respectively) and from American BRV strain (accession number AY427798) were retrieved from the GenBank database. Statistical parameters of phylogenetic trees were determined by bootstrap analysis using 1000 replicates. The nucleotide sequences for the complete HE genes from the PToV strains identified in this study (12.11, 13.11, 14.7, 52.7 and 52.11) have been submitted to the DDBJ nucleotide sequence database and are retrievable from GenBank (accession numbers GU299773, GU299774, GU299775, GU299776, GU299777, respectively).
2.11. Data analysis
All data were processed using Microsoft Excel software (version 2002; Microsoft; Corp., Redmond, Washington, USA). To group piglets by IgG profiles, MeV software was used (MeV version 4.2, TM4 software suit, Dana-Farber Cancer Institute, Boston, MA, USA) (Saeed et al., 2003). A K-means algorithm was used with Euclidean distance metric and 50 iterations. The different antibody profiles obtained were initially clustered in five groups that were further refined into four groups using Microsoft Excel (Microsoft Corp.)
3. Results
3.1. Prevalence of antibodies against PToV in Spanish farms
All available serum samples were analyzed by ELISA using the recombinant PToV-N protein as coating antigen. All serum samples from sows were positive to PToV, with a mean O.D. value of 0.89 ± 0.297 (Fig. 1B). Regarding the piglets, 110 out of 120 (92%) were positive at the first week of life (Fig. 1A), with a mean O.D. value of 0.610 ± 0.345 (Fig. 1B), indicating the maternal origin of these antibodies. At the time of weaning, at 3 weeks of age, the percentage of seropositive animals had been reduced to 58% (Fig. 1A), and the mean ELISA reactivity value was 0.322 ± 0.166 (Fig. 1B), suggesting that maternal immunity was waning. However, at 7 weeks of age the number of seropositive animals had increased again, and 109 out of 120 (91%) piglets showed positive IgG values (Fig. 1A), and the ELISA mean value raised to 0.827 ± 0.524 (Fig. 1B). The following weeks, 11 and 15, all animals were seropositive (Fig. 1A) and the mean value increased up to 1.067 ± 0.570 and 1.243 ± 0.551, respectively (Fig. 1B). No differences were observed between farms, sow parity or sex of piglets.
Fig. 1.
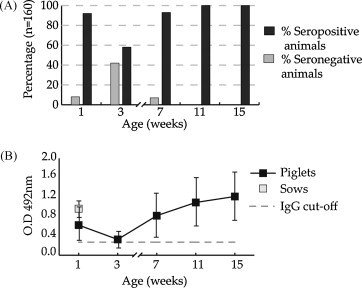
Longitudinal analysis of IgG response to PToV in pigs from three Spanish herds. Serum samples from 120 piglets and 30 sows were collected from three farms. Samples collected at weeks 1, 3, 7, 11 and 15 of piglet age and samples from sows collected at 1 week post-farrowing were analyzed for the presence of IgG to PToV-N protein by ELISA. (A) Percentages of seropositive (black bars) and seronegative (grey bars) animals over the study period. (B) Mean IgG ELISA values of sows and piglets at each sampling time are represented by grey and black squares, respectively. IgG ELISA cut-off value is indicated (- - -).
3.2. Profiling of the IgG antibody response to PToV in piglets
The ELISA results from different piglets were grouped following the dynamics of IgG antibodies against PToV, using the MeV software. As shown in Fig. 2A, four IgG profiles could be distinguished in the three farms. One group (profile 1) corresponded to piglets that showed a significant IgG increase by 7 weeks of age (44% of piglets). In that group, 18 out of 53 (34%) underwent an IgG rise detected at week 11, while in 16 out of 53 piglets (30%) a second IgG rise was observed at week 15. A second group (profile 2), representing 21% of piglets, corresponded to animals that showed an IgG increase at 11 weeks of age. A third group of piglets (11%) registered a steep rise in IgG level at week 15 while in previous weeks they displayed moderate ELISA values (profile 3). The last group (profile 4) corresponded to piglets that showed low IgG levels over the studied period, although most of them were positive all along the analysis period. That dynamics was observed in 24% of piglets.
Fig. 2.
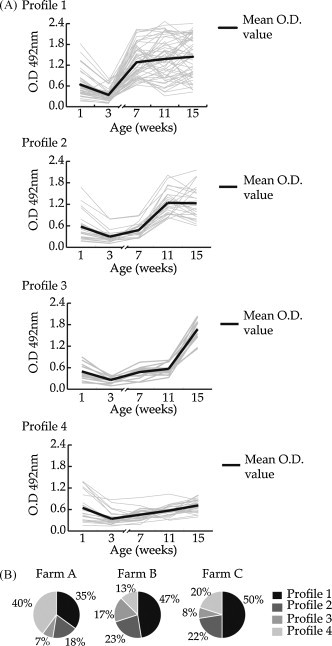
Analysis of the different patterns of IgG antibody development. The dynamics of IgG antibody rising determined by ELISA through weeks in each individual piglet were classified using MeV software using a K-means algorithm and four antibody profiles (1–4) were determined. Antibody profile for each individual piglet is represented by grey lines and the mean ELISA value between all piglets belonging to a profile is represented by a black line.
Noticeably, the percentages of animals in the four groups were slightly different between farms. As shown in Fig. 2B, in farm A, piglets showed an important immune response at week 7 (35%) (profile 1) or remained with low IgG levels for the length of the study (profile 4) (40%). In farm B, almost half of the piglets developed antibodies at week 7 (47%) (profile 1), 23% at week 11 (profile 2) and 17% at week 15 (profile 3), but only a 13% did not show high IgG levels at any time point (profile 4). In farm C, 50% of piglets showed an immune response characteristic of profile 1, but in 22% and 8% of animals the rise in IgG levels occurred at weeks 11 and 15, respectively (profiles 2 and 3), and 20% of piglets did not show a noticeable humoral immune response (profile 4).
3.3. Neutralizing antibodies to PToV
To study the presence of neutralizing antibodies against PToV in piglets and in adult sows, serum samples from 13 sows and from 9 piglets (piglets 22 and 44 from farm A, piglet 79 from farm B and piglets 3, 9, 38, 60, 12 and 94 from farm C) were analyzed. Selected piglets represented IgG profiles 1, 3 and 4 (3 piglets per profile).
Eleven out of the 13 analyzed sows showed neutralizing antibodies but, as mentioned before, they were all positive by ELISA and also by western-blot (data not shown). All 1-week-old piglets analyzed showed neutralizing antibodies, and their titers were similar to those of their corresponding sows (Table 1 ). As shown in Fig. 3 and in Table 1, at weaning, all piglets but one (no. 12, profile 4) were positive, but neutralizing antibody titers decreased in all animals. At week 7, neutralizing titers decreased slightly or were kept similar, even in piglets from profile 1 that had shown a high IgG increase by ELISA at that time (Fig. 3A). However, at week 11, piglets from profile 1 showed high neutralizing antibody titers, while all piglets from profiles 3 and 4 showed low ones (Fig. 3A). At 15 weeks of age, all piglets from profiles 1 and 3 showed high levels of neutralizing antibodies, and piglets from profile 4 showed low or moderate neutralizing titers (Fig. 3A).
Table 1.
Comparative serological analysis by ELISA and virus neutralization assay (NT) of nine selected piglets at different weeks of age and their corresponding sows.
| Piglet | Piglets age (weeks) |
Sow | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 7 | 11 | 15 | ||||
| Profile 1 | ELISAa | 9 | 0.53 | 0.33 | 1.24b | 1.88 | 0.99 | 1.41 |
| NPc | 1.98 | 0.81 | 0.59 | 1.62 | 1.86 | 1.84 | ||
| ELISA | 3 | 0.68 | 0.34 | 1.92 | 2.03 | 1.90 | 1.41 | |
| NT | 1.85 | 1.17 | 1.26 | 2.20 | 2.95 | 1.84 | ||
| ELISA | 60 | 0.61 | 0.33 | 1.84 | 2.47 | 1.89 | 0.65 | |
| NT | 1.75 | 0.87 | 1.01 | 2.19 | 2.32 | 1.60 | ||
| Profile 3 | ELISA | 38 | 0.69 | 0.27d | 0.20 | 0.33 | 1.56 | 0.52 |
| NT | 2.11 | 1.42 | 1.01 | 1.24 | 1.86 | 1.48 | ||
| ELISA | 70 | 0.35 | 0.24 | 0.53 | 0.49 | 2.01 | 0.54 | |
| NT | 2.25 | 1.57 | 1.37 | 1.67 | 2.96 | 1.70 | ||
| ELISA | 44 | 1.35 | 0.67 | 0.84 | 0.66 | 1.18 | 1.30 | |
| NT | 1.88 | 1.33 | 1.41 | 0.59 | 2.26 | 1.30 | ||
| Profile 4 | ELISA | 12 | 0.72 | 0.32 | 0.45 | 0.75 | 0.92 | 0.96 |
| NT | 0.95 | 0.45 | 0.45 | 1.18 | 1.16 | 2.09 | ||
| ELISA | 22 | 1.36 | 0.63 | 0.49 | 0.55 | 0.55 | 1.19 | |
| NT | 1.78 | 0.99 | 1.37 | 1.07 | 0.94 | 1.36 | ||
| ELISA | 94 | 0.28 | 0.26 | 0.30 | 0.55 | 0.99 | 0.73 | |
| NT | 0.83 | 0.66 | 0.50 | 0.82 | 2.53 | 1.31 | ||
ELISA (IgG) results are given as O.D. 492 values (ELISA cut-off value: OD492 nm = 0.27).
In bold are indicated sera that gave positive ELISA results but negative neutralizing values.
Neutralization titers are expressed as Log ND50 (neutralization cut-off value: Log N50− = 0.6).
In bold and italics are indicated sera that gave negative ELISA results but positive neutralizing values.
Fig. 3.
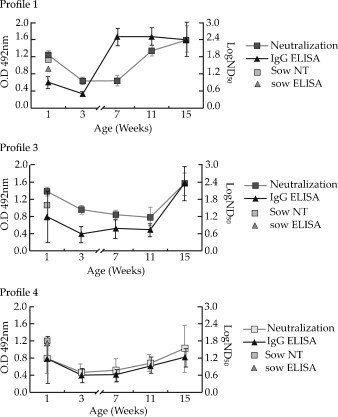
Comparison between the levels of IgG and neutralizing antibodies to PToV in nine selected piglets from different antibody dynamics profiles. Serum samples from 9 piglets representative from the groups established based on IgG profile (profiles 1, 3 and 4) were selected (3 piglets per group) and analyzed by virus neutralization test. Neutralizing antibody titers (grey square) were determined in serum samples taken at weeks 1, 3, 7, 11 and 15 of piglet age. Mean IgG ELISA values from the selected piglets from each group are shown (black triangle). Mean sows neutralizing titers (empty square) and ELISA IgG values (empty triangle) for each group is also indicated.
Overall, a fairly good correlation between ELISA and neutralization results was obtained from piglets showing IgG profiles 3 and 4 (R 2 = 0.67 and 0.60, respectively) (Fig. 3B and C). However, a low correlation was obtained (R 2 = 0.16) (Fig. 3A) for piglets corresponding to profile 1. In addition, five ELISA positive but neutralizing negative samples (Table 1, in bold), and four ELISA negative but neutralizing positive samples were obtained (Table 1, in bold and italic).
3.4. Detection of PToV by real-time RT-PCR in stool samples from piglets at different weeks of age
Seventy-five out of 180 samples (42%) were positive for PToV RNA. In 33 piglets (92%), PToV was detected in stools at least at one of the time points examined. Only in 3 piglets (8%) PToV RNA was not detected in their stools at any of time points examined; two of these piglets were from farm B and the other one from farm C.
As shown in Fig. 4 , differences on PToV prevalence were observed between farms. PToV was highly prevalent in farm A, with about 50–58% of positive pigs at weeks 1, 7 and 15, and 75% in 3 and 11-week-old pigs. In farm B, only one piglet was positive at weeks 1 and 3; however, PToV prevalence increased in the following weeks, and between 42% and 58% of piglets was positive at weeks 7, 11 and 15. In farm C, low PToV incidence was observed in young piglets (1 and 3 weeks of age), since only one (8%) piglet was positive at week 1 and no piglets were positive at 3 weeks of age. However, at weeks 7 and 11, 67% and 75% of piglets were RT-PCR positive, respectively, and none of the piglets was shedding virus at 15 weeks of age (Fig. 4).
Fig. 4.
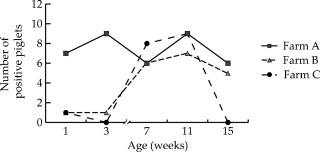
PToV detection by rt-RT-PCR in farms. Faecal samples were collected from 12 piglets from farms A (black square), B (grey triangle) and C (black dot) at weeks 1, 3, 7, 11 and 15. PToV was detected by rt-RT-PCR using primers rtNII5′ and rtNII3′ to amplify a region from N gene. Number of PToV positive piglets in each farm at different weeks is shown.
As determined by titration using RNA standard curve, low virus loads were observed in most samples. However, some samples contained up to 106 to 107 copies of viral RNA suggesting that the corresponding piglets were shedding high amounts of PToV virus at the time of sampling.
3.5. Sequencing of PToV isolates
To confirm the specificity of the fragments obtained by RT-PCR, PCR products from 32 randomly selected animals (13 samples from farm A, 12 from farm B and 7 from farm C) were purified and used directly for sequencing. All sequenced fragments were related with sequences previously described from other PToV strains and clustered in five groups (Fig. 5 ).
Fig. 5.
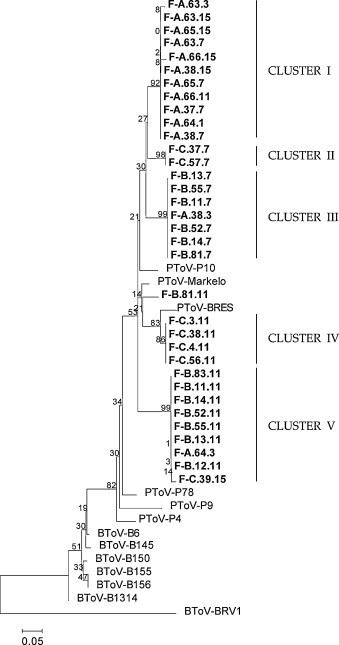
Phylogenetic analysis from partial PToV-N gene sequences obtained from Spanish farms. Gene sequences were aligned using the ClustalW method and the phylogenetic tree was performed by the neighbour joining method. Statistical bootstrap value obtained after 1000 replicates is shown in each branch.
Most of the sequences obtained from piglets from farm A, were grouped together in cluster I (Fig. 5). These sequences were phylogenetically related to P10 strain, showing 98–99% nucleotide identity. Only two sequences (F-A.38.3 and F-A.64.3) aligned in different clusters, III and V, respectively (Fig. 5). Most sequences from samples collected from farm B clustered in two different groups (clusters III and V) showing a 91–92% of sequence identity between them (Fig. 5). Cluster III comprised all sequences obtained from samples taken at week 7 and cluster V comprised all sequences obtained from samples taken at week 11, suggesting that piglets were infected by two different PToV strains over time. The only exception was the sequence of F-B.81.11 that was included in cluster IV. Sequences obtained from farm C grouped also in two clusters (II and IV). One group, cluster II, corresponded to sequences obtained from samples collected at week 7. Those sequences showed an 89.6% nucleotide identity with respect to P10 strain, but a 91.2% with respect to sequences from clusters I and III. However, sequences obtained from this farm at week 11 grouped in cluster IV and showed higher nucleotide identity with respect to the BRES strain (97%) than with respect to cluster II (89%). Only one sequence from farm C (F-C.39.15), obtained from a 15-week-old pig, was grouped in cluster V.
To further study if piglets from farm B were infected by two different PToV strains at weeks 7 and 11, the complete HE gene from RNA samples was sequenced. The HE gene was amplified from samples 12.11, 13.11, 14.7, 52.7 and 52.11 and the amplified products were cloned in a commercial vector pGemT and sequenced. Sequences from samples 12.11 and 52.11 were phylogenetically related, showing a 98% nucleotide identity, but only a 80% of similarity with respect to samples 52.7, 14.7 and 13.11 (Fig. 6 ). In addition, the HE genes from samples 12.11 and 52.11 were related to BRES (90%) and P4 (92%) strains. In contrast, the HE gene from samples 52.7, 14.7 and 13.11 were closely related to the Markelo strain (95%). Using the alternative primer pair ToV-M5′ and PToV-HE3′, a 640 bp amplification product was obtained from sample 13.7. The obtained sequence showed that the HE gene was partially deleted in this sample and only 83 nucleotides from the HE 3′-end were conserved. That region was fused to the canonical transcription-regulating signal (TRS) sequence located at the 5′-end of the gene.
Fig. 6.
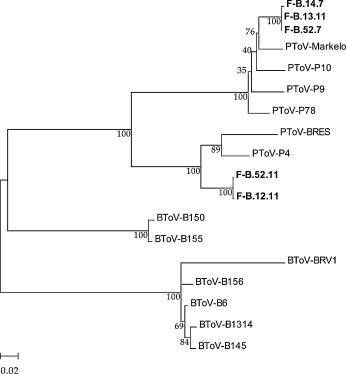
Phylogenetic analysis from complete HE gene from samples 12.11, 13.11, 14.7, 52.7 and 52.11 from farm B. Gene sequences were aligned using the ClustalW method and the phylogenetic tree was performed by the neighbour joining method. Statistical bootstrap value obtained after 1000 replicates is shown in each branch.
4. Discussion
PToV is broadly dispersed around Europe (Kroneman et al., 1998, Matiz et al., 2002, Smits et al., 2003) and has also been detected in Canada (Durham et al., 1989) and South Africa (Penrith and Gerdes, 1992). However, to date, there is little information available about PToV epidemiology. Probably, the impossibility of growing the virus in culture cells has precluded the development of diagnostic methods. Recently, it have been reported the development of an ELISA assay (Pignatelli et al., 2009) and a real-time RT-PCR method (Pignatelli et al., 2010) to detect either antibodies to PToV or viral RNA, respectively, in swine samples. Using these methods, it was possible to determine the presence of PToV in the Spanish porcine livestock. In the present work, the longitudinal monitoring of PToV infection was carried out in three Spanish farms. This work further confirmed the presence of PToV in Spanish swine herds, and allowed to determine its infection dynamics during the first 15 weeks of age.
The high prevalence (about 100%) of antibodies against PToV observed both in animals older than 7 weeks and in adult sows, and the high incidence of PToV infection in piglets from all three farms, clearly suggests that PToV is endemic in these farms and probably reflects the widespread nature of this infection. These results are in agreement with previous studies performed with a reduced number of animals from different Spanish farms (Pignatelli et al., 2009, Pignatelli et al., 2010). In addition, the present data about the occurrence of PToV shedding in faeces are similar to those observed in another longitudinal study performed in a small number of piglets from two farms in Central Europe (Kroneman et al., 1998). Similar virus infection dynamics in young animals have also been described for BToV (Duckmanton et al., 1998) and other enteric viruses such as rotavirus in Danish (Svensmark et al., 1989) and Italian (Martella et al., 2007) farms. The presence of PToV in sows was not evaluated in the present study but it appears to be low according to a previously reported study (Matiz et al., 2002).
Some insights into virus spread can be drawn from the analysis of both blood and faecal samples collected over a 15 weeks period. During the first week of life, when piglets remained with their sows, half of the piglets from farm A and some piglets from farms B and C shed PToV, indicating that those piglets had already been infected and, therefore, sows might be the viral source. As determined from the analyses by ELISA and neutralizing assays, most piglets (92%) from the three farms had received antibodies against PToV through maternal colostrum. No differences between IgG and neutralizing antibody levels were observed between piglets that shed or not the virus at first week of age. Differences were neither observed in the levels of antibodies in serum samples from sows. Hence, the results indicate that, especially in farm A, maternal antibody transfer probably provided a partial or null protection against virus infection in piglets. A potential explanation for the higher rate of infection in farm A could be that sows from this farm shed higher amounts of virus than those from other farms. Unfortunately, faecal samples from sows were not available and, thus, the virus loads in those animals could not be determined. Similar observations about newborn animal infection and virus spreading by adult animals have been previously reported for BToV (Koopmans et al., 1989) and rotaviruses (Snodgrass and Wells, 1978).
Maternal antibodies decreased gradually over time and half of the animals had lost their maternal IgG against PToV nucleocapsid protein around the weaning age, even though low neutralizing antibodies titers could remain in some of these piglets. The results of the ELISA were confirmed by western-blot against the nucleocapsid protein (data not shown). Therefore, some contradictory results between neutralization and ELISA (or western-blot) tests were observed at such age. This observation could also be related to the different antigens used in the two assays (Wang et al., 2005) and/or to the different neutralizing or anti-N antibody levels present in maternal colostrum.
After weaning, approximately at 3 weeks of age, piglets that would have lost their maternal protection were moved to another facility and grouped with other piglets. That scenario seems to be critical for a quick virus spread since by week 7 most of the piglets from the three farms had become infected, as evidenced by the presence of virus in their faeces. Similar observations have been reported for BToV in calves from different European herds (Koopmans et al., 1989) and also from studies performed using sentinel calves (Koopmans et al., 1990). The dynamics of the IgG production against the PToV-N protein after weaning in different animals were categorized in four profiles. Profiles 1, 2 and 3 corresponded to animals in which a significant rise of the IgG response was observed at weeks 7, 11 and 15, respectively. On the other hand, animals assigned to profile 4 were those that did not develop a detectable humoral immune response, at least during the studied period.
Interestingly, piglets from profile 1 developed high levels of antibodies against the N protein at week 7, as determined by ELISA and also by western-blot (data not shown), but not neutralizing antibodies. This observation would reflect an early antibody development after a first contact with the virus that was not evidenced by the neutralizing test. Again, differences between the antigens used in these two assays could also explain this finding, since the higher antigenicity of the nucleocapsid protein may provide higher ELISA sensitivity, whereas the heterologous neutralization assay could fail to detect low virus-neutralizing antibody titers.
Between 8 and 9 weeks of age pigs were moved to the fattening facilities, implying new animal groupings. The real-time RT-PCR results reflected a new burst of virus infection at week 11 in all three studied farms. Interestingly, differences about the origin of the virus were observed. Piglets from farm A shed the same virus strain over the weeks, a finding that is in line with the observation that some piglets from farm A had not developed an immune response after the first contact with the virus at weeks 1 and 3. These animals could be persistently infected with PToV and shed virus intermittently for weeks. Re-infection of intestinal cells in the presence of circulating antibodies might also occur, as it has also been reported with BToV in calves, as well as with rotavirus (Koopmans et al., 1990, Parreno et al., 2004). These findings would indicate a lack of correlation between serum IgG and mucosal protection (Koopmans et al., 1990). In torovirus, impairment of mucosal immune response has been attributed to infection by these viruses of dome M cells (Pohlenz et al., 1984, Woode et al., 1984), which play an important role in local immunity.
Overall, the present results show that in farm A the dynamics of PToV infection and the immune response against the virus were clearly different to those observed in farms B and C. The incidence of PToV infection at each time point was higher in farm A than in the other farms. Interestingly, a higher percentage of animals that did not develop a significant immune response against the virus was also found in this farm. As mentioned before, a possible explanation for these results could be that sows from farm A might spread higher amounts of virus to their offspring than those from farms B and C. This would indicate that the degree of exposure to the virus during the first week of piglet's life would influence the development of the immune response against the virus. The infection in the presence of maternal antibodies may decrease or delay the development of an active immune response in piglets, as it seems to occur in animals showing an IgG profile of type 4. This situation has been previously reported for BToV (Woode et al., 1985, Weiss and Horzinek, 1987, Koopmans et al., 1990, Koopmans et al., 1991) as well as for the bovine coronavirus (BCoV) (Heckert et al., 1991a, Heckert et al., 1991b).
On the other hand, in farm B and, probably, farm C, a second PToV strain was infecting piglets around 11 weeks of age, as it could be concluded from the sequence analysis from the N gene fragment and the HE complete gene from four animals. These observations suggest that at least two different PToV strains were circulating simultaneously in these farms, and individual animals could be infected by these two strains during their productive life. Moreover, in a previous study it was found that piglets could be simultaneously infected by the two PToV strains (Pignatelli et al., 2010). The rearrangement of piglets within the farm facilities and also the stress caused by the transport/movement of animals might contribute to PToV spread.
In addition, the obtained HE gene sequences also indicated a shift on HE lineages. This result could indicate immunological differences between HE lineages, as it has been shown to occur in the case of BToV, where two different HE lineages have been defined and antigenic differences observed (Smits et al., 2005). However, there is little information about torovirus HE functions and its antigenic properties and, thus, further studies would be required to understand these issues.
In conclusion, PToV appears to be endemic in Spanish farms. PToV is an intestinal pathogen that causes subclinical infections in neonatal and young piglets and probably in adult animals. BToV has been reported to cause gastroenteritis in gnobiotic calves under experimental conditions (Woode et al., 1982, Pohlenz et al., 1984) and under natural infections (Brown et al., 1990), by itself or in combination with other pathogens (Weiss et al., 1984, Hoet et al., 2002, Hoet et al., 2003). However, the link between PToV and disease remains unclear yet and more extensive studies are needed to establish the role of PToV as a gastroenteritis causing agent.
Acknowledgments
We thank Raquel Blanco and Juan Ramón Rodríguez for critical reading of the manuscript. We also want to thank R.J. de Groot (Utrecht University, Utrecht, The Netherlands) for providing the E. Derm cells and BEV virus. This work was supported by grants CICYT-BIO2006-10988 to Dolores Rodríguez and CONSOLIDER-PORCIVIR CSD2006-00007 to Joaquim Segalés and Dolores Rodríguez. Jaime Pignatelli and Miriam Jimenez were both recipients of contracts financed with founding from the CONSOLIDER-PORCIVIR research project.
References
- Beards G.M., Brown D.W., Green J., Flewett T.H. Preliminary characterisation of torovirus-like particles of humans: comparison with Berne virus of horses and Breda virus of calves. J. Med. Virol. 1986;20(1):67–78. doi: 10.1002/jmv.1890200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D., Morgan J.H., Bridger J.C. Survey for Breda virus in neonatal calf diarrhoea. Vet. Rec. 1990;126(14):337. [PubMed] [Google Scholar]
- Brown D.W., Beards G.M., Flewett T.H. Detection of Breda virus antigen and antibody in humans and animals by enzyme immunoassay. J. Clin. Microbiol. 1987;25(4):637–640. doi: 10.1128/jcm.25.4.637-640.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckmanton L., Carman S., Nagy E., Petric M. Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J. Clin. Microbiol. 1998;36(5):1266–1270. doi: 10.1128/jcm.36.5.1266-1270.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham P.J., Hassard L.E., Norman G.R., Yemen R.L. Viruses and virus-like particles detected during examination of feces from calves and piglets with diarrhea. Can. Vet. J. 1989;30(11):876–881. [PMC free article] [PubMed] [Google Scholar]
- Haschek B., Klein D., Benetka V., Herrera C., Sommerfeld-Stur I., Vilcek S., Moestl K., Baumgartner W. Detection of bovine torovirus in neonatal calf diarrhoea in Lower Austria and Styria (Austria) J. Vet. Med. B Infect. Dis. Vet. Pub. Health. 2006;53(4):160–165. doi: 10.1111/j.1439-0450.2006.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert R.A., Saif L.J., Myers G.W. Mucosal and systemic isotype-specific antibody responses to bovine coronavirus structural proteins in naturally infected dairy calves. Am. J. Vet. Res. 1991;52(6):852–857. [PubMed] [Google Scholar]
- Heckert R.A., Saif L.J., Myers G.W., Agnes A.G. Epidemiologic factors and isotype-specific antibody responses in serum and mucosal secretions of dairy calves with bovine coronavirus respiratory tract and enteric tract infections. Am. J. Vet. Res. 1991;52(6):845–851. [PubMed] [Google Scholar]
- Hoet A.E., Cho K.O., Chang K.O., Loerch S.C., Wittum T.E., Saif L.J. Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am. J. Vet. Res. 2002;63(3):342–348. doi: 10.2460/ajvr.2002.63.342. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Nielsen P.R., Hasoksuz M., Thomas C., Wittum T.E., Saif L.J. Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J. Vet. Diagn. Invest. 2003;15(3):205–212. doi: 10.1177/104063870301500301. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Saif L.J. Bovine torovirus (Breda virus) revisited. Anim. Health Res. Rev. 2004;5(2):157–171. doi: 10.1079/ahr200498. [DOI] [PubMed] [Google Scholar]
- Ito T., Okada N., Fukuyama S. Epidemiological analysis of bovine torovirus in Japan. Virus Res. 2007;126(1–2):32–37. doi: 10.1016/j.virusres.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M., Cremers H., Woode G., Horzinek M.C. Breda virus (Toroviridae) infection and systemic antibody response in sentinel calves. Am. J. Vet. Res. 1990;51(9):1443–1448. [PubMed] [Google Scholar]
- Koopmans M., Herrewegh A., Horzinek M.C. Diagnosis of torovirus infection. Lancet. 1991;337(8745):859. doi: 10.1016/0140-6736(91)92573-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M., van den Boom U., Woode G., Horzinek M.C. Seroepidemiology of Breda virus in cattle using ELISA. Vet. Microbiol. 1989;19(3):233–243. doi: 10.1016/0378-1135(89)90069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroneman A., Cornelissen L.A., Horzinek M.C., de Groot R.J., Egberink H.F. Identification and characterization of a porcine torovirus. J. Virol. 1998;72(5):3507–3511. doi: 10.1128/jvi.72.5.3507-3511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazza A. Diarrea neonatale dei vitelli: identificazione al M.E. in colorazione negativa di particelle virali morfologicamente riferibili al Bredavirus. Atti Soc Ital Buiatria. 1989;21:121–127. [Google Scholar]
- Lavazza A., Candotti P., Perini S., Vezzali L. Electron microscopic identification of torovirus-like particles in post-weaned piglets with enteritis. 14th IPVS Congress; Bologna, Italy; 1996. [Google Scholar]
- Liebler E.M., Kluver S., Pohlenz J., Koopmans M. The significance of Breda virus as a diarrhea agent in calf herds in Lower Saxony. Dtsch. Tierarztl. Wochenschr. 1992;99(5):195–200. [PubMed] [Google Scholar]
- Martella V., Bányai K., Lorusso E., Bellacicco A.L., Decaro N., Camero M., Bozzo G., Moschidou P., Arista S., Pezzotti G., Lavazza A., Buonavoglia C. Prevalence of group C rotaviruses in weaning and post-weaning pigs with enteritis. Vet. Microbiol. 2007;123(1–3):26. doi: 10.1016/j.vetmic.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Matiz K., Kecskemeti S., Kiss I., Adam Z., Tanyi J., Nagy B. Torovirus detection in faecal specimens of calves and pigs in Hungary: short communication. Acta Vet. Hung. 2002;50(3):293–296. doi: 10.1556/AVet.50.2002.3.5. [DOI] [PubMed] [Google Scholar]
- Park S.I., Jeong C., Kim H.H., Park S.H., Park S.J., Hyun B.H., Yang D.K., Kim S.K., Kang M.I., Cho K.O. Molecular epidemiology of bovine noroviruses in South Korea. Vet. Microbiol. 2007;124(1–2):125–133. doi: 10.1016/j.vetmic.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreno V., Bejar C., Vagnozzi A., Barrandeguy M., Costantini V., Craig M.I., Yuan L., Hodgins D., Saif L., Fernandez F. Modulation by colostrum-acquired maternal antibodies of systemic and mucosal antibody responses to rotavirus in calves experimentally challenged with bovine rotavirus. Vet. Immunol. Immunopathol. 2004;100(1–2):7–24. doi: 10.1016/j.vetimm.2004.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrith M.L., Gerdes G.H. Breda virus-like particles in pigs in South Africa. J. S. Afr. Vet. Assoc. 1992;63(3):102. [PubMed] [Google Scholar]
- Pignatelli J., Jiménez M., Grau-Roma L., Rodríguez D. Detection of porcine torovirus by real time RT-PCR in piglets from a Spanish farm. J. Virol. Methods. 2010;163:398–404. doi: 10.1016/j.jviromet.2009.10.031. [DOI] [PubMed] [Google Scholar]
- Pignatelli J., Jimenez M., Luque J., Rejas M.T., Lavazza A., Rodriguez D. Molecular characterization of a new PToV strain. Evolutionary implications. Virus Res. 2009;143:33–43. doi: 10.1016/j.virusres.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlenz J.F., Cheville N.F., Woode G.N., Mokresh A.H. Cellular lesions in intestinal mucosa of gnotobiotic calves experimentally infected with a new unclassified bovine virus (Breda virus) Vet. Pathol. 1984;21(4):407–417. doi: 10.1177/030098588402100407. [DOI] [PubMed] [Google Scholar]
- Saeed A.I.S.V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Scott A.C., Chaplin M.J., Stack M.J., Lund L.J. Porcine torovirus? Vet. Rec. 1987;120(24):583. doi: 10.1136/vr.120.24.583-a. [DOI] [PubMed] [Google Scholar]
- Smits S.L., Gerwig G.J., van Vliet A.L., Lissenberg A., Briza P., Kamerling J.P., Vlasak R., de Groot R.J. Nidovirus sialate-O-acetylesterases: evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J. Biol. Chem. 2005;280(8):6933–6941. doi: 10.1074/jbc.M409683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Lavazza A., Matiz K., Horzinek M.C., Koopmans M.P., de Groot R.J. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 2003;77(17):9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass D.R., Wells P.W. Passive immunity in rotaviral infections. J. Am. Vet. Med. Assoc. 1978;173(5 Pt 2):565–568. [PubMed] [Google Scholar]
- Svensmark B., Nielsen K., Dalsgaard K., Willeberg P. Epidemiological studies of piglet diarrhoea in intensively managed Danish sow herds. III. Rotavirus infection. Acta Vet. Scand. 1989;30(1):63–70. doi: 10.1186/BF03548069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorster J.H., Gerdes G.H. Breda virus-like particles in calves in South Africa. J. S. Afr. Vet. Assoc. 1993;64(2):58. [PubMed] [Google Scholar]
- Wang Y., Chang Z., Ouyang J., Wei H., Yang R., Chao Y., Qu J., Wang J., Hung T. Profiles of IgG antibodies to nucleocapsid and spike proteins of the SARS-associated coronavirus in SARS patients. DNA Cell Biol. 2005;24(8):521–527. doi: 10.1089/dna.2005.24.521. [DOI] [PubMed] [Google Scholar]
- Weiss M., Horzinek M.C. Morphogenesis of Berne virus (proposed family Toroviridae) J. Gen. Virol. 1986;67(Pt 7):1305–1314. doi: 10.1099/0022-1317-67-7-1305. [DOI] [PubMed] [Google Scholar]
- Weiss M., Horzinek M.C. The proposed family Toroviridae: agents of enteric infections. Brief review. Arch. Virol. 1987;92(1–2):1–15. doi: 10.1007/BF01310058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M., Steck F., Horzinek M.C. Purification and partial characterization of a new enveloped RNA virus (Berne virus) J. Gen. Virol. 1983;64(Pt 9):1849–1858. doi: 10.1099/0022-1317-64-9-1849. [DOI] [PubMed] [Google Scholar]
- Weiss M., Steck F., Kaderli R., Horzinek M.C. Antibodies to Berne virus in horses and other animals. Vet. Microbiol. 1984;9(6):523–531. doi: 10.1016/0378-1135(84)90014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woode G.N., Pohlenz J.F., Gourley N.E., Fagerland J.A. Astrovirus and Breda virus infections of dome cell epithelium of bovine ileum. J. Clin. Microbiol. 1984;19(5):623–630. doi: 10.1128/jcm.19.5.623-630.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woode G.N., Reed D.E., Runnels P.L., Herrig M.A., Hill H.T. Studies with an unclassified virus isolated from diarrheic calves. Vet. Microbiol. 1982;7(3):221–240. doi: 10.1016/0378-1135(82)90036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woode G.N., Saif L.J., Quesada M., Winand N.J., Pohlenz J.F., Gourley N.K. Comparative studies on three isolates of Breda virus of calves. Am. J. Vet. Res. 1985;46(5):1003–1010. [PubMed] [Google Scholar]


