Abstract
Helicases are one of the smallest motors of biological system, which harness the chemical free energy of ATP hydrolysis to catalyze the opening of energetically stable duplex nucleic acids and thereby are involved in almost all aspect of nucleic acid metabolism including replication, repair, recombination, transcription, translation, and ribosome biogenesis. Basically, they break the hydrogen bonding between the duplex helix and translocate unidirectionally along the bound strand. Mostly all the helicases contain some conserved signature motifs, which act as an engine to power the unwinding. After the discovery of the first prokaryotic DNA helicase from Escherichia coli bacteria in 1976 and the first eukaryotic one from the lily plant in 1978, many more (>100) have been isolated. All the helicases share some common properties, including nucleic acid binding, NTP hydrolysis and unwinding of the duplex. Many helicases have been crystallized and their structures have revealed an underlying common structural fold for their function. The defects in helicases gene have also been reported to be responsible for variety of human genetic disorders, which can lead to cancer, premature aging or mental retardation. Recently, a new role of a helicase in abiotic stress signaling in plant has been discovered. Overall, helicases act as essential molecular tools for cellular machinery and help in maintaining the integrity of genome. Here an overview of helicases has been covered which includes history, biochemical assay, properties, classification, role in human disease and mechanism of unwinding and translocation.
Keywords: ATPase, Helicase classification, Helicase-related diseases, Molecular motors, Signature motifs, Unwinding enzyme
1. Introduction
Motors are integral parts of cars running along highways and tractors on farms; they operate air conditioners in office buildings, refrigerators at home and conveyor belts in factories. Motors are everywhere, even inside each cell of our body and these motors are known as biomolecular motors. A typical biomolecular motor is a protein that uses free energy from adenosine tri-phosphate (ATP) or other nucleotide tri-phosphate (NTP) hydrolysis as its fuel to produce mechanical force. Biomolecular motor proteins are generally involved in several critical cellular processes and have functions ranging from ATP synthesis, organelle transport, muscle contraction, protein folding and nucleic acid translocation. Therefore, they are involved in cell signaling, division, and motion and these specific activities are dependent on the cell type. These motor-dependent activities initiate with DNA replication and transcription and are followed by protein synthesis and transport, forming an important part of cellular function.
The DNA molecule that makes up a chromosome is like a rubber ladder that is twisted into a coil, called a double helix. DNA of each living organism contains the genetic information, which determines the characteristics of the individuals. But the duplex DNA is energetically very stable because of the hydrogen bonding formed between the two interacting and antiparallel DNA strands. Therefore, the genetic information, which is locked in the duplex DNA, cannot be accessed for many genetical processes, until it is opened [1]. For this purpose, a special class of enzymes has evolved, whose functions are to allow access to the genetic information. These are known as DNA helicases, which catalyze the unwinding of duplex DNA and thus play an important role in many important cellular processes, including DNA replication, repair, recombination, transcription and translation [2], [3], [4], [5], [6]. They are also considered as motor proteins translocating on DNA by using nucleoside triphosphate hydrolysis as the source of power energy [7], [8]. All the DNA helicases contain ATP-dependent DNA unwinding activity and DNA-dependent ATPase activities, which is a corollary activity to duplex unwinding [5], [6], [9]. In general, helicases require a molecular mechanism for converting the chemical energy generated by the ATP hydrolysis into the mechanical energy required for strand separation [6], [9]. RNA helicases are also important proteins, which unwind the secondary structures present in the RNAs and thus are essential for almost all the RNA-related biological processes, which includes transcription, pre-mRNA splicing, ribosome biogenesis, editing, mRNA export, translation, RNA turnover and organelle function.
Multiple DNA helicases have been isolated from different organisms [2], [3], [4], [5], because of a variety of different needs for the duplex DNA to unwind in different DNA transactions. For example, 14 different DNA helicases have been isolated from a simple single cell organism like Escherichia coli and several from eukaryotic systems [2], [5], [6]. More than 100 DNA helicases have been reported so far. Most helicases contain short conserved amino acid fingerprints called helicase motifs. They are named as Q, I, Ia, Ib, II–VI motifs. Because of the presence of motif II (DEAD), they are also called as DEAD/H-box helicases family of proteins [10], [11]. High sequence conservation has been maintained in this large group of helicases, suggesting that the motifs containing helicase genes evolved from a common ancestor. The defects in helicase genes can cause several human diseases. The importance of DNA helicases was highlighted several years ago with the discovery that DNA repair helicase can correct the human DNA excision repair disorders Xeroderma pigmentosum (XP) and Cockayne's syndrome (CS) [12]. In plant, the importance of the helicase has been recently highlighted with the discovery that the overexpression of a DNA helicase gene in plant can give abiotic stress tolerance to the plants and thereby confirming its new role in stress signaling [13]. A variety of different DNA helicases have been isolated from both prokaryotic and eukaryotic systems and the number is still growing. There is an urgent need now, for the classification of helicases, since this aspect has not been extensively covered in previously published reviews. This article focuses on the general aspect of DNA helicases including historical background, biochemical assays, basic properties, classification of DNA helicases, diseases related to defect in helicases and mechanism of unwinding and translocation.
2. The historical aspects of DNA helicases
First, Watson and Crick had solved the structure of the duplex DNA in 1953 [14]. After that the DNA interacting proteins were discovered. Till 1976 there was no report of helicases protein hence the period 1953–76 was considered as the pre-helicase years.
In 1976 Hoffman-Berling and his colleagues reported the isolation of the first DNA helicase from E. coli (helicase I, a traI gene product) [15]. In 1978 Hotta and Stern reported the first eukaryotic DNA helicase from lily plant [16]. During 1982–83 a more direct assay (strand displacement assay) for measuring helicase activity was developed by Venkatesan's and Matson's groups [17], [18]. In 1982 first bacteriophage protein (T4 gene 41 protein) was reported as DNA helicase by Venkatesan and his colleagues [17]. In 1986 the first viral encoded protein, the SV40 large tumor antigen, was reported as DNA helicase [19]. This is one of the best-characterized DNA helicase reported so far. In1988, Hodgeman and Gorbalenya discovered helicase signature motifs (seven conserved amino acid domains) [15], [20]. In 1989 two helicase superfamilies (SF1 and 2) were reported [21] and also the DEAD box helicase family was identified by Linder and his colleagues [11]. In 1990 Tuteja and his colleagues reported the purification and characterization of the first human DNA helicase from HeLa cells [22] and the E. coli RecQ protein was also reported as DNA helicase [23]. In 1992 the first mitochondrial DNA helicase was reported from mammalian (bovine brain) tissue [24]. In 1994 the first human autoantigen (Ku protein) was identified as DNA helicase [25].
In 1996 Tuteja and his colleagues reported the first plant DNA helicase in the purified form from pea chloroplast [26] and Subramanya and his colleagues solved the crystal structure of the first DNA helicase (PcrA from thermophilic bacterium) [27]. In 1997 MCM proteins (MCM4/6/7 complex) from HeLa cells was reported as DNA helicase [28]. Also in 1997 the Werner syndrome protein (WRN) and the Bloom's syndrome gene product (BLM) were reported as DNA helicases [29], [30]. In 2000 cloning of the first plant gene encoding biochemically active DNA helicase was reported [31] and also the first eIF-4A from pea was reported also as DNA helicase [31]. In 2002 first biochemically active malaria parasite DNA helicase from Plasmodium cynomolgi was reported by Tuteja and colleagues [32]. In 2003 a new helicase motif (Q motif) was identified in the DEAD-box family of helicases [33]. In 2005 the first Plasmodium falciparum DNA helicase was reported in the purified form [34]. In 2005 the first transgenic tobacco plants overexpressing the helicase gene was reported and also these plants were shown to tolerate the high salinity stress [13].
3. Biochemical assay for measuring helicase activity
The most useful direct in vitro method for measuring unwinding activity catalyzed by a helicase was developed almost simultaneously by the Nossal group and the Richardson group [17], [18]. Helicase unwinds partially duplex DNA substrate (32P-labeled oligonucleotide annealed to a longer single-stranded (ss) DNA molecule) yielding two ssDNA molecules of different sizes that are resolved from the starting duplex by electrophoresis followed by autoradiography (Fig. 1A and B ). The radioactive label within the DNA permits direct visualization (Fig. 1B) and quantitation of the results [2], [9]. A unit of unwinding activity is usually defined as the amount of DNA helicase required to unwind a certain percentage of the partially duplex substrate in 30 min at 37 °C [2], [3]. Besides this conventional assay, several additional assays have been developed, which are not commonly used. For example electron microscopy assay, fluorescence-based assays, a rapid quench-flow method, filtration assay, a time-resolved fluorescence resonance energy transfer (TRET) assay, a scintillation proximity assay (SPA), assay based on Flashplate technology, homogenous time resolved fluorescence quenching assay (HTR-FQA) and electrochemiluminescence (ECL)-based helicase assay [2].
Fig. 1.
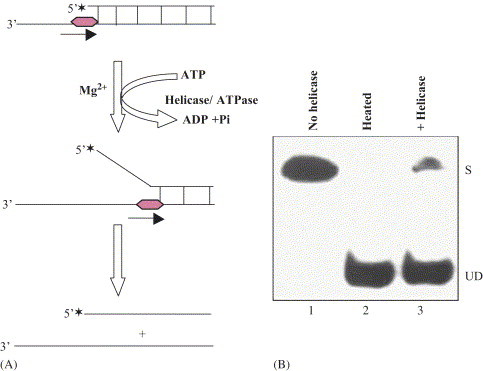
A. Scheme of biochemical assay for measuring unwinding activity of ATP/Mg2+-dependent DNA helicase. Asterisks denote the 32P-labeled end of the DNA. The partial duplex DNA helicase substrate was prepared by annealing the radiolabeled DNA oligo (small DNA) to longer ssDNA as described [2], [3]. B. Autoradiogram of the gel: lane 1, reaction without enzyme; lane 2, heat-denatured substrate and lane 3, reaction in presence of DNA helicase enzyme. S=substrate; UD=unwound DNA.
4. Basic properties of DNA helicases
Helicases share at least three common basic properties: (i) nucleic acid binding, (ii) NTP/dNTP binding and hydrolysis and (iii) NTP/dNTP hydrolysis-dependent unwinding of duplex nucleic acids [35]. Therefore, helicases contain following multiple enzymatic and other activities: (i) DNA and Mg2+-dependent ATP hydrolysis activity (ATPase). In absence of DNA its ATPase activity is very low, but in presence of ssDNA this activity is stimulated several fold, (ii) DNA binding activity (not covalent binding), (iii) translocase activity; the ATPase activity is coupled to unidirectional translocation of the helicase along the bound strand, (iv) strand displacement activity also known as DNA melting or unwinding activity; this activity is ATP dependent. Without ATP a bona fide helicase cannot unwind the duplex DNA, and (v) ATP binding activity. In their sequence, helicases contain ATP binding domain, where ATP can bind.
Helicases usually need ssDNA as a loading zone where they bind in a sequence independent manner and translocate unidirectionally. In general, they bind to ssDNA with higher affinity than to dsDNA. However, RecBCD, SV40 large antigen, and RuvB helicases preferentially bind to dsDNA [2]. Many helicases need a replication fork-like structure on the substrate for optimum unwinding. Some DNA helicases can initiate unwinding also from the ends of blunt-ended duplex DNA, such as RecBCD, UvrD, Rep and RecQ from E. coli and SV40 large T antigen [2], [6]. In general, replicative hexameric helicases bind ssDNA tightly in the presence of NTP, and weakly in the presence of NDP. This binding results in a conformational change in homo-oligomeric helicases and thereby affects the assembly state of the protein. In general, DNA helicases exhibit specific polarity, which is defined as the direction of DNA helicase movement on initially bound ssDNA template (i.e. 3′–5′ or 5′–3′) with respect to the polarity of the sugar phosphate backbone. For those helicases involved in DNA replication, the polarity of the reaction is strongly indicative of helicase placement on the leading (a 3′–5′ polarity) or lagging (5′–3′ polarity) strands. It has been reported that RecBCD contains bipolar enzyme activity, where RecB and RecD components of the complex unwind DNA from 3′ to 5′ and 5′–3′ directions, respectively [36]. Recently, few more examples have been reported for a bipolar activity of DNA helicases, such as PcrA helicase from Bacillus anthracis [37], HerA DNA helicases from Thermophilic archaea [38], a DNA helicase from Plasmodium falciparum [34], and pea DNA helicase 47 (PDH47) [39]. The fact that many helicases function as hexamers, as well as consideration of the necessity for multiple DNA-binding sites has led to the suggestion that oligomerization may be necessary for helicase function. On the basis of active assembly of the DNA helicases they can be grouped as monomeric or multimeric helicases.
5. Classification of DNA helicases
It is found that about 1% of each of the prokaryotic and eukaryotic genomes codes for helicases [40]. With the exciting progress in the field the number of helicases isolated in all the systems is continuously growing, which has created the problem of a bewildering array of different names and their classification. Therefore, it is important that a clear, scientific system for nomenclature and classification of helicases be formulated. Although this class of proteins does not, at this point, lend itself to a simple classification scheme. They can be classified on the basis of nucleic acid substrate they act upon, source of identification, oligomeric nature, directionality of unwinding, associated functions and also on the basis of families and SF (motifs). The various possible ways for the classification of the helicases are mentioned below:
5.1. Classification on the basis of kind of duplex strand unwinding
If helicases unwind the substrates consisting of partial duplex regions of DNA/DNA, RNA/RNA, RNA/DNA, and DNA/RNA hybrids to single stranded polynucleotides, they can be classified as DNA helicases, RNA helicases, RNA–DNA, and DNA–RNA helicases, respectively. The RNA–DNA helicases use the partial duplex hybrid substrate consisting of short DNA stretch annealed to long RNA strand. The RNA–DNA helicases first bind and translocate on RNA strand to displace the DNA strand. The DNA–RNA helicases use the partial duplex hybrid substrate of DNA/RNA consisting of short stretch of RNA annealed to a long DNA strand. The DNA–RNA helicases first bind and translocate on DNA strand to displace the RNA strand. There are only few proteins known that are endowed with both DNA and RNA helicase activities, for example human DNA helicase (HDH) I, IV and VIII [4], [41], SV40 large T antigen [42] and DNA helicase II from calf thymus [43]. HDH I and IV can also unwind the DNA–RNA or RNA–DNA duplexes [4]. All these helicases can come under both the category of DNA and RNA helicases and thereby might be involved in both the DNA and RNA metabolisms. There is a single example of E. coli Rho protein, a transcription termination factor, which catalyzes the unwinding of RNA–DNA hybrid and comes under RNA–DNA helicase category [5]. Although Rho protein can also unwind duplex RNA, provided that a specific sequence of ssRNA is provided for Rho protein binding, the enzyme does not unwind duplex DNA. UvrD is also an E. coli RNA–DNA helicase although it is best known as a DNA helicase. UvrD has robust activity on RNA/DNA hybrids. However, the significance of this activity, if any, is not known [44].
5.2. Classification on the basis of source of identification
A strand displacement assay described for the characterization of the gene 4 DNA helicase of the E. coli bacteriophage T7 [18] brought rapid progress in the detection of eukaryotic and viral helicases. More than one helicase is found to be present in each system. Now helicases have been isolated from many sources. They can be classified on the basis of source in which they were identified and named as prokaryotic, eukaryotic, bacteriophage and viral helicases. Table 1 summarizes the number of DNA helicases isolated from different systems. Now even from malarial parasite two different DNA helicase proteins have been purified which contains ATP and Mg2+ dependent DNA unwinding and ssDNA dependent ATPase activities [32], [34].
Table 1.
Biochemically active prokaryotic and eukaryotic DNA helicases reported so far
| S. No. | Organism | Number reported so far |
|---|---|---|
| 1. | Escherichia coli | 14 |
| 2. | Bacteriophage | 6 |
| 3. | Parasites (Plasmodium) | 2 (one from Plasmodium cynomolgi and one from Plasmodium falciparum) |
| 4. | Virus | 12 |
| 5. | Yeast | 15 |
| 6. | Plant | 10 (including three from chloroplast) |
| 7. | Calf thymus | 11 |
| 8. | Human | 25 |
| 9. | Frog | 1 |
| 10. | Mouse/rat | 3 |
| 11. | Drosophila | 4 |
5.3. Classification on the basis of oligomeric structure of protein
On the basis of structures of protein the helicases can be classified as monomeric or dimeric or hexameric or multimeric helicases. The inchworm model of DNA unwinding is consistent with any oligomeric state for the protein including monomeric [6], [45]. The bacterial helicases II, IV and PriA [5, and references cited there in], HDH IV, V and VI are the few examples of monomeric forms [4]. The E. coli Rep helicase is a monomer up to concentrations of 8 mM in the absence of DNA but it forms a dimer upon binding to DNA [5], [6]. The majority of DNA helicases for which the state of protein assembly has been examined appear to form oligomeric structure [5], [6], [8]. Multiple binding sites generally exist within single polypeptide. Oligomerization probably provides a simple mechanism by which helicases acquire multiple nucleic acid-binding sites. Various examples of oligomeric form of helicases are given in Table 2 . Hexameric helicases contain a characteristic ring-shaped structure. Except eukaryotic minichromosomal maintenance (MCM) helicase all the hexameric helicases are homohexamer and mostly involved in DNA replication [8]. Though some hexameric helicases are also involved in recombination and transcription termination [8]. The Rho and DnaB helicases from E. coli were among the first shown to form hexamer. Electron microscopy studies have confirmed that the ssDNA transverses in the centre of the hexameric ring. The hexameric G40P DNA helicase encircles the 5′ tail, interacts with the duplex DNA at the ss/ds-DNA junction, and excludes the 3′ tail of the forked DNA [46]. The DnaB, T4gp41 and T7gp4 helicases encircle ssDNA, whereas SV40 large T antigen and BLV E1 helicases can encircle either ssdNA or dsDNA. Although RuvB helicase has been suggested to encircle dsDNA. Oligomerization of many DNA helicases can be modulated by interactions with other ligands. For example, the E. coli Rep helicase is a monomer up to concentrations of 12 mM in the absence of DNA but it forms a dimer upon binding to DNA [5].
Table 2.
Oligomeric form of DNA helicases
| Helicase | Oligomeric form |
|---|---|
| E. coli helicase IIa | Dimer |
| E. coli helicase III | Dimer |
| E. coli Rep | Dimer |
| HeLa helicase α | Dimer |
| HDH II/Ku | Dimer (heterodimer) |
| Pea chloroplast helicase I | Dimer |
| HSV-1 origin binding protein | Dimer |
| Herpes helicase/primase | Dimer (heterodimer) |
| E. coli DnaB | Hexamer |
| E. coli Rho | Hexamer |
| E. coli RuvB | Hexamer |
| M. thermoautrophicum MCM | Hexamer |
| T7 gene 4 (E. coli phage) | Hexamer |
| T4 gene 41 (E. coli phage) | Hexamer (requires GTP) |
| SSP1 gene 40 (B. subtilis phage) | Hexamer |
| Plasmid encoded RSF1010 RepA | Hexamer |
| SV40 large T antigen | Hexamer |
| Bovine papillomavirus E1 | Hexamer |
| HhcsA | Hexamer |
| MCM | Hexamer |
| Bloom's syndrome helicase | Hexamer |
| E. coli RecB | Oligomer (dimer-tetramer) |
| E. coli UvrAB complex | Oligomer (hetero oligomer) |
| E. coli RecBCD | Oligomer (heterotrimer) |
Note: Examples of monomeric DNA helicases are mentioned in the text.
E. coli Helicase II (UvrD) is also active as a monomer.
5.4. Classification on the basis of polarity of translocation
On the basis of polarity the DNA helicases can be classified as 3′–5′ helicases and 5′–3′ helicases. Many examples of both categories of helicases are shown in Table 3 . Those that are classified, as 3′–5′ helicases require a free ssDNA with a 3′ end adjacent to the double stranded DNA, while those that are called 5′–3′ helicases require an adjacent free single stranded DNA with a 5′ end. Very few DNA helicases have been reported for their bipolar DNA helicase activity, for example, HerA, PcrA (Bacillus), PDH47 and a DNA helicase from Plasmodium falciparum [34], [39]. Most helicases unwind duplex DNA without a sequence preference and for translocation they use phosphodiester backbone of the DNA molecule.
Table 3.
Polarity of biochemically active DNA helicases
| 3′–5′ DNA helicases | 5′–3′ DNA helicases |
|---|---|
| Bacterial: helicase II, helicase IV, PriA, Rep, RecQ, RecB, RecG, PcrA (E. coli), PcrA (Bacillus) | Bacterial: helicase I, helicase III, DnaB, UvrAB complex, RuvAB, Rho, RecD, DinG, PcrA (Bacillus) |
| Bacteriophage: P4geneα | Bacteriophage: T4gene 41, T4 dda, T7 gene 4, SSP1-G40P |
| Viral: SV40 T-antigen, Polyoma T-antigen, HSV-1 (UL9), BPV-1 (E1 protein), AAV (Rep68, Rep 78), AAV-Rep40, Vaccinia virus A18R, MBP-Rep52 (AAV) | Viral: HSV-1 (UL5/UL8/UL52), LEF-3, SARS-CoV helicase (Coronoviridae helicase) |
| Plasmodium: P. cynomolgi DNA helicase, P. falciparum DNA helicase | Plasmodium: P. falciparum DNA helicase |
| Yeast: helicase I, Rad 25 (ssl2), Srs2, Sgs1, MCM, MER3 | Yeast: Rad3, PIF1, RFC associated helicase, helicase A, helicase B, ScHelI, helicase III, Dna2 |
| Frog: Xenopus laevis ovarian DNA helicase I | Frog: Not reported so far |
| Fruitfly: Dhel III, DmRecQ5 | Fruitfly: Dhel I, dhel II |
| Calf thymus: helicase A, helicase E (ε), helicase I, helicase II, cytosolic helicase, Bovine mitochondrial helicase (57 kDa) | Calf thymus: helicase B, helicase C, helicase D, helicase F, helicase δ |
| Human: Human DNA helicase (HDH) I, HDH II/Ku, HDH III, HDH V, HDH VI, TFIIH, [ERCC3 (XPB)], helicase α, helicase ε, RIP100, helicase Q1, WRN, BLM, MCM 4/6/7, REQ1, hFBH1, HEL308, RECQ1. | Human: HDH IV, HDH VIII, ERCC2 (XPD), helicase Q2, HchlR1, HhcsA, BACH1 |
| Mouse/Rat: Rat T1P49a | Mouse/Rat: Mouse ATPase C1, Mouse ATPase B |
| Plant: Pea chloroplast DNA helicase I and II, Pea nuclear DNA helicase (PDH) 45, PDH65, PDH120, PDH47, At-Ku70/80 | Plant: Pea nucleolar DNA helicase (PNDH), PDH47 |
5.5. Classification of helicases on the basis of motifs (superfamilies)
Based on their primary sequences, DNA helicases can be classified into families and SFs. Helicases constitute a large family of enzymes with diverse biological functions. A computer-assisted amino acid sequence analysis of helicases from many different organisms has revealed seven short conserved motifs, called ‘helicase core motifs’ [8], [10], [11], [40], [44]. With this discovery, the helicases have been classified into three SF namely SF1–3 based on the extent of similarity and the organization of these conserved motifs. High sequence conservation has been maintained in this large group of helicases, suggesting that the motifs containing helicases genes evolved from a common ancestor. Hence, these helicase motifs can be used efficiently for the detection and the prediction of new helicases in the genome databases. SF1 and SF2 are the largest and related and contain nine conserved motifs (Q, I, Ia, Ib, II–VI) while SF3 has just three motifs (A, B and C) [35]. Motif III differs between SF1 and SF2 family proteins.
In general the SF1 family members (Rep, UvrD, PcrA-Bacillus, T4-Dda, and HSV-UL5) are considered to be ssDNA translocases while SF2 (eIF4A, RecQ, UvrB, RecG, PriA-E. coli, and HCV-NS3, UL9 and NPH-II) are dsDNA translocases [35], [47]. This property may be the distinction between the SF1 and SF2 family. Eukaryotic eIF4A is a prototype member of SF2. Within SF2, an Snf2 like family of eukaryotic proteins defines a variety of proteins with similarities to yeast protein Snf2 (also called Swi2) [48]. Because of the variations in motif II, the SF2 family of helicases are further subgrouped as DEAD, DEAH, DEXH box proteins. The examples of SF3 family of helicases are RuvB, MCM and some viral encoded proteins [35], [47]. Another group called family 4 (F4) contains five motifs (1, Ia, 2–4) [35]. The Rho helicase is a RNA–DNA helicase and belong to family 5 (F5). The motifs of SF1 and 2 are usually clustered in a region of 200–700 amino acids and can be called as a core-region. The important characteristics of these motifs are described in the following sections.
-
i.
Q Motif: This contains 9 amino acids and the consensus sequence for ‘Q motif’ is GFccPTPIQ, where c is a charged side chain residue [33]. Specific mutations of the Q motif and upstream aromatic group in the translation initiation factor eIF4A in the yeast suggest that conserved residues are important for in vivo viability and for ATPase activity of the purified proteins. It has been shown that this ‘Q motif’ is part of a highly conserved structure, which consists of a loop-helix-loop, which in turn is capable of interactions with motif I and a bound ATP [33].
-
ii.
Motif I: It is present in many nucleotide-binding proteins also and forms a phosphate-binding loop or ‘P loop’ [49]. This motif is also known as ‘A’ motif of ATPase or ‘Walker motif A’. The consensus sequence of this motif for RNA helicases of SF 1 is AxxGxGKT [21]. It is known that the last three residues ‘GKT’ are required for the interaction of the protein with Mg2+ and ATP [50]. The amino group of Lys (K) of GKT interacts with the β and γ phosphates of the ATP molecule [50]. The role of conserved Gly (G) is structural and it helps to maintain the flexible loop conformation [51].
-
iii.
Motif Ia: The consensus sequence for this motif is Pro–Thr–Arg–Glu–Leu–Ala and its secondary structure prediction suggests that it may form an element of the β/α core of a number of proteins of ‘DEAD-box’ family [10]. It has been suggested that this motif is involved in ssDNA binding and it forms the groove for facilitating this binding [52].
-
iv.
Motif II: This motif is also known as ‘B’ motif of ATPase or ‘Walker motif B’ [17]. The consensus sequence for this motif is Asp–Glu–Ala–Asp (DEAD), therefore the proteins containing this motif are also known as ‘DEAD-box’ proteins [11]. The DE of DEAD sequence is highly conserved and is also present in many proteins, which play a role in DNA and RNA replication [21]. The residue D of DE has been shown to interact with Mg2+, which is required for binding ATP. The mutation in D of DE also affects the ATPase and helicase activities. The ATPase motif seems to be responsible for coupling of ATPase and helicase activity [53]. With the growing number of identified helicases, some variations in the motif II has been observed and these are arranged in different subgroups of SF2 such as DEAD, DEAH, DEXH and DEAH* box proteins. The proteins of the DEAH* subgroup show the strongest variations in the conserved regions and are characterized by a DNA helicase activity.
-
v.
Motif III: This motif III of SF2 family plays an important role in the unwinding activity. In UL5 of HSV-I, a motif III mutant exhibited apparent uncoupling of ATPase and DNA helicase activities as the mutant enzyme retained ATPase activity but lacked significant helicase activity [47]. The mutation in this domain (SAT to AAA) leads to loss of RNA helicase activity in eIF-4A while its ATP binding and hydrolysis as well as RNA binding activities were intact [53]. This result suggests that motif III of SF2 is involved in the coupling of ATPase reaction to unwinding. These mutational data collectively suggest the importance of the interactions between the amino acid residues of various domains, which may be responsible for yet unknown mechanism of coupling the NTPase activity with the RNA unwinding. It has also been suggested that the SAT residues might be involved in the transmission of energy derived from ATP hydrolysis to unwind the duplex RNA.
-
vi.
Motif VI: The consensus sequence for this motif is His–x–x–Gly Arg–x–x–R and it is the third most conserved segment in the proteins of this family [21]. This motif has been suggested to be involved in interaction with RNA. It has been proposed that this motif is a part of the ATP-binding cleft and is also involved in coupling between helicase and NTPase activities of the protein. Several helicases exhibited nucleic acid binding defects when the motif VI residues were altered [53]. Point mutation in E. coli UvrD DNA helicase (SF1 family) has a negative impact on a variety of activities including ssDNA-binding, rate of ATP hydrolysis and ligand induced conformational changes. An apparent uncoupling of ATPase and helicase activities was observed with motif VI mutant of HSV1-UL5. Changing the basic residues H or R to the uncharged glutamine abolishes RNA binding and reduces ATP hydrolysis, which also results in reduced helicase activity. All these suggest that motif VI, which mediates conformational changes associated with nucleotide binding by virtue of its close proximity to both the NTP and the DNA-binding site is required for the helicase to move along the DNA substrate. These studies suggest that motif VI mediates ligand-induced conformational changes, which are required for the helicase to move along the nucleic acid substrate.
5.6. Classification of helicases on the basis of associated function
The universal presence of helicases reflects their fundamental importance in DNA metabolism such as replication, repair, recombination and transcription [2], [3], [5], [35], [47]. Therefore on the basis of their associated functions, the DNA helicases can be classified as replicative helicases, repair helicases, recombination helicases and other functions helicases.
6. Human DNA helicases and helicase-related diseases
The very first HDH was purified and well characterized by Tuteja et al. [22]. The name of this helicase was given as HDH I. This helicase was found to be also functioning as RNA helicase and could unwind all kinds of hybrid duplexes (DNA–DNA, RNA–RNA, DNA–RNA, RNA–DNA) [4], [22]. Interestingly, the same protein was also found to be present in plant. By using the anti-HDH I antibodies, a plant homolog of HDH I was purified from pea nuclei [54]. Also by using the same antibodies in pea cell, the protein was found to localized in the dense fibrillar component of the nucleolus, where the rDNA transcription was active [54]. After the first report, now at least a total of 25 different DNA helicases have been isolated from human cells. These all are summarized in Table 4 . Interestingly, the HDH II and HDH IV were identified as human Ku autoantigen and nucleolin, respectively [4].
Table 4.
Human DNA helicases
| S. No. | Name of helicase | Mol. mass (kDa) | Nucleotide cofactors | Remarks |
|---|---|---|---|---|
| 1. | HDH I | 65 | ATP, dATP | First human DNA helicase reported in purified form and also functions as RNA helicase. |
| 2. | HDH II/Ku | 72, 87 | ATP, dATP | DNA end binding protein and involved in dsDNA break repair. |
| 3. | HDH III | 46 | ATP, dATP | Stimulated by fork-like structure of substrates. |
| 4. | HDH IV | 100 | ATP, dATP | Can also function as RNA helicase. |
| 5. | HDH V | 92 | ATP, dATP | Has highest turn over rate. |
| 6. | HDH VI | 128 | ATP, dATP | Stimulated by fork-like structure of substrates |
| 7. | HDH VII | 36 | nd | Trimer of one molecule of hnRNP A1 and two molecules of annexin II |
| 8. | HDH VIII | 68 | ATP | Functions both as DNA and RNA helicase. |
| 9. | HDH IX | 45 | nd | It is a Gly Arg rich protein. |
| 10. | XPD/ERCC2 | 87 | ATP, dATP | Functions in nucleotide excision repair; component of BTF2-TFIIH transcription factor. |
| 11. | XPB/ERCC3 | 89 | ATP | Functions in nucleotide excision repair; component of BTF2-TFIIH transcription factor. |
| 12. | Helicase ε | 72 | ATP, dATP, CTP | Helicase activity is dependent on HRP-A. |
| 13. | Helicase ∝ | 110 | ATP, dATP, | Stimulated by 5′-tailed fork and SSB. |
| 90 | CTP, dCTP | |||
| 14. | RIP 100 | 100 | ATP, dATP | Associated with RIP60 protein. |
| 15. | Helicase Q1 | 73 | ATP, dATP | Gene homologous to E. coli RecQ gene. |
| 16. | Helicase Q2 | 100 | ATP | Identical to DNA helicase IV. |
| 17. | HchlR1 | 112 | ATP | Can unwind RNA/DNA substrates. |
| 18. | HhcsA | 116 | ATP, dATP | Hexameric protein. |
| 19. | WRN helicase | 163 | ATP, dATP> | Mutated in Werner syndrome, homologous to RecQ and contains 3′–5′ exonuclease activity. |
| dCTP, CTP | ||||
| 20. | BLM helicase | ∼160 | ATP | Mutated in cells of Blooms syndrome patient and belongs to RecQ family. |
| 21. | Mcm4/6/7 complex | ∼600 | ATP, dATP | Stimulated by SSB and forked DNA structures; can function as a replication helicase. |
| 22. | HEL308 | 124.5 | ATP, dATP | Stimulated by RPA. |
| 23. | hFBH1 | ∼120 | ATP | First F-box protein with helicase activity. |
| 24 | Human RECQ1 | 75 | ATP | Stimulated by hRPA and can also unwind blunt end substrate with bubble of 25 nt. |
| 25 | BACH1 | 130 | ATP | Involved in dsDNA break repair. |
Because of the action of various mutagens, the DNA can be mutated and these DNA mutations in DNA could be genotoxic or cytotoxic to the cell [55], [56]. The exact DNA repair is also essential to ensure faithful transmission of genetic information from one generation to the next. If not properly repaired the biological consequence could be very harmful, which can lead to many diseases. Since DNA helicases play very important role in repair, therefore the efficient helicase functions is essential. The defect in helicase functions can lead to several human disorders like XP, CS, and Trichothiodystrophy (TTD), Werner's syndrome, Bloom's syndrome and alpha-thalassemia mental retardation on the X chromosome [4], [55], [57]. However, few inherited diseases are associated with altered processing of double-strand breaks such as Ataxia telangiectasia and Nijmegen breakage syndrome [55], [57]. The patients with these diseases can suffer from cancer, premature aging, immunodeficiency and mental retardation.
The XPB and XPD are repair helicases, which are required for local unwinding of the damage area of duplex DNA in NER in mammalian system [55]. Most of the diseases caused by defect in helicase genes are mainly characterized by genome instability, which is the consequence of defect in DNA repair, replication and recombination processes. Table 5 summarizes the helicase-related diseases.
Table 5.
Human diseases related to the mutations in the DNA helicase gene
| S. No. | Helicase gene | Disease | Symptoms |
|---|---|---|---|
| 1. | WRN (RECQL2) | Werner syndrome | Premature aging, scleroderma, cataract, diabetes, hypogonadism |
| 2. | BLM (RECQL3) | Bloom syndrome | Skin sensitive to UV light, dwarfism, immunodeficiency |
| 3. | XPB, XPD | Xeroderma pigmentosum | Light sensitivity, developmental, disabilities, blindness and deafness |
| 4. | XPB, XPD | Cockayne syndrome | Premature aging, light sensitivity, short statue |
| 5. | XPB, XPD | Trichothiodystrophy | Brittle hair |
| 6. | RTS (RECQL4) | Rothmund–Thomson syndrome | Malformations of bone, teeth, nails, skin atrophy |
| 7. | ATRX | X-linked α-thalassemia/mental retardation | Psychomotor retardation, genetial abnormalities, characteristic facial feature |
| 8. | ATRX | Juber–Marsidi syndrome | Mental retardation, growth failure, characteristic facies, growth failure |
| 9. | RAD54 | Various cancers | Symptoms of cancer |
| 10. | BRIP1 (BACH1) | Breast and ovarian cancer | Symptoms of cancer |
| 11. | HAGE | Tumor-specific expression | Symptoms of cancer |
7. Mechanism of DNA unwinding and translocation
Although the detailed molecular mechanism of DNA unwinding by helicases is still not well known. However, it is assumed that certain features of unwinding and translocation must be common to all helicases. The mechanism could be classified either as passive, or active [58]. In the passive mechanism the unwinding protein interacts with ssDNA and translocates unidirectionally. Active mechanisms are described for the oligomeric helicases, which bind both ss- and ds-DNA. There are two models for a general mechanism for helicases, described as an “active rolling” and “inchworm” models (Fig. 2A and B ) [58], [59], [60]. In the “active rolling” model (Fig. 2A), initially both the subunits of the dimer are bound to ssDNA. As a consequence of binding ATP, one of the subunits releases the ssDNA and binds to the duplex region at the fork. This is followed by helix destabilization and the release of one of the DNA strands in a process that accompanies the hydrolysis of ATP [58]. It was proposed that Rep dimer unwinds DNA by an “active rolling” mechanism in which the two subunits alternate in binding duplex DNA and 3′-ssDNA at the ss–dsDNA junction. In this model translocation along ssDNA is coupled to ATP binding, whereas ATP hydrolysis drives the unwinding of multiple DNA base pairs for each catalytic events (Fig. 2A). The Rep protein exists as a stable monomer in the absence of DNA. Once Rep monomer binds to DNA it changes its conformation and forms homodimer and becomes functionally active for translocation and unwinding [61]. It is proposed that ATP hydrolysis alternates between one subunit and the other as an integral part of the translocation and unwinding reactions [62].
Fig. 2.
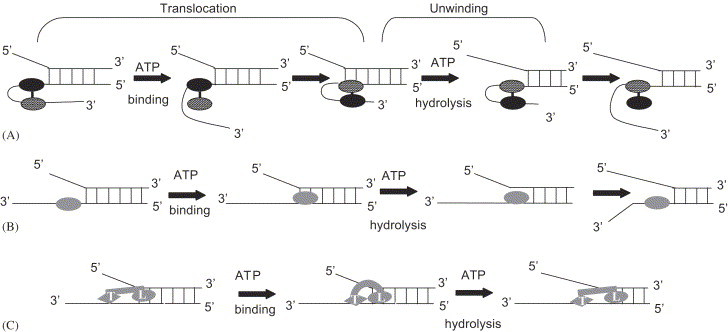
Models for mechanism of DNA helicase unwinding and translocation. The helicase protein contacts and translocates on the sugar-phosphate backbone of the DNA strand and the hydrolysis of ATP is required [47]. A. Active rolling model: The two subunits of dimeric helicase are shown as oval shape in which one is black. The dimeric helicase unwinds by interacting directly with both duplex and ssDNA. Each subunit alternates binding to duplex DNA as the dimer translocates when one subunit releases ssDNA and rebinds to duplex DNA [6]. In this model translocation along ssDNA is coupled to ATP binding, whereas ATP hydrolysis drives the unwinding of multiple DNA base pairs for each catalytic event. B. Inchworm model: This model is consistent with monomeric or oligomeric state for the protein (shown as oval shape). The enzyme monomer first binds to ssDNA and then translocates along the DNA strand and then binds to the duplex region at fork followed by unwinding and release of one of the ssDNA strand [60]. If the enzyme is hexamer as SV40 large T antigen, then one of the subunit (monomer 1) remains associated with the fork through the unwinding cycle [62]. C. Modified inchwarm model: This model is proposed for monomeric UvrD helicase [64]. In this model, it is assumed that the monomeric helicase contain two DNA binding sites: the leading site (L) binds to ds- and ssDNA both and the trailing site (T) binds to only ssDNA. Upon ATP binding the enzyme changes its conformation from extended to compact state, in which the T site is shifted forward along the ssDNA towards the L site. Upon ATP hydrolysis the enzyme changes its conformation from compact to extended state. In the extended state the T site is bound to ssDNA while L site is extended forward in the duplex region and unwinds the DNA. In all the models the shape of the helicase and location of the DNA are arbitrary [47].
For the “inchworm” model (Fig. 2B) the enzyme is bound to ssDNA and then translocates along the DNA strand to the fork region, probably upon binding of ATP. Helix destabilization and release of one of the ssDNA strands takes place as ATP is hydrolyzed [59]. This model is consistent with any monomeric or oligomeric state of the protein. Based on the crystal structure of the PcrA protein [60] a refined model for translocation of the PcrA helicase along ssDNA has been proposed. This model supports the initial “inchworm” model proposed for the Rep helicase and suggests that in the initial stage of translocation cycle, ssDNA is bound to both domains 1A and 2A of the protein. After ATP binds to the complex, the cleft between domains 1A and 2A closes. It has been suggested that domain 1A releases its grip on ssDNA and slides along it, while the domain 2A maintains a tight grip on ssDNA. Upon ATP hydrolysis, the protein returns to its initial conformation, the inter-domain cleft opens and domain 2A translocates along the ssDNA.
The distinction between “inchworm” and “active rolling” models is: one model (inchworm) involves two binding sites, a ss- and ds-DNA binding site either belonging to a single monomeric enzyme or present on two different subunits of a dimeric enzyme while the second model (rolling) involves two subunits of a dimeric enzyme which bind alternately to ss- and ds-DNA. Both mechanisms require the hydrolysis of ATP, but it is not certain at which step this hydrolysis takes place, although ATP binding appears to be associated with an increased affinity of the enzyme for duplex DNA or RNA.
Matson's group has proposed a modified version of inchworm model for DNA unwinding by an active monomeric DNA helicases (Fig. 2C). This model assumes at least two non-equivalent DNA-binding sites on the monomeric protein. The leading site (L) has an affinity for duplex DNA and may also bind ssDNA, whereas the trailing DNA-binding site (T) only has an affinity for ssDNA. A cycle of unwinding begins with the enzyme in an “extended” conformation in which the T site is bound to ssDNA, and the L site is extended forward in the vicinity of the ssDNA/dsDNA junction. Binding of ATP to the enzyme induces a conformational change in the protein to a more compact state in which the T site is shifted forward along the DNA lattice with respect to the L site. Upon ATP hydrolysis a distinct number of base pairs are disrupted at the ssDNA/dsDNA junction, and product release is associated with a return of the enzyme to its original conformation by extension of the L site forward with respect to the T site. At this point, the L site is in close proximity to the new ssDNA/dsDNA junction, and the cycle is repeated to catalyze further unwinding (Fig. 2C) [63]. Bianco and Kowalczykowski [64] have suggested another inchworm model called “quantum inchworm” mechanism for translocation and unwinding of duplex DNA by RecBC DNA helicase. Translocation and unwinding are two separate and consecutive events in the mechanism and are bought about by two different domains (leading [L] and trailing [T]) within the RecBC enzyme. In this model the L domain anchors the enzyme to only one strand of duplex DNA and translocates along it while T domain is responsible for unwinding. During the translocation and unwinding reaction the L domain binds up to 23 nt ahead of the T domain while the T domain uses energy derived from ATP binding and hydrolysis to open the duplex [64], [65].
8. Conclusions and perspectives
The role of helicases in almost every aspect of DNA and RNA metabolisms makes them a very important molecule of the cell, and thereby has several implication of general interest. Despite the diversity of their functions and a large range of organisms in which these proteins have been identified, high sequence conservation has been maintained in the large group of helicases, suggesting that all these helicase genes evolved from a common ancestor. With the completion of genome sequences of a number of organisms, it is interesting to note that each genome contains a large number of putative helicases. Although now many DNA helicases have been isolated from different sources, but the clear-cut role of only helicases has been reported. Still there is a need to find out the exact function of many eukaryotic helicases. Since the basic of DNA metabolism are almost same in eukaryotes from yeast to mammals, the combination of genetics in the yeasts together with in vitro systems and biochemical characterization will help to clarify the in vivo roles of the various DNA helicases. It now appears that oligomeric helicases (mainly hexameric) are often integral components of larger protein complexes, and their role within the complex is to provide molecular motor function. Therefore, it is naïve to think of helicases simply as nucleic acid unwinding enzymes [7]. Though the DEAD-box helicases contain common conserved sequences yet they differ mainly in N-and C-terminal sequences, which contain different targeting signals. Furthermore, many mechanisms of regulation both at the level of expression and at the post-transcriptional level explain the wide spectrum of functions involving DEAD-box helicases.
In the near future, it will be important to identify and characterize specific substrates, interacting proteins, and cofactors of DNA helicases to precisely elucidate their specific functions in nucleic acid metabolism. It will be important to understand the detailed energetics of translocation process and the mechanism of unwinding which is still not well understood. Presently, low-resolution structural analysis, revealed by electron microscopy, is beginning to provide clues to some of the functions and mechanism of action of these remarkable proteins in prokaryotes. A better understanding of these proteins in plant will still have to await the necessary breakthrough that hopefully is provided by the high-resolution structure of a helicase to be solved by X-ray crystallography. We still have to understand the molecular details of how other proteins modulate the activity of helicases or how helicases function within larger macromolecular complexes such as the primosome. The crystal structures of SF1 and SF2 helicases have shown that the DNA helicase motifs are clustered together in the tertiary structure, forming NTP-binding pocket and a portion of the nucleic acid-binding site [35]. According to Hall and Matson, [35] the conserved helicase motifs can be envisioned as the engine of a helicases generating energy by the consumption of fuel (NTPs) and using the energy to do work. The non-conserved portion of helicase structure may contain some specific domain such as protein–protein interaction domain, cellular localization signals, site-specific DNA recognition domains and oligomerization interfaces, which could be unique to individual helicases. Despite the crystal structures of many helicases had been solved still there are many questions that remain unanswered.
Many helicases have now been found to be associated with several human diseases. Therefore, a proper functioning of DNA helicases in vivo is very important. Recently, the viral and other parasitic helicases are attractive targets for drug therapy; therefore, a better understanding of structure and mechanism of these helicases will be helpful in the discovery of a new drug. Very interestingly, a new and important role of a DNA helicase in the salinity stress tolerance in plant suggests a novel pathway to engineer to maximize crop yield in sub-optimal conditions. This discovery will have a great biotechnological application of helicases in the future.
Acknowledgements
This work was partially supported by the grants from Department of Biotechnology, Government of India and Defence Research Development Organization, Government of India.
References
- 1.Soultanas P., Wigley D.B. Trends Biochem. Sci. 2001;26:47. doi: 10.1016/s0968-0004(00)01734-5. [DOI] [PubMed] [Google Scholar]
- 2.Tuteja N., Tuteja R. Eur. J. Biochem. 2004;271:1835. doi: 10.1111/j.1432-1033.2004.04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuteja N. J. Exp. Bot. 2003;54:2201. doi: 10.1093/jxb/erg246. [DOI] [PubMed] [Google Scholar]
- 4.Tuteja N., Tuteja R. Nat. Genet. 1996;13:11. doi: 10.1038/ng0596-11. [DOI] [PubMed] [Google Scholar]
- 5.Matson S.W. Prog. Nucleic Acids Res. Mol. Biol. 1991;40:289. doi: 10.1016/s0079-6603(08)60845-4. [DOI] [PubMed] [Google Scholar]
- 6.Lohman T.M. Mol. Microbiol. 1992;6:5. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 7.West S.C. Cell. 1996;86:177. doi: 10.1016/s0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 8.Patel S.S., Picha K.M. Annu. Rev. Biochem. 2000;69:651. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 9.Caruthers J.M., McKay D.B. Curr. Opin. Struct. Biol. 2002;12:123. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 10.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. FEBS Lett. 1988;235:16. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linder P., Lasko P.F., Ashburner M., Leroy P., Nielsen P.J., Nishi K., Schneir J., Slonimski P.P. Nature. 1989;337:121. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 12.Schaeffer L., Roy R., Humbert S., Moncollin V., Vermeulen W., Hoeijmakers J.H.J., Chambon P., Egly J.-M. Science. 1993;260:58. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 13.Sanan-Mishra N., Pham X.H., Sopory S.K., Tuteja N. Proc. Natl. Acad. Sci. USA. 2005;102:509. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson J.D., Crick F.H.C. Nature. 1953;191:737. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Monem M., Durwald H., Hoffmann-Berling H. Eur. J. Biochem. 1976;65:441. doi: 10.1111/j.1432-1033.1976.tb10359.x. [DOI] [PubMed] [Google Scholar]
- 16.Hotta Y., Stern H. Biochemistry. 1978;17:1872. doi: 10.1021/bi00603a011. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesan M., Silver I.L., Nossal N.G. J. Biol. Chem. 1982;257:12426. [PubMed] [Google Scholar]
- 18.Matson S.W., Tabor S., Richardson C.C. J. Biol. Chem. 1983;258:14017. [PubMed] [Google Scholar]
- 19.Stahl H., Droge P., Knippers R. EMBO J. 1986;5:1939. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodgeman T.C. Nature. 1988;333:22. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 21.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Nucleic Acids Res. 1989;17:4713. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuteja N., Tuteja R., Rahman K., Kang L.-Y., Falaschi A. Nucleic Acids Res. 1990;18:6785. doi: 10.1093/nar/18.23.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umezu K., Nakayama K., Nakayama H. Proc. Natl. Acad. Sci. USA. 1990;87:5363. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hehman G.L., Hauswirth W.W. Proc. Natl. Acad. Sci. USA. 1992;89:8562. doi: 10.1073/pnas.89.18.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuteja N., Tuteja R., Ochem A., Taneja P., Huang N.W., Simoncsits A., Susic S., Rahman K., Marusic L., Chen J., Zhang J., Wang S., Pongor S., Falaschi A. EMBO J. 1994;13:4991. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuteja N., Phan T.N., Tewari K.K. Eur. J. Biochem. 1996;238:54. doi: 10.1111/j.1432-1033.1996.0054q.x. [DOI] [PubMed] [Google Scholar]
- 27.Subramanya H.S., Bird L.E., Brannigan J.A., Wigley D.B. Nature. 1996;384:379. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 28.Ishimi Y. J. Biol. Chem. 1997;272:24508. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 29.Gray M.D., Shen J.C., Kamath-Loeb A.S., Blank A., Sopher B.L., Martin G.M., Oshma J., Loeb L.A. Nature Genet. 1997;17:100. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 30.Karow J.K., Chakraverty R.K., Hickson R.D. J. Biol. Chem. 1997;272:30611. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 31.Pham X.H., Reddy M.K., Ehtesham N.Z., Matta B., Tuteja N. Plant J. 2000;24:219. doi: 10.1046/j.1365-313x.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 32.Tuteja R., Malhotra P., Song P., Tuteja N., Chauhan V.S. Mol. Biol. Parasitol. 2002;124:79. [PubMed] [Google Scholar]
- 33.Tanner N.K., Cordin O., Banroques J., Doère M., Linder P. Mol. Cell. 2003;11:127. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 34.Pradhan A., Chauhan V.S., Tuteja R. Mol. Biochem. Parasitol. 2005;140:55. doi: 10.1016/j.molbiopara.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Hall M.C., Matson S.W. Mol. Microbiol. 1999;34:867. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 36.Dillingham M.S., Spies M., Kowalczykowski S.C. Nature. 2003;423:893. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 37.Naqvi A., Tinsley E., Khan S.A. J. Bacteriol. 2003;185:6633. doi: 10.1128/JB.185.22.6633-6639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Constantinesco F., Forterre P., Koonin E.V., Aravind L., Elie C.A. Nucleic Acids Res. 2004;32:1439. doi: 10.1093/nar/gkh283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vashisht A., Pradhan A., Tuteja R., Tuteja N. Plant J. 2005;44:76. doi: 10.1111/j.1365-313X.2005.02511.x. [DOI] [PubMed] [Google Scholar]
- 40.Gorbalenya A.E., Koonin E.V. Curr. Opin. Struct. Biol. 1993;3:419. [Google Scholar]
- 41.Costa M., Ochem A., Staub A., Falaschi A. Nucleic Acids Res. 1999;27:817. doi: 10.1093/nar/27.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SenGupta D.J., Borowiec J.A. Science. 1992;256:1656. doi: 10.1126/science.256.5064.1656. [DOI] [PubMed] [Google Scholar]
- 43.Zhang S., Grosse F. Biochemistry. 1994;33:3906. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 44.Matson S.W., Bean D.W., George J.W. Bioessays. 1994;16:13. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 45.Tuteja N. Crit. Rev. Plant Sci. 2000;19:449. [Google Scholar]
- 46.Ayora S., Weise F., Mesa P., Stasiak A., Alonso J.C. Nucleic Acids Res. 2002;30:2280. doi: 10.1093/nar/30.11.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuteja N., Tuteja R. Eur. J. Biochem. 2004;271:1849. doi: 10.1111/j.1432-1033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- 48.Laurent B.C., Treitel M.A., Carlson M. Proc. Natl. Acad. Sci. USA. 1991;88:2687. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanner N.K., Linder P. Mol. Cell. 2001;8:251. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 50.Walker J.E., Saraste M., Runswick M.J., Gay N.J. EMBO J. 1982;1:945. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorbalenya A.E., Koonin E.V. Nucleic Acids Res. 1989;17:8413. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Cell. 1999;97:75. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 53.Pause A., Sonenberg N. EMBO J. 1992;11:2643. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuteja N., Beven A.F., Shaw P.J., Tuteja R. Plant J. 2001;25:9. doi: 10.1046/j.1365-313x.2001.00918.x. [DOI] [PubMed] [Google Scholar]
- 55.Tuteja N., Tuteja R. Crit. Rev. Biochem. Mol. Biol. 2001;36:261. doi: 10.1080/20014091074192. [DOI] [PubMed] [Google Scholar]
- 56.Tuteja N., Singh M.B., Misra M.K., Bhalla P.L., Tuteja R. Crit. Rev. Biochem. Mol. Biol. 2001;36:337. doi: 10.1080/20014091074219. [DOI] [PubMed] [Google Scholar]
- 57.Lindahl T., Wood R.D. Science. 1999;286:1897. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 58.Lohman T.M., Bjornson K.P. Annu. Rev. Biochem. 1996;65:169. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 59.Yarranton G.T., Gefter M.L. Proc. Natl. Acad. Sci. USA. 1979;76:1658. doi: 10.1073/pnas.76.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Cell. 1999;97:75. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 61.Delagoutte E., von Hippel P.H. Q. Rev. Biophys. 2002;35:431. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 62.Hsieh J., Moore K.J., Lohman T.M. J. Mol. Biol. 1999;288:255. doi: 10.1006/jmbi.1999.2666. [DOI] [PubMed] [Google Scholar]
- 63.Mechanic L.E., Hall M.C., Matson S.W. J. Biol. Chem. 1999;274:12288. doi: 10.1074/jbc.274.18.12488. [DOI] [PubMed] [Google Scholar]
- 64.Bianco P.R., Kowalczykowski S.C. Nature. 2000;405:368. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- 65.Delagoutte E., von Hippel P.H. Q. Rev. Biophys. 2003;36:1. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]


