Abstract
In pathogen recognition by C-type lectins, several levels of complexity can be distinguished; these might modulate the immune response in different ways. Firstly, the pathogen-associated molecular pattern repertoire expressed at the microbial surface determines the interactions with specific receptors. Secondly, each immune cell type possesses a specific set of pathogen-recognition receptors. Thirdly, changes in the cell-surface distribution of C-type lectins regulate carbohydrate binding by modulating receptor affinity for different ligands. Crosstalk between these receptors results in a network of multimolecular complexes, adding a further level of complexity in pathogen recognition.
Introduction
C-type lectin receptors (CLRs) are proteins that contain carbohydrate recognition domains (CRDs) and bind carbohydrate structures in a Ca2+-dependent manner. Ca2+ ions are directly involved in ligand binding, as well as in maintaining the structural integrity of the CRD that is necessary for the lectin activity [1]. Depending on the amino acid sequence, the CRD has specificity for mannose, galactose or fucose structures. Moreover, interaction of these carbohydrate structures with the different CLRs is dependent on carbohydrate branching, spacing and multivalency.
CLRs are either produced as transmembrane proteins or are secreted as soluble proteins (Table 1 ). They have been shown to act both as adhesion and as pathogen-recognition receptors [2]. The broad selectivity of the monosaccharide-binding site and the geometrical arrangement of multiple CRDs provide a first basis for discriminating between self and non-self [3]. Thus, CLRs recognize carbohydrate patterns expressed by the invading microorganisms and have a role in both innate and adaptive immunity [4]. Several microorganisms have developed strategies for exploiting the CLR-mediated uptake to their benefit, and evade the host immune defenses [5, 6].
Table 1.
Overview of two subfamilies of C-type lectins and their most recently identified ligands.
| Groupa | Molecular structure | C-type lectin | Newly identified pathogens [specific ligand] | References |
|---|---|---|---|---|
| Collectins | Soluble | MBL | Neisseria meningitidis [nonglycosylated outer membrane opacity protein and porin] | [14•] |
| SP-A | Mycobacterium avium [lipoarabinomannan] | [38•] | ||
| SP-D | Mycobacterium avium [lipoarabinomannan] | [38•] | ||
| Schistosoma mansoni [α1-3-linked fucose] | [13] | |||
| All three members (MBL, SP-A and SP-D) bind free DNA and apoptotic cell-derived DNA | [42•] | |||
| Type II receptors | Type II transmembrane | DC-SIGN | Hepatitis C virus [E1 and E2 glycoproteins] | [43] |
| Marburg virus [GP-glycoprotein] | [44] | |||
| SARS-CoVb [S-glycoprotein] | [44] | |||
| Helicobacter pylori [Lewis X/Y LPS] | [26••] | |||
| Aspergillus fumigatus [galactomannan] | [45] | |||
| Mycobacterium tuberculosis [(man)3-ara] | [11] | |||
| L-SIGN | Hepatitis C virus [E1 and E2 glycoproteins] | [43] | ||
| Marburg virus [GP-glycoprotein] | [44] | |||
| SARS-CoV [S-glycoprotein] | [44, 46] | |||
| Mycobacterium tuberculosis [(man)3-ara] | [11] | |||
| Schistosoma mansoni | [10•] | |||
| mSIGN-R1 | Streptococcus pneumoniae [capsular polysaccharide] | [47] | ||
| Mycobacterium tuberculosis [(man)3-ara] | [11] | |||
| Langerin | Mycobacterium leprae [nonpeptide antigen] | [33•] | ||
Group is determined by nomenclature in use at ‘A genomics resource for animal lectins’ (URL: http://ctld.glycob.ox.ac.uk).
SARS-CoV, severe acute respiratory syndrome coronavirus. Note that this table is limited to novel ligands identified in 2004; a more extensive description is given in [48].
In this review, we focus on the role of CLRs in the recognition of pathogens that leads either to immune protection or immune evasion. We discuss binding specificity as a consequence of differences in ligand glycosylation, the molecular and structural elements that regulate the interaction with pathogen-associated molecular patterns (PAMPs) and, finally, the increasing evidence of interactions with other immune receptor families, which adds to the complexity of CLR-mediated pathogen recognition.
Identification of new carbohydrate structures
Recent findings reported by several investigators indicate that CLRs recognize subtle differences in the arrangement and branching of the carbohydrate residues. Despite the fact that several CLRs share a CRD and bind mannose-containing structures, different branching and spacing of these structures create unique sets of carbohydrate-recognition profiles for each receptor.
Therapeutic manipulation of carbohydrate–protein interactions requires detailed knowledge of the specific spectrum of carbohydrate structures recognized by each CLR. Oligosaccharide microarray technologies are currently being applied to facilitate a systematic and high-throughput analysis of protein–carbohydrate interactions.
A well-known example is the neoglycolipid technology that generates lipid-linked oligosaccharide arrays from glycoproteins, glycolipids, proteoglycans, polysaccharides, whole organs or chemically synthesized oligosaccharides [7]. By glycan array profiling, discrimination between high- and low-affinity carbohydrate ligands for the murine CLRs specific intercellular adhesion molecule (ICAM)-3-grabbing nonintegrin (SIGN)-R1, SIGN-R3 and langerin has been achieved [8•].
Alternative glycan arrays have also been documented. Recently, Guo et al. [9•] showed that dendritic cell (DC)-SIGN and its homolog L-SIGN have distinct ligand-binding properties. Whereas L-SIGN was only able to bind to mannose-containing ligands, DC-SIGN reacted with many more glycans, including fucose moieties on Lewis blood group antigens [9•]. The observation that DC-SIGN and L-SIGN differ in their carbohydrate-binding profiles was also reported by van Liempt et al. [10•]. By examining the binding of various mutants of DC-SIGN and L-SIGN to Schistosoma mansoni egg antigen, they showed that the difference in one single amino acid at the binding site is responsible for the fucose specificity of DC-SIGN [10•]. More specifically, they indicate that although L-SIGN seems unable to bind to Lewis-X, it still interacts with other fucosylated glycans such as Lewis A, -B and –Y [10•].
The specific carbohydrate structure recognized by DC-SIGN and its homologs L-SIGN and murine SIGN-R1 on mycobacterial mannosylated lipoarabinomannan was also identified, all receptors showing similar affinity for the mannose-cap oligosaccharide (man)3-ara (Table 1) [11].
Finally, biochemical studies using mutants of DC-SIGN demonstrated that binding to the HIV envelope protein gp120 is mediated by a distinct but overlapping epitope when compared with ICAM-2 and ICAM-3, providing a rationale for specific targeting of the gp120–DC-SIGN interaction, without affecting other physiologically important ICAM-2 and ICAM-3 interactions with DC-SIGN [12].
Several new carbohydrate structures have been identified for other CLRs, such as surfactant protein (SP)-D (Table 1). Despite the fact that this soluble CLR is a key component of the innate immunity in lung alveoli, limited information is available on its binding to complex carbohydrate structures. Recently, by using surface plasmon resonance spectroscopy, van de Wetering et al. [13] identified terminal α1-3-linked fucose residues as strong ligands for SP-D. Furthermore, for the first time, SP-D was shown to bind multicellular larval stages of the parasitic worm Schistosoma mansoni, transiently residing in the lung, by interacting with these fucosylated glycans exposed at the cell surface [13].
Novel binding targets were also identified in the interaction between the collectin mannose-binding lectin (MBL) and the bacterium Neisseria meningitidis (Table 1). Surprisingly, MBL was found to bind meningococci via two nonglycosylated major outer membrane proteins, opacity protein and porin, of N. meningitidis [14•]. Binding of CLRs to nonsugar targets on the surface of pathogens might further increase the complexity of pathogen–CLR interactions.
C-type lectin oligomerization: multivalent microdomains bind to microbial surfaces
The capacity of CLRs to detect microorganisms depends on the density of the PAMP present on the microbial surface, as well as on the degree of oligomerization of the lectin receptor. In fact, the assembly of several CRDs in multimers points the binding sites towards a common direction, increasing the binding valency and allowing interactions with the dense carbohydrate arrays on microbial surfaces.
The collectins form trimers, which further assemble into larger oligomers, appearing as a ‘bouquet of flowers’ [15]. Mutations that hamper the assembly of human MBL into oligomers result in a reduced capability to activate components of the complement system, thus increasing both the risk and severity of infections and leading to autoimmunity [16].
Moreover, transmembrane CLRs have developed several strategies to increase their strength of binding to PAMPs. Proteomic analysis of membrane-purified DC-SIGN complexes showed that DC-SIGN exists as tetramers at the surface of immature monocyte-derived DCs, and that this assembly is required for high-affinity binding of glycoproteins such as HIV gp120 [17•]. Moreover, biochemical and hydrodynamic studies on truncated DC-SIGN demonstrated that the portion of the neck of each molecule adjacent to the CRD is sufficient to mediate the formation of dimers, whereas the neck regions near the amino-terminal are required to stabilize tetramers [18]. In the same study, it was also shown that the CRDs are flexibly linked to the neck regions, which project CRDs from the cell surface and enable DC-SIGN to bind to various glycans on microbial surfaces [18].
Recently, we observed that DC-SIGN showed higher levels of organization (clustering) on the surface of DCs, depending upon the differentiation state of the DCs when they developed from monocytic precursors [19••]. High-resolution electron microscopy images demonstrated a direct relationship between DC-SIGN function as a viral receptor and its microlocalization on the plasma membrane. During development of human monocyte-derived DCs, DC-SIGN organization changes from a random distribution pattern into well-defined microdomains on the cell membrane. These submicron-sized microdomains have an average diameter of 100–200 nm, as established by electron microscopy and near-field scanning optical microscopy [20]. The organization of DC-SIGN in microdomains on the plasma membrane is important for the binding and internalization of virus particles, suggesting that these multimolecular assemblies of DC-SIGN act as a docking site for pathogens such as HIV-1 to invade the host [19••].
Further investigation is necessary to establish whether also other CLRs are organized in similar multi-molecular assemblies at the cell membrane.
Pathogen recognition by C-type lectins: protection or evasion?
There is increasing evidence to suggest that pathogens use several strategies to evade host immune surveillance [5, 6].
DC-SIGN was first identified as an HIV-1 receptor that enhanced infection of T lymphocytes in trans [21]. Recently, Arrighi et al. [22•] demonstrated that DC-SIGN is required for the formation of an infectious synapse between HIV-1-bearing DCs and resting CD4+ T cells. More specifically, the use of small RNAi-expressing lentiviral vectors specifically to knock down DC-SIGN on DCs showed that DC-SIGN− DCs are still able to internalize HIV-1, although the binding of virions was found to be reduced, but they cannot transfer HIV-1 infection to target cells [22•, 23]. It is becoming clear that other viruses besides HIV-1 target DC-SIGN to promote their dissemination and modulate DC function for establishing chronic infections. Hepatitis C virus has recently been shown to target DC-SIGN and L-SIGN to escape lysosomal degradation [24]. Similarly, severe acute respiratory syndrome coronavirus binds to DC-SIGN and is transferred by DCs to susceptible target cells through a synapse-like structure [25].
In addition to viruses, Helicobacter pylori has also been demonstrated to target DC-SIGN to block a polarized Th1 response [26••]. This bacterium binds to DC-SIGN via its lipopolysaccharide (LPS), which exposes Lewis blood group antigens. In particular, reversible on–off switching of specific fucosyltransferases regulates the expression of Lewis-X and -Y on the LPS, thus causing LPS phase variation (Lewis-X+/Y+ and Lewis-X+/Y−) within a strain. Interestingly, whereas Lewis-X+/Y− H. pylori escapes binding to DCs, the Lewis-X+/Y+ phase variant exploits binding to DC-SIGN to increase interleukin-10 levels and block the skewing of naïve T cells to Th1 cells [26••].
A similar evasion strategy has also been developed by the yeast-like fungus Cryptococcus neoformans, which infects the respiratory and nervous systems, to escape aggregation by SP-D [27]. Only acapsular C. neoformans is aggregated by SP-D and subsequently removed by microciliary clearance. Therefore, after deposition in the lung, C. neoformans readily starts producing a capsule, thus also releasing soluble capsular components, including glucuronoxylomannan (GXM). SP-D binds with high affinity to soluble glucuronoxylomannan, and this interaction has been shown to inhibit SP-D aggregation of acapsular C. neoformans, thus interfering with pathogen clearance [27].
Bordetella pertussis, the causative agent of human whooping cough, uses a slightly different mechanism for resisting the bactericidal effects of SP-A [28]. SP-A binds via its CRD to the lipid A component of LPS. However, B. pertussis LPS has a terminal trisaccharide which apparently shields the bacteria from SP-A-mediated clearance by sterically limiting access of SP-A to the lipid A region [28].
Despite these examples of immune evasion by pathogens through CLRs, we must not forget that these receptors have a fundamental role in limiting the early proliferation of infectious microorganisms. The importance of MBL in restricting the complications associated with Staphylococcus aureus infection is highlighted by studies involving MBL-null mice [29]. This in vivo study demonstrated that 100% of the MBL-null mice died 48 hours after exposure to an intravenous inoculation of S. aureus compared with a 45% mortality rate in wild-type mice [29]. Similarly, SP-D knockout mice displayed a delayed clearance of Pneumocystis carinii infection, increased inflammation and altered nitric oxide metabolism [30].
Interestingly, although DC-SIGN is exploited by several viruses to escape lysosomal degradation, Moris et al. [31] recently demonstrated that DC-SIGN does not always protect captured virions against degradation. In fact, a fraction of the incoming viral material is processed by the proteasome and leads to activation of anti-HIV-specific cytotoxic T lymphocytes by DC-SIGN-expressing cells [31]. This work suggests that the different routing of the incoming viral material might be directly related to the viral load. Additional evidence for a protective in vivo role for a SIGN family member is provided by Lanoue et al. [32•]. Mice lacking SIGN-R1 are significantly more susceptible to Streptococcus pneumoniae infection and fail to clear the bacteria from the circulation, showing an important role for SIGN-R1 in the protection against septicemia [32•].
The Langerhans cell-specific C-type lectin langerin has also been shown to have a crucial role in the generation of CD1a-restricted T cell clones against a Mycobacterium leprae lipid antigen [33•].
Recent findings have demonstrated that Pneumocystis induces nuclear translocation of nuclear factor-κB in human alveolar macrophages primarily through interactions with the macrophage mannose receptor (MMR) [34]. This identifies the MMR as a pattern recognition receptor (PRR) which, besides mediating phagocytosis, is also capable of signaling in response to infectious non-self challenge.
Levels of complexity in pathogen recognition
In the past few years, evidence has accumulated illustrating the important role of the C-type lectins for the normal functioning of the immune system. From this, a picture is emerging that suggests that microbial recognition is based on networks between C-type lectins and other innate immune recognition receptors.
For example, the lectin pathway of complement activation is essential in innate antimicrobial defense. This is initiated by multimolecular complexes composed of MBL and MBL-associated serine proteases, which subsequently cleave complement C4 and C2, thus triggering the complement cascade. Studies with both MBL-null mice and MBL-deficient individuals have demonstrated that the absence of MBL-mediated complement activation severely increases susceptibility to herpes simplex virus-2 infection [35]. Similarly, a direct interaction of MBL with Candida albicans has been documented; this was shown to result in agglutination and accelerated complement activation and to lead to fungal growth inhibition, even in the absence of opsonophagocytosis by DCs [36].
Interestingly, cooperation among members of the C-type lectin family has also been reported. Taylor et al. [37•] demonstrated that in murine peritoneal macrophages, SIGN-R1 exhibits additive cooperation with the β-glucan receptor dectin-1 in the nonopsonic recognition of yeast.
In addition, SP-A and SP-D, but not MBL, have been shown to enhance phagocytosis of Mycobacterium avium by macrophages through upregulation of MMR cell-surface expression and activity [38•].
Furthermore, SP-A has been reported to augment the phagocytosis of S. pneumoniae by alveolar macrophages through an increase in cell-surface localization of scavenger receptor A [39].
These recent observations, together with previous reports documenting crosstalk between C-type lectins and TLRs [40, 41], clearly indicate that specific interactions among different immune receptors might differentially regulate the outcome of an immune response against a specific pathogen.
Conclusions
In the recognition of pathogens, several levels of complexity can be distinguished that might determine the destiny of the pathogen and the outcome of an immune response (Figure 1 ). Firstly, the pathogen itself expresses at its surface a specific PAMP, or a combination of different PAMPs, which dictates the recognition by specific receptors. Secondly, the set of PRRs expressed varies among different cells, resulting in a specific set of pathogen-recognition receptors which is different for each cell type of the immune system. Thirdly, changes in PRR cell-surface distribution adds yet another level of complexity, modulating the binding to certain carbohydrate moieties, and thus regulating the affinity for different ligands. Finally, these receptors have been shown to interact with each other, thereby giving rise to a network of multimolecular complexes that add a fourth level of complexity to pathogen recognition. Unraveling the precise mechanisms regulating the formation and exact function of these receptor networks is the present challenge.
Figure 1.
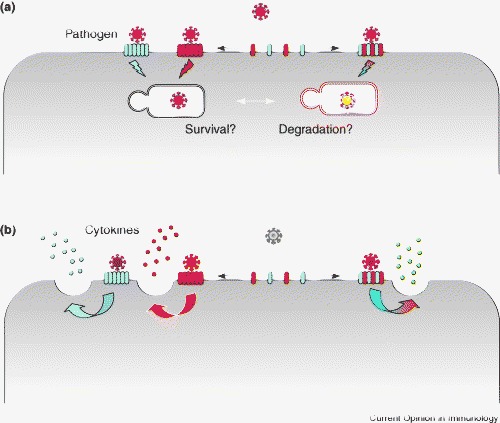
Levels of complexity in lectin-mediated pathogen recognition determine pathogen destiny and the immune response. Differences in PAMPS expressed by pathogens and PRRs on host cells determine a first and second level of complexity in recognition. (a) Changes in receptor distribution at the cell surface might further influence the binding to a pathogen and the subsequent intracellular routing of the pathogen, and determine pathogen survival or lysosomal degradation. (b) The formation of mixed multimolecular receptor assemblies (for example, between CLRs and TLRs, complement receptors or other CLRs) might further extend the PAMP profile. Moreover, the outcome of a cell response to a pathogen (for example, in terms of the release of different cytokines) might differ depending on the triggering of single receptors or receptor complexes.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
References
- 1.Drickamer K. C-type lectin-like domains. Curr Opin Struct Biol. 1999;9:585–590. doi: 10.1016/s0959-440x(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 2.Cambi A., Figdor C.G. Dual function of C-type lectin-like receptors in the immune system. Curr Opin Cell Biol. 2003;15:539–546. doi: 10.1016/j.ceb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Geijtenbeek T.B., van Vliet S.J., Engering A., ‘t Hart B.A., van Kooyk Y. Self- and nonself-recognition by C-type lectins on dendritic cells. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- 4.Figdor C.G., van Kooyk Y., Adema G.J. C-type lectin receptors on dendritic cells and Langerhans cells. Nat Rev Immunol. 2002;2:77–84. doi: 10.1038/nri723. [DOI] [PubMed] [Google Scholar]
- 5.van Kooyk Y., Geijtenbeek T.B. DC-SIGN: escape mechanism for pathogens. Nat Rev Immunol. 2003;3:697–709. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- 6.van Kooyk Y., Engering A., Lekkerkerker A.N., Ludwig I.S., Geijtenbeek T.B. Pathogens use carbohydrates to escape immunity induced by dendritic cells. Curr Opin Immunol. 2004;16:488–493. doi: 10.1016/j.coi.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Feizi T., Chai W. Oligosaccharide microarrays to decipher the glyco code. Nat Rev Mol Cell Biol. 2004;5:582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 8•.Galustian C., Park C.G., Chai W., Kiso M., Bruening S.A., Kang Y.S., Steinman R.M., Feizi T. High and low affinity carbohydrate ligands revealed for murine SIGN-R1 by carbohydrate array and cell binding approaches, and differing specificities for SIGN-R3 and langerin. Int Immunol. 2004;16:853–866. doi: 10.1093/intimm/dxh089. [DOI] [PubMed] [Google Scholar]; Both this study and that described in reference 9 highlight the importance of a detailed knowledge of the carbohydrate-binding profile and discuss two high-throughput methods that allow the identification of novel carbohydrate structures recognized by receptors.
- 9•.Guo Y., Feinberg H., Conroy E., Mitchell D.A., Alvarez R., Blixt O., Taylor M.E., Weis W.I., Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat Struct Mol Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]; See annotation to [8•].
- 10•.Van Liempt E., Imberty A., Bank C.M., Van Vliet S.J., Van Kooyk Y., Geijtenbeek T.B., Van Die I. Molecular basis of the differences in binding properties of the highly related C-type lectins DC-SIGN and L-SIGN to Lewis X trisaccharide and Schistosoma mansoni egg antigens. J Biol Chem. 2004;279:33161–33167. doi: 10.1074/jbc.M404988200. [DOI] [PubMed] [Google Scholar]; This study shows for the first time a clear correlation between specific carbohydrate recognition and amino acid sequence, and clearly explains the different carbohydrate-binding profiles of the two homologs DC-SIGN and L-SIGN.
- 11.Koppel E.A., Ludwig I.S., Hernandez M.S., Lowary T.L., Gadikota R.R., Tuzikov A.B., Vandenbroucke-Grauls C.M., van Kooyk Y., Appelmelk B.J., Geijtenbeek T.B. Identification of the mycobacterial carbohydrate structure that binds the C-type lectins DC-SIGN, L-SIGN and SIGNR1. Immunobiology. 2004;209:117–127. doi: 10.1016/j.imbio.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Su S.V., Hong P., Baik S., Negrete O.A., Gurney K.B., Lee B. DC-SIGN binds to HIV-1 glycoprotein 120 in a distinct but overlapping fashion compared with ICAM-2 and ICAM-3. J Biol Chem. 2004;279:19122–19132. doi: 10.1074/jbc.M400184200. [DOI] [PubMed] [Google Scholar]
- 13.van de Wetering J.K., van Remoortere A., Vaandrager A.B., Batenburg J.J., van Golde L.M., Hokke C.H., van Hellemond J.J. Surfactant protein D binding to terminal alpha1-3-linked fucose residues and to Schistosoma mansoni. Am J Respir Cell Mol Biol. 2004;31:565–572. doi: 10.1165/rcmb.2004-0105OC. [DOI] [PubMed] [Google Scholar]
- 14•.Estabrook M.M., Jack D.L., Klein N.J., Jarvis G.A. Mannose-binding lectin binds to two major outer membrane proteins, opacity protein and porin, of Neisseria meningitidis. J Immunol. 2004;172:3784–3792. doi: 10.4049/jimmunol.172.6.3784. [DOI] [PubMed] [Google Scholar]; Both this study and that described in reference 33 indicate the existence of an interaction between C-type lectins and nonsugar moieties.
- 15.Lu J., Teh C., Kishore U., Reid K.B. Collectins and ficolins: sugar pattern recognition molecules of the mammalian innate immune system. Biochim Biophys Acta. 2002;1572:387–400. doi: 10.1016/s0304-4165(02)00320-3. [DOI] [PubMed] [Google Scholar]
- 16.Larsen F., Madsen H.O., Sim R.B., Koch C., Garred P. Disease-associated mutations in human mannose-binding lectin compromise oligomerization and activity of the final protein. J Biol Chem. 2004;279:21302–21311. doi: 10.1074/jbc.M400520200. [DOI] [PubMed] [Google Scholar]
- 17•.Bernhard O.K., Lai J., Wilkinson J., Sheil M.M., Cunningham A.L. Proteomic analysis of DC-SIGN on dendritic cells detects tetramers required for ligand binding but no association with CD4. J Biol Chem. 2004;279:51828–51835. doi: 10.1074/jbc.M402741200. [DOI] [PubMed] [Google Scholar]; This study provides a clear demonstration that tetramerization of DC-SIGN does occur at the cell membrane.
- 18.Feinberg H., Guo Y., Mitchell D.A., Drickamer K., Weis W.I. Extended neck regions stabilize tetramers of the receptors DC-SIGN and DC-SIGNR. J Biol Chem. 2005;280:1327–1335. doi: 10.1074/jbc.M409925200. [DOI] [PubMed] [Google Scholar]
- 19••.Cambi A., de Lange F., van Maarseveen N.M., Nijhuis M., Joosten B., van Dijk E.M., de Bakker B.I., Fransen J.A., Bovee-Geurts P.H., van Leeuwen F.N. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J Cell Biol. 2004;164:145–155. doi: 10.1083/jcb.200306112. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows a further level of organization of the membrane-associated C-type lectin DC-SIGN in submicron-sized microdomains associating with lipid rafts and acting as scaffolds for virus binding.
- 20.Koopman M., Cambi A., de Bakker B.I., Joosten B., Figdor C.G., van Hulst N.F., Garcia-Parajo M.F. Near-field scanning optical microscopy in liquid for high resolution single molecule detection on dendritic cells. FEBS Lett. 2004;573:6–10. doi: 10.1016/j.febslet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 21.Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J., Cornelissen I.L., Nottet H.S., KewalRamani V.N., Littman D.R. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 22•.Arrighi J.F., Pion M., Garcia E., Escola J.M., van Kooyk Y., Geijtenbeek T.B., Piguet V. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J Exp Med. 2004;200:1279–1288. doi: 10.1084/jem.20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the specific involvement of DC-SIGN in the formation of an infectious synapse between DCs and T cells during HIV-1 transmission.
- 23.Arrighi J.F., Pion M., Wiznerowicz M., Geijtenbeek T.B., Garcia E., Abraham S., Leuba F., Dutoit V., Ducrey-Rundquist O., van Kooyk Y. Lentivirus-mediated RNA interference of DC-SIGN expression inhibits human immunodeficiency virus transmission from dendritic cells to T cells. J Virol. 2004;78:10848–10855. doi: 10.1128/JVI.78.20.10848-10855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig I.S., Lekkerkerker A.N., Depla E., Bosman F., Musters R.J., Depraetere S., van Kooyk Y., Geijtenbeek T.B. Hepatitis C virus targets DC-SIGN and L-SIGN to escape lysosomal degradation. J Virol. 2004;78:8322–8332. doi: 10.1128/JVI.78.15.8322-8332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Bergman M.P., Engering A., Smits H.H., van Vliet S.J., van Bodegraven A.A., Wirth H.P., Kapsenberg M.L., Vandenbroucke-Grauls C.M., van Kooyk Y., Appelmelk B.J. Helicobacter pylori modulates the T helper cell 1/T helper cell 2 balance through phase-variable interaction between lipopolysaccharide and DC-SIGN. J Exp Med. 2004;200:979–990. doi: 10.1084/jem.20041061. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study clearly demonstrates how the pathogen–host interaction can be influenced by the differential expression of specific PAMPs, which can directly modulate the immune response.
- 27.van de Wetering J.K., Coenjaerts F.E., Vaandrager A.B., van Golde L.M., Batenburg J.J. Aggregation of Cryptococcus neoformans by surfactant protein D is inhibited by its capsular component glucuronoxylomannan. Infect Immun. 2004;72:145–153. doi: 10.1128/IAI.72.1.145-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaeffer L.M., McCormack F.X., Wu H., Weiss A.A. Bordetella pertussis lipopolysaccharide resists the bactericidal effects of pulmonary surfactant protein A. J Immunol. 2004;173:1959–1965. doi: 10.4049/jimmunol.173.3.1959. [DOI] [PubMed] [Google Scholar]
- 29.Shi L., Takahashi K., Dundee J., Shahroor-Karni S., Thiel S., Jensenius J.C., Gad F., Hamblin M.R., Sastry K.N., Ezekowitz R.A. Mannose-binding lectin-deficient mice are susceptible to infection with Staphylococcus aureus. J Exp Med. 2004;199:1379–1390. doi: 10.1084/jem.20032207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atochina E.N., Gow A.J., Beck J.M., Haczku A., Inch A., Kadire H., Tomer Y., Davis C., Preston A.M., Poulain F. Delayed clearance of Pneumocystis carinii infection, increased inflammation, and altered nitric oxide metabolism in lungs of surfactant protein-D knockout mice. J Infect Dis. 2004;189:1528–1539. doi: 10.1086/383130. [DOI] [PubMed] [Google Scholar]
- 31.Moris A., Nobile C., Buseyne F., Porrot F., Abastado J.P., Schwartz O. DC-SIGN promotes exogenous MHC-I-restricted HIV-1 antigen presentation. Blood. 2004;103:2648–2654. doi: 10.1182/blood-2003-07-2532. [DOI] [PubMed] [Google Scholar]
- 32•.Lanoue A., Clatworthy M.R., Smith P., Green S., Townsend M.J., Jolin H.E., Smith K.G., Fallon P.G., McKenzie A.N. SIGN-R1 contributes to protection against lethal pneumococcal infection in mice. J Exp Med. 2004;200:1383–1393. doi: 10.1084/jem.20040795. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides the first example of a member of the SIGN family being able to exert a protective function in the innate immune response.
- 33•.Hunger R.E., Sieling P.A., Ochoa M.T., Sugaya M., Burdick A.E., Rea T.H., Brennan P.J., Belisle J.T., Blauvelt A., Porcelli S.A. Langerhans cells utilize CD1a and langerin to efficiently present nonpeptide antigens to T cells. J Clin Invest. 2004;113:701–708. doi: 10.1172/JCI19655. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to [14•].
- 34.Zhang J., Zhu J., Imrich A., Cushion M., Kinane T.B., Koziel H. Pneumocystis activates human alveolar macrophage NF-kappaB signaling through mannose receptors. Infect Immun. 2004;72:3147–3160. doi: 10.1128/IAI.72.6.3147-3160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gadjeva M., Paludan S.R., Thiel S., Slavov V., Ruseva M., Eriksson K., Lowhagen G.B., Shi L., Takahashi K., Ezekowitz A. Mannan-binding lectin modulates the response to HSV-2 infection. Clin Exp Immunol. 2004;138:304–311. doi: 10.1111/j.1365-2249.2004.02616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ip W.K., Lau Y.L. Role of mannose-binding lectin in the innate defense against Candida albicans: enhancement of complement activation, but lack of opsonic function, in phagocytosis by human dendritic cells. J Infect Dis. 2004;190:632–640. doi: 10.1086/422397. [DOI] [PubMed] [Google Scholar]
- 37•.Taylor P.R., Brown G.D., Herre J., Williams D.L., Willment J.A., Gordon S. The role of SIGNR1 and the beta-glucan receptor (dectin-1) in the nonopsonic recognition of yeast by specific macrophages. J Immunol. 2004;172:1157–1162. doi: 10.4049/jimmunol.172.2.1157. [DOI] [PubMed] [Google Scholar]; This study provides a clear demonstration that cooperation between C-type lectins takes place in the immune system.
- 38•.Kudo K., Sano H., Takahashi H., Kuronuma K., Yokota S., Fujii N., Shimada K., Yano I., Kumazawa Y., Voelker D.R. Pulmonary collectins enhance phagocytosis of Mycobacterium avium through increased activity of mannose receptor. J Immunol. 2004;172:7592–7602. doi: 10.4049/jimmunol.172.12.7592. [DOI] [PubMed] [Google Scholar]; See annotation to [37•].
- 39.Kuronuma K., Sano H., Kato K., Kudo K., Hyakushima N., Yokota S., Takahashi H., Fujii N., Suzuki H., Kodama T. Pulmonary surfactant protein A augments the phagocytosis of Streptococcus pneumoniae by alveolar macrophages through a casein kinase 2-dependent increase of cell surface localization of scavenger receptor A. J Biol Chem. 2004;279:21421–21430. doi: 10.1074/jbc.M312490200. [DOI] [PubMed] [Google Scholar]
- 40.Brown G.D., Herre J., Williams D.L., Willment J.A., Marshall A.S., Gordon S. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geijtenbeek T.B., Van Vliet S.J., Koppel E.A., Sanchez-Hernandez M., Vandenbroucke-Grauls C.M., Appelmelk B., Van Kooyk Y. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med. 2003;197:7–17. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Palaniyar N., Nadesalingam J., Clark H., Shih M.J., Dodds A.W., Reid K.B. Nucleic acid is a novel ligand for innate, immune pattern recognition collectins surfactant proteins A and D and mannose-binding lectin. J Biol Chem. 2004;279:32728–32736. doi: 10.1074/jbc.M403763200. [DOI] [PubMed] [Google Scholar]; This study identifies nucleic acids as novel ligand structures for C-type lectins and indicates a possible role for these receptors in the clearance of apoptotic cells.
- 43.Cormier E.G., Durso R.J., Tsamis F., Boussemart L., Manix C., Olson W.C., Gardner J.P., Dragic T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc Natl Acad Sci USA. 2004;101:14067–14072. doi: 10.1073/pnas.0405695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano-Gomez D., Dominguez-Soto A., Ancochea J., Jimenez-Heffernan J.A., Leal J.A., Corbi A.L. Dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin mediates binding and internalization of Aspergillus fumigatus conidia by dendritic cells and macrophages. J Immunol. 2004;173:5635–5643. doi: 10.4049/jimmunol.173.9.5635. [DOI] [PubMed] [Google Scholar]
- 46.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang Y.S., Kim J.Y., Bruening S.A., Pack M., Charalambous A., Pritsker A., Moran T.M., Loeffler J.M., Steinman R.M., Park C.G. The C-type lectin SIGN-R1 mediates uptake of the capsular polysaccharide of Streptococcus pneumoniae in the marginal zone of mouse spleen. Proc Natl Acad Sci USA. 2004;101:215–220. doi: 10.1073/pnas.0307124101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cambi A., Koopman M., Figdor C.G. How C-type lectins detect pathogens. Cell Microbiol. 2005;7:481–488. doi: 10.1111/j.1462-5822.2005.00506.x. [DOI] [PubMed] [Google Scholar]


