Summary
Neandertals were an anatomically distinct hominoid species inhabiting a vast geographical area ranging from Portugal to western Siberia and from northern Europe to the Middle East. The species became extinct 28,000 years ago, coinciding with the arrival of anatomically modern humans (AMHs) in Europe 40,000 years ago. There has been considerable debate surrounding the main causes of the extinction of Neandertals. After at least 200,000 years of successful adaption to the climate, flora and fauna of Eurasia, it is not clear why they suddenly failed to survive. For many years, climate change or competition with anatomically modern human (AMH) have been the leading hypotheses. Recently these hypotheses have somewhat fallen out of favour due to the recognition that Neandertals were a highly developed species with complex social structure, culture and technical skills.
Were AMHs lucky and survived some catastrophe that eradicated the Neandertals? It seems unlikely that this is the case considering the close timing of the arrival of AMHs and the disappearance of Neandertals. Perhaps the arrival of AMHs also brought additional new non-human microscopic inhabitants to the regions where Neandertals lived and these new inhabitants contributed to the disappearance of the species.
We introduce a medical hypothesis that complements other recent explanations for the extinction of Neandertals. After the ancestors of Neandertals left Africa, their immune system adapted gradually to the pathogens in their new Eurasian environment. In contrast, AMHs continued to co-evolve with east African pathogens. More than 200,000 years later, AMHs carried pathogens that would have been alien to pre-historic Europe. First contact between long separated populations can be devastating. Recent European and American history provides evidence for similar events, where introduction of viral, protozoan or bacterial pathogens to immunologically naïve populations lead to mass mortality and local population extinction. We propose that a virus, possibly from the family Herpesviridae, contributed to Neandertal extinction.
Background
Ancestors of the Neandertals spread across Eurasia more than 700,000 years ago evolving into Neandertals 370,000 years ago. AMHs evolved in Africa, becoming anatomically modern by 200,000 years ago, left Africa approximately 100,000 years ago and reached Europe some 40,000 years ago [1], [2], [3], [4], [5].
In Europe, the latter part of the last glaciation, 50,000–12,000 years ago, was characterized by rapid climate changes and frequent cold conditions, leading to a vegetation and faunal turnover. This was true of the entire Quaternary period. Climate change and the expansion of AMHs coincided with the disappearance of the entire Neandertal population between 28,000 years ago [6].
Until recently, the intellectual superiority of AMHs over Neandertals was cited as the major cause of Neandertal extinction. This hypothesis is of questionable merit as recent discoveries show that Neandertals were capable of a behaviour that must be regarded as modern human like [7]. Bearing this in mind, it is of special interest why we are the descendants of AMHs and not of Neandertals. Why was the Neandertal, which was adapted to its environment, driven to extinction while AMHs were able to survive in a novel environment for the species and to which they were not adapted?
Complicating the picture, it is widely accepted that Neandertals were physically stronger than AMHs. They were better adapted to European conditions during the Ice-Age, having lived there for 200,000 years. Modern humans in contrast, had migrated from subtropical environments in North Eastern Africa, and were better suited for warmer conditions. Neandertals were much more competent in using tools and preparing weapons than previously thought [8]. The more we learn about Neandertals, the harder it is to believe that we simply out-smarted them. The Neandertals were indeed extremely successful hunters and were taking down game ranging from large woolly mammoths, to quick and agile deer. Analysis of mtDNA of populations in Asia supports the view that there was only a single dispersal out of Africa by AMHs, and additionally this population has split early into at least one European and one Asian branch although the evidence is not conclusive for this scenario [9]. It is therefore not very likely, assuming similar reproduction rates that AMHs outnumbered and overwhelmed the Neandertals.
It was recently proposed that the Neandertals were unable to adapt their hunting skills to a rapidly changing environment [10]. However, they survived in a harsh and variable environment for more than 200,000 years and concrete evidence is lacking as to why they completely failed to do so in the Late Pleistocene. It was recently shown that a Neandertal population survived at the southernmost point of Europe 28,000 years ago in isolated refuges well after the arrival of AMHs in Europe [11]. At least 15,000 years of co-existence of both AMHs and Neandertals in Europe provide evidence against hypotheses proposing extinction by violent conflicts between the two homo populations.
Even if we accept that it was mainly chance and not inherent superiority of the AMHs that decided upon the fate of the Neandertals, something lead to the death of many thousands individuals throughout Europe. Such an explanation would also be helpful in elucidating why a putative assimilation of Neandertals into early AMH populations was apparently modest [12].
Alternative and/or additional hypotheses have been presented to explain the extinction of Neandertals. Diamond has suggested that either violent conflicts or a putative pathogen might have lead to a decrease of the Neandertal population [13]. Recently, Stormer and Mysterud described two different mechanisms by which long term exposure to smoke pollutants in caves and rock shelters might have contributed to Neandertal extinction. Open burning releases large amounts of air pollutants, including dioxins, that can induce short term and long term effects on human health, especially in children [14]. Underdown suggested a contributory role for Transmissible Spongiform Encephalopathies (TSEs) that could have infected Neandertal groups as a result of general cannibalistic activity and brain tissue consumption in particular [15].
The hypothesis
The evolutionary origin of several viral pathogens correlates with the genetic and paleontological data of first contact and near first contact of AMHs and Neandertals. We propose that AMHs introduced an infectious agent, deriving from Eastern Africa, into the population of the Neandertals. We would expect Neandertals to be extremely vulnerable to most introduced diseases. First, in Africa, AMHs and their ancestors had gone through intense selection and co-evolution with pathogens in their sub-tropical environment. The survivors who withstood those particular diseases emigrated to Europe. We believe that Neandertals would have encountered fewer human pathogens in their evolutionary history, at least during the past 200,000 years before AMH arrival in Europe. And those infectious diseases that might have been present in Europe at that time, would then have been of a very different nature than those in Africa. Why should have Neandertals experienced fewer plagues and infectious diseases before the arrival of AMHs? The main reason is that most diseases affecting humans are ultimately zoonotic diseases that originated in animals. For a zoonotic pathogen it is normally much easier to breach the species barrier if the species are closely related. Therefore African primates could have served as a reservoir or transmitter of pathogens for developing humans. Examples for this can also be observed in recent history (HIV jumped from primates to humans some 70–100 years ago). Even zoonoses from other species would be facilitated in Africa by the greater species diversity of rodents, bats and insects that serve as reservoirs for many viruses and other pathogens (reviewed in Refs. [16], [17], [18]). This environmental background may have led to a higher genetic resistance against infectious diseases in AMHs compared to Neandertals. The third reason why Neandertals were more vulnerable is they were descended from a very small group and long-term effective population size of Neandertals was smaller than that of modern humans and extant great apes at the time of AMH arrival. Recent studies including mitochondrial DNA analysis propose that it was as low as 10,000 individuals [19]. There is also evidence that the genetic variability of the Neandertals was very low. The mtDNA genetic diversity in Neandertals between 38,000 and 70,000 years ago was estimated to be one-third of that in contemporary modern humans [19] and, more importantly, much lower than that of the AMHs who more recently descended from the largest human gene pool in Africa. Greater diversity of the immune system increases the number of pathogens that can be recognized. Given the overall lack of genetic diversity of Neandertals, they likely lacked immune gene variability as well putting them at a higher risk of developing disease relative to AMHs.
These aspects taken together support the hypothesis that Neandertals could have been more susceptible to infectious diseases in general and especially to newly introduced ones. What characteristics must a pathogen fulfil, at least theoretically, to be a threat of the type we envisage here? We speculate that such a pathogen may have exhibited the following characteristics:
-
(1)
The pathogen was abundant in Africa, and AMHs frequently and easily came into contact with it before leaving Africa for Europe.
-
(2)
The infectious agent did not depend on a vector for transmission, such as arthropods. Such vectors would not have been able to co-migrate with the AMHs from Africa to glacial Europe.
-
(3)
The infectious agent was circulating among AMHs during the migration from Africa to Europe, without being eliminated from the population, i.e., persistent or chronic infection developed in AMHs.
-
(4)
AMHs adapted to the infectious agent in a way that did not strongly affect the health of the entire population, although some individuals may have developed a lethal infection.
-
(5)
The infectious agent could be transmitted by social contact.
-
(6)
The pathogen appeared, or at least continued to evolve, in Africa after Neandertal ancestors had left the continent.
Protozoan infections, such as malaria, sleeping sickness, chagas and others almost always require vectors for transmission. Bacterial infections rarely develop into both chronic but still highly contagious infections. Fungal infections only rarely lead to lethal infections in mammals. In contrast many viral infections fulfil all the above requirements. These points lead us to hypothesize that viral infections may have been carried by AMHs as they left Africa, and that these infections may have spread to other hominid species they encountered in Eurasia—Homo neanderthalensis in particular.
We speculate that a virus from the order Herpesvirales and the family Herpesviridae could have been that contagious agent. Other viruses are also possible (see Table 1 ) though we feel herpesviruses may have some properties that make them more likely candidates. We further speculate that the agent could have been a herpesvirus belonging to the subfamily Alphaherpesvirinae and the genus Varicellovirus, such as present-days Varizella-Zoster virus (Human Herpesvirus 3).
Table 1.
Overview over viral pathogens: likelihood ranking: whether a viral pathogen is a (+): very likely candidate, (o) possible but not very likely candidate, or a (−) highly unlikely candidate for contributing to Neandertal extinction. Requires vector “yes”, means that humans do not normally transmit the disease to other humans, and other vectors (e.g., arthropods, mammals) are required for this. y.a., years ago.
| Virus family or subfamily | Representative(s) (common names) | Emergence of family/clades in human hosts | Transmission | Persistence (humans) | Pathogenicity (humans) | Requires vector? | Genome | Likelihood ranking |
|---|---|---|---|---|---|---|---|---|
| Poxviridae | Smallpox | ∼16,000 y.a.? [41] | Social sontact | No | Very high | No | dsDNA | − |
| Herpesviridae | Herpes simplex, VZV, CMV, EBV | Millions of y.a., 100,000 y.a. | Airborn (droplet) | Yes | Low–high | No | dsDNA | + |
| Adenoviridae | Adenovirus A-F | Co-evolved with vertebrates [42] | Fecal-oral | No | Low–high | No | dsDNA | − |
| Papillomaviridae | HPV | Co-evolved with primates/humans [43] | Social contact, sexual contact | Yes | Low–medium | No | dsDNA | o |
| Polyomaviridae | BKV, JCV, SV-40 | Out of Africa 100,000 y.a., twice? [29] | Droplets, urine, fecal-oral | Yes | Low–medium | No | dsDNA | + |
| Retroviridae | HIV, HTLV | Millions of y.a.–hundreds y.a. (HIV) [44] | Bites, sexual contact | Yes | Low–medium | No | ssRNA− | o |
| RT | ||||||||
| Hepadnaviridae | HBV | Co-speciated with primates/humans [27], [28] | Bites, sexual contact | Yes | Low–medium | No | dsDNA− | o |
| RT | ||||||||
| Paramyxoviridae | Measles, RSV | Co-evolved in animals, in humans not more than 4–5000 y.a. [45] | Airborn droplets | No | Medium–high (infants) | No | ssRNA− | − |
| Orthomyxoviridae | Influenza A | Emerged in birds, human Influenza several thousand y.a. [46], [47] | Airborn droplets | No | Low–high | No | ssRNA− | − |
| Bunyaviridae | Rift-Valley Fever, Hanta | Co-evolved with arthropods (RVF) and rodents (hanta), in humans ∼15,000 y.a. [48] | Bites, saliva, urine | No | Medium–high | Yes | ssRNA− | − |
| Arenaviridae | Lassa Fever | Co-evolved with rodents, in humans ∼15,000 y.a. [49] | Saliva, fecal-oral, bites, blood, sexual contact | No | Low–very high | No | ssRNA− | − |
| Filoviridae | Ebola, Marburg | Origin unclear, Ebola and Marburg diverged 7–8000 y.a. [50] | Droplets, bites, sexual contact, blood | No | Low–very high | No | ssRNA− | − |
| Reoviridae | Rotavirus | Close relationship between animal and human rotaviruses [51] | Fecal-oral | No | High (infants) | No | dsRNA | − |
| Flaviviridae | Yellow fever, dengue, West-Nile fever | Co-evolved with primates/humans [27], [28], [52], dengue 1000 y.a. [53] | Arthropod bites, blood | No | Low–high | Yes | ssRNA+ | − |
| Coronaviridae | SARS | SARS in 2002 [54], other coronavirus ∼1941 [55] | Droplets, fecal-oral | No | Low–high | No | ssRNA+ | − |
| Togaviridae | Rubella, chikungunya, RRV | Of old world origin [56], ancestor of chikungunya emerged 300–1300 y.a. [57], origin of rubella unknown | Airborn droplets, insect bites | No | Low–high | Yes/no | ssRNA+ | o |
| Picornaviridae | Polio | Co-evolved with primates and humans? [58], [59] | Fecal-oral | No | Low–medium | No | ssRNA+ | − |
| Parvoviridae | Parvovirus B19 | Evolution linked to host species [60], [61], not much known about human parvoviruses [62], [63] | Airborn droplets, blood | Yes | Low–high (pregnancy) | No | ssDNA | o |
Varizella-Zoster virus (VZV), the most important human pathogenic representative of the Varicelloviruses, is the causative agent of varicella (chickenpox) and herpes zoster (shingles). VZV is able to persistently infect humans over the entire lifespan. Primary infection with Varizella-Zoster virus can lead to chickenpox. Chickenpox is extremely contagious as over 90% of unvaccinated people become infected [20], while herpes zoster occurs only in individuals who have had a primary VZV infection. Today, most cases of herpes zoster occur in individuals who are more than 45 years old.
In addition to the role played by advancing age, immunosuppression in general dramatically increases the risk for development of herpes zoster. Herpes zoster is common in patients treated with immunosuppressive drugs for malignant diseases or to prevent rejection of bone marrow or organ transplants [21]. Patients who are immunocompromised have earlier and increased manifestations of herpes zoster infection than those who are not immunocompromised [22]. Disseminated disease, with a mortality rate of more than 40%, is a complication observed in immunosuppressed patients. Herpes zoster is also the earliest opportunistic infection observed in patients positive for human immunodeficiency virus (HIV).
In serious cases of herpes zoster, severe neurological complications occur, including encephalitis, meningitis, myelitis and herpes zoster ophthalmicus. Even after aggressive management, patients may have long-term sequelae, including vision loss [22]. Therefore, even if primarily lethal infections with VZV were rare in Neandertals, long term effects such as neurological disorders may have severely compromised the infected individual and if sufficient numbers were infected whole populations could have suffered.
VZV is an example that we chose because we found that it fulfilled all requirements of a possibly harmful Neandertal pathogen. Other herpesviruses, e.g., Simplexvirus, Cytomegalovirus, Roseolovirus, Lymphocryptovirus, Rhadinovirus with the most important representatives Herpes simplex 1 and 2, Human Cytomegalovirus, Human Herpesvirus 6, Epstein-Barr virus and Human Herpesvirus 8 (HHV-8) are also candidates. Viruses from other orders, such as polyomaviruses, hepadnaviruses, retroviruses, togaviruses or parvoviruses could also be candidates, although for various reasons each of these seems less likely than herpesviruses. For example, retroviruses are often transmitted by sexual intercourse or biting, but judging from the genetic evidence so far available these activities are unlikely to have occurred at a high enough frequency between human and Neandertals to establish the virus in the Neandertal population. Other viruses exclude themselves from the pool of candidates by either not supporting a persistent infection (e.g., rubella virus), by requiring a vector (e.g., West-Nile virus and Yellow fever) or by simply arriving too late (e.g., dengue virus and Influenza A) to be relevant to Neandertals (see Table 1).
Evaluation of the hypothesis
Can VZV, or a virus in general, fulfil the requirements for a pathogen that could be successfully introduced by AMHs to Neandertal population in Europe? There are several characteristics and properties of viruses that support our hypothesis.
-
(1)
Today we know that the majority of emerging viral diseases are transmitted from animals to humans. Many of them have developed in tropical and sub-tropical regions of the world and crossed the species barrier when humans came in close contact with wild or domesticated animals.
-
(2)
There are a number of examples of chronic and recurring viral infections in humans that reach a very high seroprevalence in susceptible populations. Infections caused by retroviruses, herpesviruses, hepadnaviruses and many others are able to develop into persistent infections lasting for years.
-
(3)
In immunocompetent humans many viral infections are not lethal, especially if a population has had enough time to adapt or to co-evolve with that pathogen. After a more or less pronounced acute phase after primary infection, symptoms often decline. Examples are HIV infections, hepatitis B and C and Herpes simplex.
-
(4)
If a viral infection is introduced into an immunologically naïve population that has not encountered the virus for generations, the outcome may be dramatic. The introduction of measles and small pox into the Americas by Europeans resulted in millions of deaths in Native American populations.
-
(5)
Even if many viruses in fact depend on vectors, a large number of viruses can be transmitted by direct human-to-human contact as the viruses are often contained in body fluids and excrement. Smallpox, chickenpox, kissing disease and SARS are examples of viruses which can infect by means of direct human to human transmission.
-
(6)
For many viruses, it has been shown that their genetic subtypes are scattered over the entire globe with no strong local or regional pattern. This means that they were already circulating in the human population that left Africa 100,000 years ago.
We investigated whether the evolutionary history of AMHs and the Neandertals matches the time and place of origin and evolution of some of the viral pathogens that could have played a role in Neandertal extinction. Molecular analysis of herpesvirus genomes indicates that herpesviruses emerged in parallel with primate and human evolution, probably many millions of years ago [23], [24]. As far as VZV is concerned, it is likely that differentiation of VZV clades initiated at the time of AMH dispersal out of Africa around 100,000 years ago [25] (Fig. 1, Fig. 2 ). There is evidence that the same is true for members of the herpesvirus subfamily Gammaherpesvirinae, such as HHV-8 and Epstein-Barr virus [26].
Fig. 1.

Simplified routes of Neandertals, AMHs and viral pathogens out of Africa.
Fig. 2.
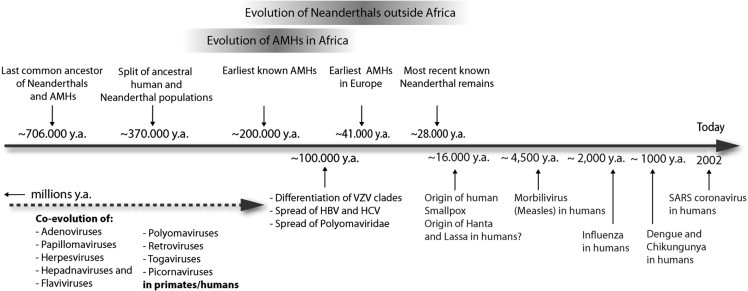
Timeline (lower figure part): AMH and Neandertal evolution (modified from Refs. [40], [5]) in relationship to virus evolution. Bar and arrow lengths are schematic and not to scale. y.a., years ago.
For hepatitis C virus (HCV), hepatitis B virus (HBV) and polyomaviruses it was also suggested that it was already present in the human population when AMHs started to spread around the world 100,000–150,000 years ago [27], [28], [29]. In contrast, smallpox virus, dengue virus, chikungunya virus and morbillivirus are believed to have originated after the development of farming in the Middle East around 10,000–16,000 years ago or even later in history [28] being far to late to have infected Neandertals.
Among the herpesviruses, VZV displays a unique survival strategy [30]. The mode of VZV transmission in isolated populations over extended periods of time has been demonstrated on several occasions, e.g., small tribes in the Amazon basin in Brazil that had little or no contact with neighbouring peoples but nevertheless tribe members were seropositive for past VZV infections [31]. Another example comes from the Shetland Islands, off the Scottish coast. In the early 1900s the island inhabitants lived in small villages. Each island population was separated from the next island. Even in such isolated communities, several cases of varicella in children followed a single case of shingles in an adult [30]. An efficient mode of transmission in isolated societies depends upon reactivation of the virus as zoster in adults and its subsequent transmission to children as varicella. The group of AMHs that left Africa to (re)-colonize Europe were at least a partially isolated group on their way north.
It remains unclear whether AMHs and Neandertals came in close contact with each other and if so, how frequent and intensive this contact was [3]. Also a certain degree of contact between individual Neandertal groups would be required to further spread the pathogen within the entire European Neandertal population. In support of our hypothesis, it may be noted that Neandertals did not disappear immediately after the arrival of AMHs in Europe. They coexisted for at least 15,000 years, providing a time for the spread of a virus from an AMH group to a Neandertal group and between different Neandertal groups. Favouring co-existence, the last known localities of Neandertal habitation were in Spain including a cave in Gibraltar. Humans appear in the fossil record 28,000 years ago and subsequently Neandertals disappeared. However, this was 15,000 years after AMHs entered Europe and thus, contact between the two groups across Europe is highly likely during that interval.
There are currently two major approaches to test our hypothesis. The approaches rely on recent advances in extracting and sequencing DNA from ancient Neandertal samples. First, it might be possible to detect viral sequences in DNA samples extracted form bones if marrow is present. If present, short viral sequences of 60–80 bp could be easily and unambiguously detected and distinguished from Neandertal DNA. At present, no viruses or other pathogens without a DNA genome would be detected, as RNA does not survive thousands of years in ancient samples. Naturally, to enable detection, the corresponding pathogen, or its DNA, should be present in skeletal samples per se, and not merely represented in the soil or environment. Herpesviruses have a genome of double-stranded DNA and therefore it should be possible to look for specific virus sequences in ancient DNA. The VZV genome consists of approximately 125,000 bp of DNA with more than 70 open reading frames (ORFs) and is arranged in long and short unique segments with terminal repeat regions [21]. In principle it should be possible to find unique sequence elements in the VZV DNA that could be used for assay development.
Alternatively, sequences may be identifiable in the genome sequence trace files generated from whole-genome sequencing projects. Microbial sequences tend to be ignored or discarded as contaminants from ancient DNA metagenomic studies though there are exceptions [32]. A targeted search for herpesvirus sequences could be attempted from relevant genomic data, with the caveat that, much like exogenous contamination, modern human herpesvirus contamination could confound analysis.
As far as the presence of distinct herpes virus in bones is concerned, there is currently not enough data to determine if a virus inhabiting spinal nerves would be detectable by PCR in a given fossil bone. Unfortunately, direct detection of RNA-viruses such as HCV is not possible with current methods, as RNA is not preserved in most ancient samples. However, advances in protein sequencing could potentially allow for the sequencing of short viral peptides that could at least identify the pathogen. However, this has not been tested yet in a fossil context for viruses. Biomolecular methods have been used for the first time to show a correlation between extinction and disease in mammals, although in that study, skin was available for study [33]. This is at least a proof of principle for pathogen detection in ancient samples, permitting empirical correlation of pathogen presence and subsequent extinction. Nevertheless future developments in molecular analysis and discoveries of better preserved fossils may help to detect even small amounts of genetic material that would have been overlooked a few years ago.
A second line of investigation is currently feasible. An analysis of known immunologically relevant genes of Neandertals, and a subsequent comparison with the corresponding genes from AMHs, could be performed. At least one fully sequenced Neandertal genome would be necessary. It is well known that there are projects ongoing to achieve exactly this [34], [35], [36]. As a starting point, chromosome 6 could be analyzed. It contains the family of major histocompatibility complex (MHC) genes and harbours the most gene-dense region of the entire human genome. The MHC region on chromosome 6 contains approximately 140 genes and many of these have a known function in the immune system or in autoimmunity. An analysis of the allelic diversity of the MHC class I complex, for example could yield a wealth of information in respect to susceptibility of Neandertals to a variety of diseases. However, to fully understand the degree of allelic diversity of MHC genes one needs the chromosome 6 sequence of a large group of Neandertal individuals with both a geographical and temporal sampling protocol that would allow one to examine MHC diversity over space and time. Fixation of specific alleles in the Neandertal during the overlap with the arrival of AMHs could indicate selection on the MHC by a pathogen.
Consequences of the hypothesis and discussion
The hypothesis that a pathogen introduced by AMHs later played a role in the extinction of a different species (in this case, Neandertals) is not novel. The Hyperdisease Hypothesis of MacPhee and Marx [37] developed this scenario to explain megafaunal extinctions during the Pleistocene–Holocene transition. On the other hand, a disease-mediated extinction hypothesis for Neandertals that has some possibility of empirical testing has not been presented previously. Earlier reports concerning possible rickets infections of Neandertals did not aim at explaining their extinction, but rather tried to explain Neandertal bone morphology [38]. A possible reason disease has not been considered is that it was a long held belief that Neandertals were not culturally sophisticated and were therefore displaced by more advanced AMHs. It is now clear that lack of cultural sophistication was certainly not applicable to Neandertals.
We present a concrete hypothesis that on the one hand makes predictions about the social and biological interactions between AMHs and Neandertals and on the other hand can, in principle, be tested. Additionally, our hypothesis could theoretically be extended to extinction events affecting other relatives of Homo sapiens, such as H. floresiensis [39] just as the Hyperdisease Hypothesis for megafaunal extinctions can be extended to a wide variety of unrelated megafaunal species.
Conclusion
The hypothesis presented here is not the only possible explanation for Neandetal extinction but adds an additional consideration to the debate that should be taken into account. It could contribute to understanding the puzzling extinction of Neandertals. Climate change and direct conflict with modern humans could clearly have contributed or caused Neandertal extinction without pathogen involvement. However, removing pathogens from the list of possible suspects has proven wrong in the past.
Conflict of interest statement
None declared.
Acknowledgement
The authors thank Ross MacPhee for helpful discussions and improvements to the manuscript.
References
- 1.Stringer C.B., Andrews P. Genetic and fossil evidence for the origin of modern humans. Science. 1988;239:1263–1268. doi: 10.1126/science.3125610. [DOI] [PubMed] [Google Scholar]
- 2.Green R.E., Malaspinas A.S., Krause J. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrera K.J., Somarelli J.A., Lowery R.K., Herrera R.J. To what extent did Neanderthals and modern humans interact? Biol Rev Camb Philos Soc. 2009;84:245–257. doi: 10.1111/j.1469-185x.2008.00071.x. [DOI] [PubMed] [Google Scholar]
- 4.Krings M., Stone A., Schmitz R.W., Krainitzki H., Stoneking M., Paabo S. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 5.Hublin J.J. Out of Africa: modern human origins special feature: the origin of Neandertals. Proc Natl Acad Sci USA. 2009;106:16022–16027. doi: 10.1073/pnas.0904119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzedakis P.C., Hughen K.A., Cacho I., Harvati K. Placing late Neanderthals in a climatic context. Nature. 2007;449:206–208. doi: 10.1038/nature06117. [DOI] [PubMed] [Google Scholar]
- 7.Balter M. Paleoanthropology. Dressed for success: Neandertal culture wins respect. Science. 2004;306:40–41. doi: 10.1126/science.306.5693.40. [DOI] [PubMed] [Google Scholar]
- 8.David F., D’Iatchenko V., Enloe J.G. New Neandertal remains from the Grotte du Bison at Arcy-sur-Cure, France. J Hum Evol. 2009;57:805–809. doi: 10.1016/j.jhevol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Macaulay V., Hill C., Achilli A. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 10.Finlayson C., Carrion J.S. Rapid ecological turnover and its impact on Neanderthal and other human populations. Trends Ecol Evol. 2007;22:213–222. doi: 10.1016/j.tree.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Finlayson C., Pacheco F.G., Rodriguez-Vidal J. Late survival of Neanderthals at the southernmost extreme of Europe. Nature. 2006;443:850–853. doi: 10.1038/nature05195. [DOI] [PubMed] [Google Scholar]
- 12.Trinkaus E. European early modern humans and the fate of the Neandertals. Proc Natl Acad Sci USA. 2007;104:7367–7372. doi: 10.1073/pnas.0702214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond JM. The third chimpanzee: the evolution and future of the human animal harper perennial; 2006.
- 14.Stormer F.C., Mysterud I. Cave smoke: air pollution poisoning involved in Neanderthal extinction? Med Hypotheses. 2007;68:723–724. doi: 10.1016/j.mehy.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Underdown S. A potential role for transmissible spongiform encephalopathies in Neanderthal extinction. Med Hypotheses. 2008;71:4–7. doi: 10.1016/j.mehy.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Wong S., Lau S., Woo P., Yuen K.Y. Bats as a continuing source of emerging infections in humans. Rev Med Virol. 2007;17:67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolle M.A. Mosquito-borne diseases. Curr Probl Pediatr Adolesc Health Care. 2009;39:97–140. doi: 10.1016/j.cppeds.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Mahy B.W. Zoonoses and haemorrhagic fever. Dev Biol Stand. 1998;93:31–36. [PubMed] [Google Scholar]
- 19.Briggs A.W., Good J.M., Green R.E. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science. 2009;325:318–321. doi: 10.1126/science.1174462. [DOI] [PubMed] [Google Scholar]
- 20.Swingler GH. Chickenpox. Clin Evid 2007;2007 [online].
- 21.Arvin A.M. Varicella-zoster virus. Clin Microbiol Rev. 1996;9:361–381. doi: 10.1128/cmr.9.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver B.A. Herpes zoster overview: natural history and incidence. J Am Osteopath Assoc. 2009;109:S2–6. [PubMed] [Google Scholar]
- 23.McGeoch D.J., Cook S. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- 24.McGeoch D.J., Cook S., Dolan A., Jamieson F.E., Telford E.A. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 25.Muir W.B., Nichols R., Breuer J. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J Virol. 2002;76:1971–1979. doi: 10.1128/JVI.76.4.1971-1979.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGeoch D.J. Molecular evolution of the gamma-herpesvirinae. Philos Trans R Soc Lond B Biol Sci. 2001;356:421–435. doi: 10.1098/rstb.2000.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmonds P. Reconstructing the origins of human hepatitis viruses. Philos Trans R Soc Lond B Biol Sci. 2001;356:1013–1026. doi: 10.1098/rstb.2001.0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82:693–712. doi: 10.1099/0022-1317-82-4-693. [DOI] [PubMed] [Google Scholar]
- 29.Pavesi A. Utility of JC polyomavirus in tracing the pattern of human migrations dating to prehistoric times. J Gen Virol. 2005;86:1315–1326. doi: 10.1099/vir.0.80650-0. [DOI] [PubMed] [Google Scholar]
- 30.Grose C. Varicella zoster virus: out of Africa and into the research laboratory. Herpes. 2006;13:32–36. [PubMed] [Google Scholar]
- 31.Schuette M.C. A qualitative analysis of a model for the transmission of varicella-zoster virus. Math Biosci. 2003;182:113–126. doi: 10.1016/s0025-5564(02)00219-5. [DOI] [PubMed] [Google Scholar]
- 32.Zhao F., Qi J., Schuster S.C. Tracking the past: interspersed repeats in an extinct Afrotherian mammal, Mammuthus primigenius. Genome Res. 2009;19:1384–1392. doi: 10.1101/gr.091363.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyatt K.B., Campos P.F., Gilbert M.T. Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS One. 2008;3:e3602. doi: 10.1371/journal.pone.0003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briggs A.W., Stenzel U., Johnson P.L. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci USA. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green R.E., Briggs A.W., Krause J. The Neandertal genome and ancient DNA authenticity. EMBO J. 2009;28:2494–2502. doi: 10.1038/emboj.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green R.E., Krause J., Ptak S.E. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 37.MacPhee R.D., Marx P.A. The 40,000-year plague: humans, hyperdisease, and first-contact extincations. In: Goodman S.M., Patterson B.D., editors. Natural change and human impact in madagascar. Smithsonian Institution Press; Washington, London: 1997. pp. 169–217. [Google Scholar]
- 38.Wright D.J. Syphilis and Neanderthal man. Nature. 1971;229:409. doi: 10.1038/229409a0. [DOI] [PubMed] [Google Scholar]
- 39.Morwood M.J., Jungers W.L. Conclusions: implications of the Liang Bua excavations for hominin evolution and biogeography. J Hum Evol. 2009;57:640–648. doi: 10.1016/j.jhevol.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Noonan J.P., Coop G., Kudaravalli S. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Carroll D.S., Gardner S.N., Walsh M.C., Vitalis E.A., Damon I.K. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci USA. 2007;104:15787–15792. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benko M., Harrach B. Molecular evolution of adenoviruses. Curr Top Microbiol Immunol. 2003;272:3–35. doi: 10.1007/978-3-662-05597-7_1. [DOI] [PubMed] [Google Scholar]
- 43.Bernard H.-U. Genome diversity and evolution of papillomaviruses. In: Domingo E., Parrish C.R., Holland J.J., editors. Origin and evolution of viruses. Academic Press; London: 2008. pp. 417–430. [Google Scholar]
- 44.Woodman Z., Williamson C. HIV molecular epidemiology: transmission and adaptation to human populations. Curr Opin HIV AIDS. 2009;4:247–252. doi: 10.1097/COH.0b013e32832c0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norrby E., Kovamees J., Blixenkrone-Moller M., Sharma B., Orvell C. Humanized animal viruses with special reference to the primate adaptation of morbillivirus. Vet Microbiol. 1992;33:275–286. doi: 10.1016/0378-1135(92)90055-x. [DOI] [PubMed] [Google Scholar]
- 46.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki Y., Nei M. Origin and evolution of influenza virus hemagglutinin genes. Mol Biol Evol. 2002;19:501–509. doi: 10.1093/oxfordjournals.molbev.a004105. [DOI] [PubMed] [Google Scholar]
- 48.Zhao X., Hay J. The evolution of hantaviruses. Immunol Invest. 1997;26:191–197. doi: 10.3109/08820139709048926. [DOI] [PubMed] [Google Scholar]
- 49.Hugot J.P., Gonzalez J.P., Denys C. Evolution of the Old World Arenaviridae and their rodent hosts: generalized host-transfer or association by descent? Infect Genet Evol. 2001;1:13–20. doi: 10.1016/s1567-1348(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki Y., Gojobori T. The origin and evolution of Ebola and Marburg viruses. Mol Biol Evol. 1997;14:800–806. doi: 10.1093/oxfordjournals.molbev.a025820. [DOI] [PubMed] [Google Scholar]
- 51.Cunliffe N.A., Woods P.A., Leite J.P. Sequence analysis of NSP4 gene of human rotavirus allows classification into two main genetic groups. J Med Virol. 1997;53:41–50. [PubMed] [Google Scholar]
- 52.Smith D.B., Pathirana S., Davidson F. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78(Pt 2):321–328. doi: 10.1099/0022-1317-78-2-321. [DOI] [PubMed] [Google Scholar]
- 53.Holmes E.C., Twiddy S.S. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol. 2003;3:19–28. doi: 10.1016/s1567-1348(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 54.Lu H., Zhao Y., Zhang J. Date of origin of the SARS coronavirus strains. BMC Infect Dis. 2004;4:3. doi: 10.1186/1471-2334-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez C.M., Gebauer F., Sune C., Mendez A., Dopazo J., Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190:92–105. doi: 10.1016/0042-6822(92)91195-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavergne A., de Thoisy B., Lacoste V. Mayaro virus: complete nucleotide sequence and phylogenetic relationships with other alphaviruses. Virus Res. 2006;117:283–290. doi: 10.1016/j.virusres.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 57.Powers A.M., Brault A.C., Tesh R.B., Weaver S.C. Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol. 2000;81:471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 58.Koonin E.V., Wolf Y.I., Nagasaki K., Dolja V.V. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 59.Gromeier M., Wimmer E., Gorbalenya A.E. Genetics, pathogenesis and evolution of picornaviruses. In: Domingo E., Parrish C.R., Holland J.J., editors. Origin and evolution of viruses. Academic Press; London: 1999. pp. 287–343. [Google Scholar]
- 60.Hoelzer K., Parrish C.R. Evolution and variation of the parvoviruses. In: Domingo E., Parrish C.R., Holland J.J., editors. Origin and evolution of viruses. Academic Press; London: 2008. pp. 393–416. [Google Scholar]
- 61.Shadan F.F., Villarreal L.P. The evolution of small DNA viruses of eukaryotes: past and present considerations. Virus Genes. 1995;11:239–257. doi: 10.1007/BF01728663. [DOI] [PubMed] [Google Scholar]
- 62.Pattison J.R. B19 virus – a pathogenic human parvovirus. Blood Rev. 1987;1:58–64. doi: 10.1016/0268-960x(87)90020-8. [DOI] [PubMed] [Google Scholar]
- 63.Tijssen P. Molecular and structural basis of the evolution of parvovirus tropism. Acta Vet Hung. 1999;47:379–394. doi: 10.1556/AVet.47.1999.3.11. [DOI] [PubMed] [Google Scholar]


