Abstract
Bovine interferon-omega3 (BoIFN-ω3) gene was amplified from bovine liver genomic DNA, which encodes a 195-amino acid protein containing a 23-amino acid signal peptide. Analysis of the molecular characteristics revealed that BoIFN-ω3 evolving from IFN-ω, contained four cysteine residues and five alpha helices, showing that BoIFN-ω3 presented the typical molecular characteristics of type I interferon. BoIFN-ω3 exhibited antiviral and antiproliferative activities, which exerted a protective effect against VSV in several mammalian cell lines, as well as against BEV, IBRV, and BVDV in MDBK cell. Moreover, BoIFN-ω3 was shown to be highly sensitive to trypsin, but remaining stable despite changes in pH and temperature. Additionally, BoIFN-ω3 induced the transcription of Mx1, ISG15, and ISG56 genes, as well as the expression of Mx1 protein in a time-dependent manner. These findings will be useful to further study BoIFN-ω in host's defence against infectious diseases, particularly viral infections. Furthermore, results will facilitate further research on the bovine interferon family.
Keywords: Bovine, IFN-ω3, Molecular characteristic, Antiviral activity, Interferon-stimulated genes
Highlights
-
•
BoIFN-ω3 presents antiviral activity on several mammalian cell lines and protective effect against VSV, BEV, IBRV, and BVDV.
-
•
BoIFN-ω3 exhibits antiproliferative activity and insensitivity to pH and temperature.
-
•
BoIFN-ω3 can activate the transcription of ISGs gene, as well as the expression of Mx1 in a time-dependent manner.
1. Introduction
Interferons (IFNs), which are produced in response to viral infections, contribute to host defence by establishing an antiviral state in target cells wherein viral replication is blocked or impaired as a result of the synthesis of a number of enzymes interfering with cellular and viral processes (Staeheli, 1990, Sen, 2001). Type I IFNs are a family of cytokines with pleiotropic activities including inhibition of viral replication and cell proliferation, as well as activation of the immune system (Stark et al., 1998). Their administration has been proposed as an immunomodulatory therapy (Domenech et al., 2011) to treat several viral and immunomediated diseases. Type I IFNs, including IFN-α, IFN-β, IFN-ω, IFN-δ, IFN-τ, IFN-ε, IFN-ν, and IFN-κ, are produced by virus-infected cells and exert nonspecific antiviral activities on adjacent non-infected cells (Krause and Pestka, 2005). They display a general mechanism of action based on their interaction with specific cell-surface receptors and the subsequent induction of IFN-stimulated genes (ISGs) expression, thereby encoding direct antiviral effectors or molecules with the potential to positively and negatively regulate IFN signaling and other host responses, including enzymes, signaling proteins, chemokines, antigen-presenting proteins, transcription factors, and apoptotic proteins (Der et al., 1998). Overall, a number of ISGs that act to enhance pathogen detection and innate immune signaling (Schneider et al., 2014), such as ISG15, ISG56, and GTPase Mx1, have been shown to function as antiviral effectors. The Mx1 gene product, which is one of the first described inhibitors of virus entry, is broadly inhibitory and acts prior to genome replication at an early postentry step of the virus life cycle (Schneider et al., 2014). ISG15 demonstrates numerous antiviral functions, including inhibition of virus release, lysis of both viral and host proteins, and immunomodulatory cytokine-like properties in its unconjugated form (Schoggins, 2014). ISG56, which is induced in response to type I IFNs, dsRNAs, and viruses (Terenzi et al., 2006), has been implicated in antiviral actions of IFNs against West Nile virus and lymphocytic choriomeningitis virus (Wacher et al., 2007).
IFN-ω, which was first discovered by Hauptmann and Swetly (Hauptmann and Swetly, 1985), is a type I IFN secreted by virus-infected leukocytes, that are encoded by multiple IFN- or IFN-like genes present across mammalian groups, including cats, porcine and rabbit (Hauptmann and Swetly, 1985, Roberts et al., 1998, Charlier et al., 1993a, Mege et al., 1991). Like other type I IFNs, IFN-ω presents species-restricted biological activity in vitro and can bind to the same type I IFN receptor complex as other type I IFNs, thus exerting similar antiviral, antiproliferative, and immunomodulatory effects (Adolf, 1995). However, its antigenic structure is distantly related to IFN-α, -β, and -γ, as it does not cross react with antibodies against them (Adolf, 1995). Interferon-ω has been used for antiviral treatment in humans and many other mammalian species (Tiefenthaler et al., 1997, Hagelstein et al., 1998, Gil et al., 2014, Leal et al., 2014, Martin et al., 2002). Although human IFN-ω (HuIFN-ω) can exert in vivo antitumor effects in several models of human cancer (Horton et al., 1999), Feline IFN-ω (FeIFN-ω) shows a proven antiviral effect, both in vitro (Mochizuki et al., 1994, Ohe et al., 2008, Litzlbauer et al., 2014) and in vivo (Gil et al., 2014, Leal et al., 2014, Martin et al., 2002, Horton et al., 1999, Mochizuki et al., 1994, Ohe et al., 2008, Litzlbauer et al., 2014), against canine and feline parvovirus, herpesvirus, calicivirus, coronavirus, and rotavirus, and has been licensed for use in veterinary medicine (Virbagen®, Virbac) in Europe, Japan, Australia, New Zealand and Mexico (Domenech et al., 2011). Research on BoIFN-ω subtypes was not much reported, in 1998, BoIFN-ω1 was reported to have biological activation, and can sufficiently prevent luteolysis, without being deleterious to embryonic survival (Rodriguez et al., 1998a); in 2015, BoIFN-ω24 has been characterized and can be used as a candidate antiviral therapeutic reagent (Luo et al., 2015). The newfound BoIFN-ω3, which presents a potential as a novel antiviral therapeutic agent, exerts antiviral activity against VSV in several mammalian cell lines and a protective effect against bovine virus that including BEV, IBRV and BVDV. In this study, we present a novel type I IFN named BoIFN-ω3 on the basis of the location in the bovine genome. BoIFN-ω3 demonstrates typical characteristics of type I IFN, exerts antiviral activity in several mammalian cell lines, and displays low cytotoxicity in vitro. Additionally, BoIFN-ω3 can induce the transcription of Mx1, ISG15, and ISG56 genes, as well as the expression of Mx1 protein in a time-dependent manner. The current findings will be useful to further study BoIFN-ω in host's defence against infectious diseases, particularly viral infections, and should facilitate research on the bovine interferon family.
2. Materials and methods
2.1. Cells, viruses and antibody
Liver was collected from Holstein cows in a dairy farm in Harbin, Heilongjiang in Northeast China. Madin–Darby bovine kidney (MDBK) cells, primary bovine testicular (BT) cells, primary embryo bovine lung (BL) cells, feline kidney (F81) cells, Madin-Darby Canine Kidney (MDCK) cells, baby hamster Syrian kidney (BHK-21) cells, porcine kidney (PK-15) cells and rabbit kidney (RK-13) cells were preserved in our laboratory. Vesicular stomatitis virus (VSV) was purchased from the China Institute of Veterinary Drug Control. Bovine intestinal virus (BEV), bovine infectious bovine rhinotracheitis virus (IBRV), bovine viral diarrhea virus (BVDV) were preserved in our laboratory. Rabbit polyclonal antibody (PAb) against BoIFN-ω24 were prepared and preserved in our laboratory (Luo et al., 2015). Rabbit PAb against Mx1 (GTX110256) and GAPDH (GTX100118) were purchased from GeneTex (CA, USA). HRP-conjugated goat anti-rabbit IgG was purchased from ZSGB (Beijing, China).
2.2. Clone of BoIFN-ω3
Genomic DNA extracted from bovine liver (Sambrook and Green, 2012) was used as the PCR template. Degenerate primers BoIFNWS and BoIFNWA (Table 1 ) designed according to the alignment of BoIFN-ω subtypes, were used as the PCR primers with the following thermal profile: initial denaturation at 94 °C for 5 min, 30 amplification cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 40 s, followed by a final extension at 72 °C for 10 min. The PCR products obtained were cloned into the pMD18-T cloning vector (TaKaRa, Japana) and sequenced.
Table 1.
Sequences of the primers.
| Primers | Sequences(5′–3′) |
|---|---|
| BoIFNWS | GATCCCTGGGCTGTGACYTGTCT |
| BoIFNWA | CTTCTCTTKCAGGTAGAYATGGAT |
| BoIFN-ω3S | TCGGGATCCTGTGACCTGTCTCAGAACCA |
| BoIFN-ω3A | AGGCTCGAGTCAAGGTGAGTTCAGATCTC |
| BoGAPDHS | TTCAACGGCACAGTCAAGG |
| BoGAPDHA | ACATACTCAGCACCAGCATCAC |
| BoMx-1S | TCAACCTCCACCGAACTG |
| BoMx-1A | TCTTCTTCTGCCTCCTTCTC |
| BoISG15S | GCAGCCAACCAGTGTCTG |
| BoISG15A | CCTAGCATCTTCACCGTCAG |
| BoISG56S | TGGACTGTGAGGAAGGATGG |
| BoISG56A | AGGCGATAGACAACGATTGC |
The restriction enzyme sites that were introduced in primers are underlined.
2.3. Sequence analysis of BoIFN-ω3
Sequences analysis was performed on the DNAStar program (DNASTAR Inc., USA). The obtained sequences were identified by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the putative amino acid sequence was compared with its counterparts of other animals by using the program ClustalX. Signal peptide was predicted with SignalP (http://www.cbs.dtu.dk/services/SignalP/). The glycosylation sites were analyzed by NetGlycate 1.0 Server (http://www.cbs.dtu.dk/services/NetGlycate/) online. Multiple alignments and phylogenetic tree were constructed with ClustalX and MEGA5.0 using UPGMA method, and the availability of the branch stem was verified with 500 bootstrap replicates. Secondary structure elements were predicted using the algorithms available from NPS (http://www.npsa-pbil.ibcp.fr).
2.4. Expression and purification of recombinant BoIFN-ω3
pET32a (Invitrogen, CA, USA) was used as the expression vector. Express primers BoIFN-ω3S and BoIFN-ω3A (Table 1) containing the BamH I and Xho I sites were designed. Mature BoIFN-ω3 gene was cloned into the pET-32a vector to generate pET32a-BoIFN-ω3, which contains an N-terminal His tag for facilitating protein purification. Recombinant plasmid was transformed into Escherichia coli Rosetta (DE3) Lys cells and then recombinant protein His-BoIFN-ω3 was induced with IPTG (Sigma-Aldrich, St. Louis, MO), the recombinant His-BoIFN-ω3 was analyzed through 12% reduced SDS-PAGE. The whole cells were collected and treated with ultrasonic, supernatants and sedimentations were isolated by centrifuge and analyzed by 12% reduced SDS-PAGE, which were used to identify the solubility of His-BoIFN-ω3. Then the recombinant protein His-BoIFN-ω3 was purified using a nickel-chelated column (GenScript, Nanjing, China) according to the manufacturer's instructions. The purified protein was renatured through dialysis with TGE (50 mM Tris–HCl, 0.5 mM EDTA, 50 mM NaCl, 5% glycerinum, pH 8.0) under a urea gradient to reduce the concentration from 6 M to 0 M. After denaturation and renaturation, soluble homogeneous protein was obtained and concentration was quantified with the Bradford Protein Assay kit (Beyotime, Shanghai China) according to the instructions.
2.5. Antiviral activity and antibody blocking assay in vitro
First, titers of viruses described above were determined by an endpoint dilution assay and the titers were expressed as the tissue culture infectious dose 50 (TCID50) per milliliter using the Reed-Muench method (Guo et al., 2015). Virus titers were calculated by determining the dilution giving 50% of wells containing cells that displayed cytopathic effect. Antiviral activity was determined with a standard cytopathic effect assay (Rubinstein et al., 1981) with some modifications (Boue et al., 2000, Bracklein et al., 2006, Rodriguez et al., 1998b, Shao et al., 2015a). Briefly, the monolayers cells were seeded in 96 well plates, then inoculated with four-fold serial diluted BoIFNs. After overnight cultivation, 100 TCID50 viruses was added to each well and the plate was then re-incubated under the conditions of 37 °C in a humidified 5% CO2 atmosphere for 18–24 h. Eight wells without the viruses were used as the cell controls, and the other eight wells without BoIFNs were used as the virus controls. One antiviral activity unit (1 U) was defined as a 50% reduction in the virus induced destruction of the cell monolayer. Based on the antiviral activity measured on MDBK cells, 100 U represents 117 ng, 32.2 ng and 168 ng of BoIFN-ω3, BoIFN-ω24 and BoIFN-αA respectively.
To check whether this antiviral activity is specific, the neutralization antiviral activity of BoIFN-ω3 was measured according to the method as previously described (Shao et al., 2015b) with some modifications. MDBK cells were treated with BoIFN-ω3 (100 U) for 24 h and 2-fold serial dilutions of anti-BoIFN-ω24 rabbit serum for 2 h at 37 °C. Then, all cells were challenged with 100 TCID50 of VSV. The preimmune rabbit serum was used as negative control.
2.6. Antiproliferative assay
The antiproliferative activity of BoIFN-ω3 against MDBK cells was determined by MTT assay in vitro (Luo et al., 2015) with some modification. The monolayers of MDBK cells were seeded in 96-well plates and treated with BoIFN-ω3, BoIFN-ω24 or BoIFN-αA at 37 °C for 72 h. Then, 10 μL of MTT (5 mg/mL) was added to each well, and the plates were further incubated at 37 °C for 4 h. The culture medium was removed, and 150 μL of DMSO was added to each well followed by agitation of the plate on an orbital shaker for 15 min. Light absorbance of the reaction wells was measured at 490 nm.
2.7. Primary physicochemical characteristics of BoIFN-ω3
2.7.1. Trypsin sensitivity assay of BoIFN-ω3
BoIFN-ω3 was combined with trypsin at a final concentration of 0.25% and incubated in the water bath at 37 °C for 1 h. Then antiviral activity of BoIFN-ω3 in the MDBK/VSV system was determined. The samples without treatment were used as control (Shao et al., 2015b).
2.7.2. Stability assay of BoIFN-ω3 at pH 2
BoIFN-ω3 were adjusted to pH 2.0 using HCl and incubated at 4 °C for 24 h, and then the treated samples were reverted to the original pH (7.0) (Cheng et al., 2006, Zhao et al., 2009). Antiviral activity was determined in the MDBK/VSV system. The untreated samples were used as control.
2.7.3. Temperature sensitivity assay of BoIFN-ω3
BoIFN-ω3 were separately incubated in water bath at 42 °C, 56 °C, and 63 °C for 2 h, then the treated samples were cooled down in an icebox. Antiviral activity was determined in the MDBK/VSV system. The untreated samples were used as control.
2.8. BoIFN-ω3-induced the transcription of Mx1, ISG15 and ISG56
MDBK cells were seeded in six-well plates and treated with BoIFN-ω3 for 0, 3, 6, 9, 12, and 24 h. Subsequently, the cells total RNA was prepared using RNAprep pure Cell/Bacteria Kit (TIANGEN, Beijing, China), and then reverse transcribed into cDNA using a reverse transcription system (Promega, USA). Quantitative real-time PCR (qRT-PCR) was carried out by using an ABI 7500 Real-Time PCR system with SYBR Premix Ex Taq (Takara). The PCR was set up under the following thermal cycling conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s. The qRT-PCR was performed with the following primers: BoGAPDHS and BoGAPDHA, BoMx-1S and BoMx-1A, BoISG15S and BoISG15A, BoISG56S and BoISG56A (Table 1). GAPDH was used as the internal reference, the transcription of Mx1, ISG15 and ISG56 mRNA were analyzed with the method 2− ΔΔCT (Livak and Schmittgen, 2001).
2.9. BoIFN-ω3-induced the expression of Mx1
MDBK cells were seeded in six-well plates and treated with BoIFN-ω3 for 0, 3, 6, 9, 12, and 24 h. Subsequently, cells were lysed in Mammalian Protein Extraction Reagent (Thermo, MA, USA). Lysates were collected and separated by SDS-PAGE, and then transferred onto Biotrace PVDF membrane followed by blocking the membrane with 5% skimmed milk in PBST at 4 °C for 12 h. After incubating the membrane with PAb against Mx-1 (1:5000 dilution) and GAPDH (1:8000 dilution) simultaneously at 4 °C for 12 h, the membrane was washed three times with PBST before incubation with goat anti-rabbit IgG antibodies at 1:10,000 dilution in PBST at 37 °C for 1 h. After three times washing with PBST, the membrane was developed with Western blot ECL PLUS (Beyotime, Beijing, China) in the dark.
2.10. Statistical analysis
Data were presented as means ± standard deviations. Student's t-test was used for statistical comparisons. Statistical significance was expressed as the following P values: P ≤ 0.05 (*); P ≤ 0.01 (**).
3. Results
3.1. Sequence analysis of BoIFN-ω3
The complete BoIFN-ω3 included an open reading frame of 588 nucleotides, which encoded a mature peptide consisting of 172 amino acids and a signal peptide consisting of 23 amino acids. A 588 bp mature BoIFN-ω3 gene was then amplified (Fig. 1A). BoIFN-ω3 contained four cysteine residues at positions 1, 29, 99, and 139 and one arginine (Arg) at position 161 (Fig. 1B), which is the conserved sequence among IFN-ω. The three-dimensional structure of BoIFN-ω3 included five alpha helices labeled A to E (Fig. 1C). Only one putative O-glycosylation site was found at amino acid residue 140 of mature BoIFN-ω3.
Fig. 1.
Sequence cloning, analysis and three-dimensional prediction of the BoIFN-ω3. (A) PCR identification of mature BoIFN-ω3 gene. Lane M: Trans 2 K DNA marker; Lane 1: mature BoIFN-ω3 gene. (B) The sequence underlined with a wavy line is the signal sequence, while the arrow denotes the signal sequence cleavage site. Cysteine residues forming disulfide bonds are marked with gray circles. The conserved Arg residues (at position 161 of mature BoIFN-ω3) are marked with a gray box. Letters A–E in red refer to α-helices in BoIFN-ω3. (C) Predicted three-dimensional structures of BoIFN-ω3. The five α-helices were labeled A–E.
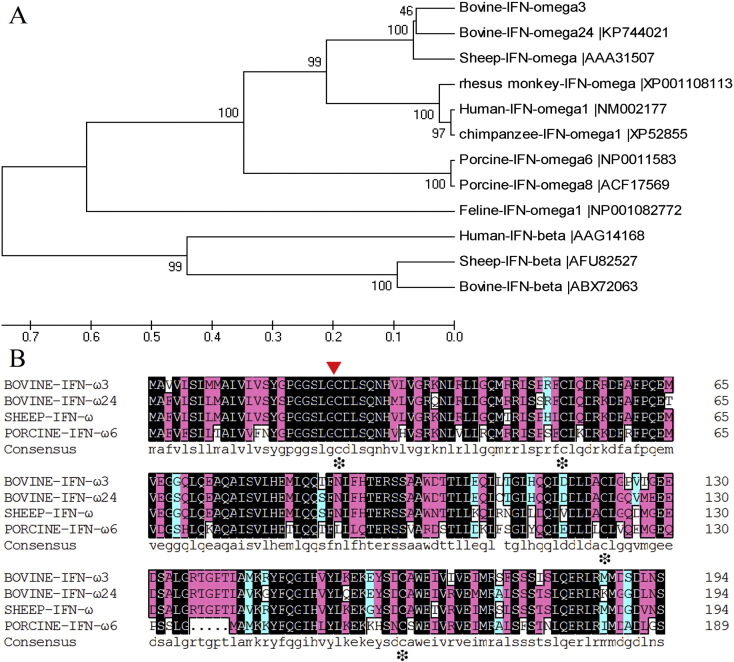
A phylogenetic tree was constructed to determine the evolutionary position of BoIFN-ω3. Subsequently the phylogenetic relationships among bovine, sheep, human, chimpanzee, rhesus monkey, porcine, and feline IFN-ω types showed that BoIFN-ω3 was clustered more closely with sheep IFN-ω (Fig. 2A). Moreover, the amino acid alignment of BoIFN-ω3 with other IFNs showed that BoIFN-ω3 64.10% identical to HuIFN-ω, 87.18% identical to BoIFN-ω24, 68.21% identical to PoIFN-ω6, 51.79% identical to BoIFN-α1, and 28.72% identical to BoIFN-β (Fig. 2B).
Fig. 2.
Phylogenetic tree construction of the BoIFN-ω3 and IFN-ω amino acid alignment. (A) The phylogenetic tree was made with MEGA 5.0 using the UPGMA method, while the availability of the branch stem was verified with the Bootstrap using 500 parameters. The scale bar represents the genetic distance. The credibility value for each node is shown. (B) IFN-ω amino acid alignment of bovine, sheep, and porcine. The arrow denotes the signal sequence cleavage site. Identical residues are in black boxes, 75%-conserved residues are in pink boxes, and semi-conserved residues were in light blue boxes. White boxes are amino acids that differ between the sequences. The conserved cysteine residues are marked with (*).
3.2. Expression and identification of BoIFN-ω3
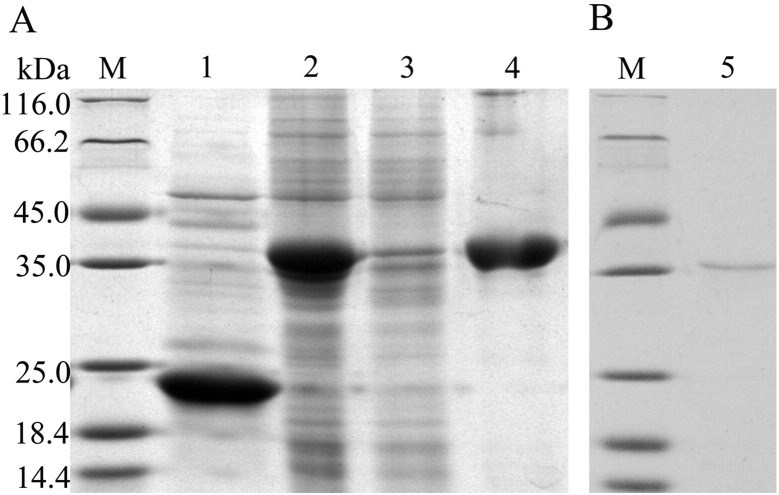
SDS-PAGE analysis revealed that the recombinant protein His-BoIFN-ω3 was mainly expressed as an inclusion body exhibiting an apparent molecular weight of 36 kDa (Fig. 3A). A purified protein His-BoIFN-ω3 band was then observed after purification (Fig. 3B).
Fig. 3.
Reduced SDS-PAGE analysis of the expressed and purified BoIFN-ω3 protein. Lane M: unstained protein marker; Lane 1: pET-32a (+) vector after induction; Lane 2: His-BoIFN-ω3 after induction; Lane 3: supernatants of His-BoIFN-ω3; Lane 4: sedimentations of His-BoIFN-ω3; and Lane 5: purified His-BoIFN-ω3.
3.3. Antiviral activity and antibody blocking assay in vitro
BoIFN-ω3 exerted a protective effect against VSV infection in MDBK, BL, BT, BHK21, F81, RK-13, and PK-15 cells, but not in MDCK cells, while also exerting a protective effect against BEV, IBRV, and BVDV (Table 2 ). Unlike BoIFN-αA, BoIFN-ω3 exerted a protective effect in MDCK-VSV and BHK21-VSV systems. Additionally, the antiviral activity of BoIFN-ω3 measured by the MDBK-VSV system was completely abrogated by PAb against BoIFN-ω24 at a dilution of 1:8. When, the antibody dilution was 1:32, the blocking activity was almost non-existent, whereas no blocking activity was observed with the addition of pre-immune rabbit serum (data not shown).
Table 2.
Comparison of antiviral activity among BoIFN-ω3, BoIFN-ω24 and BoIFN-αA.
| Measurement system | Antiviral activity (U/mg) |
||
|---|---|---|---|
| BoIFN-ω3 | BoIFN-ω24 | BoIFN-αA | |
| MDBK-VSV | 8.56 × 105 | 3.10 × 106 | 5.95 × 105 |
| BL-VSV | 2.18 × 107 | 3.86 × 106 | 7.70 × 104 |
| BT-VSV | 1.38 × 106 | 6.19 × 105 | 1.94 × 105 |
| F81-VSV | 3.41 × 105 | 2.45 × 105 | 0 |
| PK-15-VSV | 2.39 × 104 | 3.17 × 105 | 1.72 × 105 |
| RK-13-VSV | 4.75 × 105 | 3.87 × 105 | 7.70 × 104 |
| MDCK-VSV | 0 | 0 | 0 |
| BHK-21-VSV | 2.21 × 105 | 0 | 0 |
| MDBK-BEV | 6.83 × 105 | 1.02 × 106 | 5.04 × 106 |
| MDBK-IBRV | 2.74 × 105 | 3.87 × 105 | 1.26 × 106 |
| MDBK-BVDV | 1.71 × 104 | 2.22 × 105 | 6.30 × 105 |
3.4. Analysis of antiproliferative activity
MTT assay showed that BoIFN-ω3, BoIFN-ω24 and BoIFN-αA exerted dose-dependent antiproliferative effects in MDBK cells (Fig. 4 ). The antiproliferative activity of BoIFN-ω3 and BoIFN-ω24 was significantly different form that of BoIFN-αA at a concentration of 40 μg/mL (P ≤ 0.05) but was only slightly different when the concentrations were lower than 4 ng/mL. Moreover, the antiproliferative activity analysis showed that BoIFN-ω3 exerted low cytotoxicity in MDBK cells.
Fig. 4.
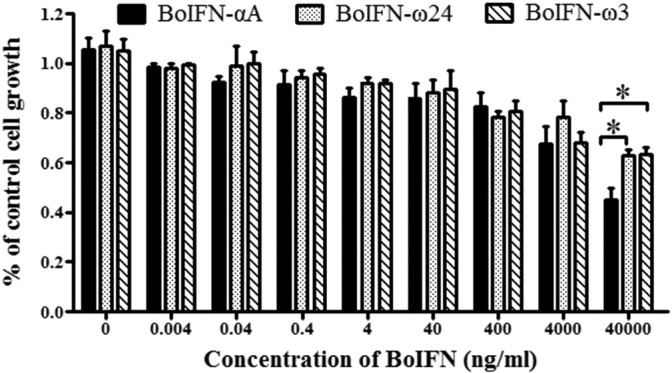
Antiproliferative activities of BoIFN-ω3, BoIFN-ω24 and BoIFN-αA in MDBK cells by an MTT assay. BoIFN-ω3, BoIFN-ω24 and BoIFN-αA were incubated in MDBK cells for 72 h, and antiproliferative activities were analyzed. Relative proliferation was determined as a percentage of the control (0 μg/mL), whereas BoIFN-αA and BoIFN-ω24 were used as the reference. The Y-axis represents the mean of triplicate experiments for percentage of the control cell growth. Antiproliferative activities of both BoIFN-ω3 and BoIFN-ω24 were significantly different from that of BoIFN-αA at concentrations of 40 μg/mL (P ≤ 0.05). Data are presented in mean ± SD (*, P ≤ 0.05).
3.5. Primary physicochemical characteristics of BoIFN-ω3
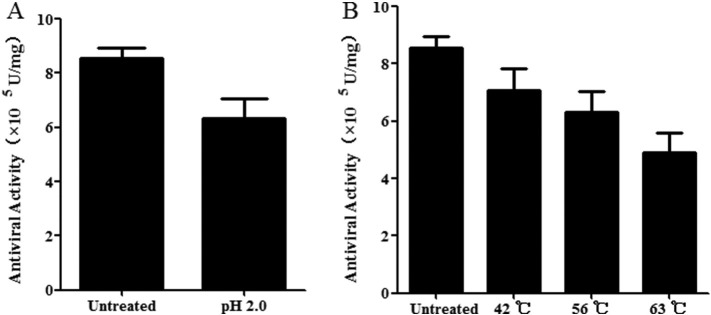
BoIFN-ω3 completely lost its antiviral activity when treated with 0.25% trypsin, and this finding demonstrated that BoIFN-ω3 was highly sensitive to trypsin. After acid treatment, BoIFN-ω3 retained its antiviral activity which was not significantly different from that of the untreated sample; this result confirmed that BoIFN-ω3 was characteristically stable at pH 2 (Fig. 5A). After treatment at 42 °C, 56 °C, and 63 °C, the antiviral activities of BoIFN-ω3 decreased by 0.17, 0.26, and 0.42, respectively, with no significant difference from that of the control group. This finding illustrated that BoIFN-ω3 was not sensitive to temperature (Fig. 5B).
Fig. 5.
(A) pH and (B) temperature sensitivities of BoIFN-ω3. The purified BoIFN-ω3 was adjusted to pH 2.0, 42 °C, 56 °C, 63 °C and then finally reverted to the original pH (7.0) or cooled down in an icebox. Afterwards, antiviral activities were separately analyzed in MDBK-VSV system. After acid and high temperature treatment, BoIFN-ω3 retained its antiviral activity with no significant difference from that of the control group.
3.6. BoIFN-ω3-induced the production of ISGs
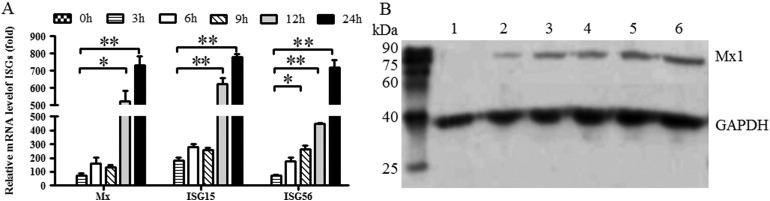
To assess the effect of BoIFN-ω3 on type I IFN response, the transcription of ISGs (including Mx-1, ISG15 and ISG56) was quantified. Quantitative real-time PCR analysis showed that the transcription of Mx-1, ISG15 and ISG56 increased sharply at 12 h in BoIFN-ω3-treated MDBK cells and sustained high expression levels until 24 h, and the expression of 24 h was significantly (P ≤ 0.01) than that of 3 h (Fig. 6A).
Fig. 6.
Analysis of ISGs transcription and Mx-1 protein expression in MDBK cells after BoIFN-ω3 treatment. (A) Transcription analysis of ISGs in MDBK cells after BoIFN-ω3 treatment. Quantitative real-time RT-PCR was used to detect the changes in Mx1, ISG15 and ISG56 transcription levels. MDBK cells without BoIFN-ω3 treatment were used as control, while GAPDH was used as the internal reference. Data are presented in mean ± SD (*, P ≤ 0.05; **, P ≤ 0.01). (B) Expression analysis of Mx-1 protein in MDBK cells after BoIFN-ω3 treatment. MDBK cells without BoIFN-ω3 treatment were used as control, while GAPDH was used as the internal reference. Lane M: EasySee Western marker; Lane 1: Control cells; and Lane 2–6: cell lysates collected at 3, 6, 9, 12, and 24 h after treatment with BoIFN-ω3.
Immunoblotting analysis showed that the Mx-1 protein was detected as a single major protein band of approximately 78 kDa, whereas the amounts of Mx-1 protein gradually increased from 3 to 24 h after treatment. In addition, Mx-1 protein expression was much higher at 24 h than at 3 h (Fig. 6B). GAPDH was used as an endogenous control, and a 36 kDa protein band was detected in all the samples.
4. Discussion
IFNs are a family of regulatory proteins with antiviral, antitumour, and multiple immunomodulatory effects. Bovine type I IFN contains 10–12 IFN-α, 5–6 IFN-β, 3–5 IFN-τ, and 15–24 IFN-ω subtypes (including pseudogenes) (Velan et al., 1985, Chaplin et al., 1996, Capon et al., 1985), which are located on chromosome 8 (Ryan and Womack, 1993). Since the discovery of IFN-ω by Hauptmann and Swetly, IFN-ω has been gradually found in human, feline, equine, porcine, rabbit, and several cattle species (Sambrook and Green, 2012, Himmler et al., 1986, Charlier et al., 1993b, Yang et al., 2007). With fluorescence in situ hybridization technology, bovine IFN-ω was found to locate on chromosome 8q15 (Sen, 2001). In this study, a novel bovine IFN-ω was cloned from bovine liver, which was named as BoIFN-ω3 on the basis of its position in the bovine genome.
The molecular characteristics of BoIFN-ω3 were analyzed using bioinformatics software. BoIFN-ω3 displayed the highest similarity to bovine IFN-ω24 at 87.18% identity. Phylogenetic analysis revealed that BoIFN-ω3 was closely related to BoIFN-ω24 and sheep IFN-ω, suggesting that the IFN-ω gene may have originated from gene duplication in a common ancestor during evolution. Cys-1, Cys-99, Cys-29 and Cys-139 of mature BoIFN-ω3 formed two disulfide bonds crucial for its activity, which is conserved in some type I IFNs (Wetzel, 1981).
To explore the biological activities of BoIFN-ω3, antiviral and antiproliferative activities were examined with the recombinant protein His-BoIFN-ω3. The results indicated that BoIFN-ω3 exerted a considerable antiviral activity against VSV infection in MDBK, BL, BT, BHK21, F81, RK-13, and PK-15 cells, but not in MDCK cells, whereas BoIFN-αA, BoIFN-ε, and BoIFN-ω24 demonstrated no antiviral activity against VSV infection in BHK-21 cells (Sambrook and Green, 2012, Bracklein et al., 2006). BoIFN-ω3 also exerted a protective effect against BEV, IBRV, and BVDV in MDBK cells. Similar to BoIFN-ω24, BoIFN-ω3 displayed low cytotoxicity in MDBK cells. In addition, BoIFN-ω3 was verified to be highly sensitive to trypsin, insensitive to temperature, and stable at pH 2. Given the substantial antiviral activity and low cytotoxicity of BoIFN-ω3, it is a potential candidate for a novel, effective therapeutic agent.
ISGs take on a wide range of activities. Many ISGs control viral, bacterial, and parasitic infections by directly targeting pathways and functions required during pathogen life cycles (Lehner et al., 2008). BoIFN-ω3 can induce the time-dependent transcription of ISGs (including Mx-1, ISG15 and ISG56). The transcription of Mx-1 increased 3 h after BoIFN-ω3 treatment, and peaked 24 h thereafter. This event was consistent with the expression of Mx-1 protein. The transcription of ISG15 and ISG56 showed a trend similar to that of Mx-1. Given that ISGs were downstream in the IFN signaling pathways, BoIFN-ω3 can obviously induce ISGs expression in a time-dependent manner, suggesting that BoIFN-ω3 can further inhibit viral replication and exert antiviral activity.
In this study, the characterization and biological activity of BoIFN-ω3 were analyzed. BoIFN-ω3 presented moderate biological activity, low cytotoxicity, and stable physicochemical characteristics, as well as exerted antiviral activity on several mammalian cell lines and a protective effect against VSV, BEV, IBRV, and BVDV. Additionally, BoIFN-ω3 can activate the expression of the ISGs gene of IFN signaling pathway downstream. Overall, our study on BoIFN-ω3 enriched the current knowledge about the bovine IFN-ω family.
Conflict of interest
No competing financial interests exist.
Acknowledgements
China Agriculture Research System (CARS-37), the Synergetic Innovation Center of Food Safety and Nutrition in Northeast Agricultural University, the sub-project of National 12th 5-year Support Key Projects (2012BAD12B03, 2012BAD12B05) and the Key Technologies Research and Development Program of Heilongjiang Province (GA09B302), support this study.
Contributor Information
Mingchun Gao, Email: gaomingchun@163.com.
Junwei Wang, Email: jwwang@neau.edu.cn.
References
- Adolf G.R. Human interferon omega–a review [J] Mult. Scler. 1995;1(Suppl. 1):S44–S47. [PubMed] [Google Scholar]
- Boue O., Garcia J.L. Antiviral and antiluteolytic activity of recombinant bovine IFN-omega1 obtained from Pichia pastoris [J] J. Interf. Cytokine Res. 2000;20(8):677–683. doi: 10.1089/10799900050116372. [DOI] [PubMed] [Google Scholar]
- Bracklein T., Theise S., Metzler A. Activity of feline interferon-omega after ocular or oral administration in cats as indicated by Mx protein expression in conjunctival and white blood cells [J] Am. J. Vet. Res. 2006;67(6):1025–1032. doi: 10.2460/ajvr.67.6.1025. [DOI] [PubMed] [Google Scholar]
- Capon D.J., Shepard H.M., Goeddel D.V. Two distinct families of human and bovine interferon-alpha genes are coordinately expressed and encode functional polypeptides [J] Mol. Cell. Biol. 1985;5(4):768–779. doi: 10.1128/mcb.5.4.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin P.J., Entrican G., Gelder K.I. Cloning and biologic activities of a bovine interferon-alpha isolated from the epithelium of a rotavirus-infected calf [J] J. Interf. Cytokine Res. 1996;16(1):25–30. doi: 10.1089/jir.1996.16.25. [DOI] [PubMed] [Google Scholar]
- Charlier M., L'Haridon R., Boisnard M. Cloning and structural analysis of four genes encoding interferon-omega in rabbit. [J] J. Interf. Res. 1993;13(5) doi: 10.1089/jir.1993.13.313. [DOI] [PubMed] [Google Scholar]
- Charlier M., L'Haridon R., Boisnard M. Cloning and structural analysis of four genes encoding interferon-omega in rabbit [J] J. Interf. Res. 1993;13(5):313–322. doi: 10.1089/jir.1993.13.313. [DOI] [PubMed] [Google Scholar]
- Cheng G., Chen W., Li Z. Characterization of the porcine alpha interferon multigene family [J] Gene. 2006;382:28–38. doi: 10.1016/j.gene.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Der S.D., Zhou A., Williams B.R. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays [J] Proc. Natl. Acad. Sci. U. S. A. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech A., Miro G., Collado V.M. Use of recombinant interferon omega in feline retrovirosis: from theory to practice [J] Vet. Immunol. Immunopathol. 2011;143(3–4SI):301–306. doi: 10.1016/j.vetimm.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil S., Leal R.O., Mcgahie D. Oral recombinant feline interferon-omega as an alternative immune modulation therapy in FIV positive cats: clinical and laboratory evaluation [J] Res. Vet. Sci. 2014;96(1):79–85. doi: 10.1016/j.rvsc.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Gao M., Bao J. Molecular cloning and characterization of a novel bovine IFN-epsilon [J] Gene. 2015;558(1):25–30. doi: 10.1016/j.gene.2014.12.031. [DOI] [PubMed] [Google Scholar]
- Hagelstein J., Kist A., Stremmel W. Antiviral potential of interferon-omega on hepatitis B virus replication in human hepatoma cells [J] Arzneimittelforschung. 1998;48(3):343–347. [PubMed] [Google Scholar]
- Hauptmann R., Swetly P. A novel class of human type I interferons [J] Nucleic Acids Res. 1985;13(13):4739–4749. doi: 10.1093/nar/13.13.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmler A., Hauptmann R., Adolf G.R. Molecular cloning and expression in Escherichia coli of equine type I interferons [J] DNA. 1986;5(5):345–356. doi: 10.1089/dna.1986.5.345. [DOI] [PubMed] [Google Scholar]
- Horton H.M., Hernandez P., Parker S.E. Antitumor effects of interferon-omega: in vivo therapy of human tumor xenografts in nude mice.[J] Cancer Res. 1999;59(16) [PubMed] [Google Scholar]
- Krause C.D., Pestka S. Evolution of the Class 2 cytokines and receptors, and discovery of new friends and relatives [J] Pharmacol. Ther. 2005;106(3):299–346. doi: 10.1016/j.pharmthera.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Leal R.O., Gil S., Sepulveda N. Monitoring acute phase proteins in retrovirus infected cats undergoing feline interferon-omega therapy [J] J. Small Anim. Pract. 2014;55(1):39–45. doi: 10.1111/jsap.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner T., Wang Y., Pido-Lopez J. The emerging role of innate immunity in protection against HIV-1 infection [J] Vaccine. 2008;26(24):2997–3001. doi: 10.1016/j.vaccine.2007.11.060. [DOI] [PubMed] [Google Scholar]
- Litzlbauer P., Weber K., Mueller R.S. Oral and subcutaneous therapy of canine atopic dermatitis with recombinant feline interferon omega [J] Cytokine. 2014;66(1):54–59. doi: 10.1016/j.cyto.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402e8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo X., Guo Y., Bao J. Characterization and antivirus activities of a novel bovine IFN-omega24 [J] Mol. Immunol. 2015;66(2):357–363. doi: 10.1016/j.molimm.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Martin V., Najbar W., Gueguen S. Treatment of canine parvoviral enteritis with interferon-omega in a placebo-controlled challenge trial [J] Vet. Microbiol. 2002;89(2–3) doi: 10.1016/s0378-1135(02)00173-6. [DOI] [PubMed] [Google Scholar]
- Mege D., Lefevre F., Labonnardiere C. The porcine family of interferon-omega: cloning, structural analysis, and functional studies of five related genes [J] J. Interf. Res. 1991;11(6) doi: 10.1089/jir.1991.11.341. [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Nakatani H., Yoshida M. Inhibitory effects of recombinant feline interferon on the replication of feline enteropathogenic viruses in vitro [J] Vet. Microbiol. 1994;39(1–2):145–152. doi: 10.1016/0378-1135(94)90095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohe K., Takahashi T., Hara D. Sensitivity of FCV to recombinant feline interferon (rFeIFN) [J] Vet. Res. Commun. 2008;32(2):167–174. doi: 10.1007/s11259-007-9019-5. [DOI] [PubMed] [Google Scholar]
- Roberts R.M., Liu L., Guo Q. The evolution of the type I interferons [J] J. Interf. Cytokine Res. 1998;18(10):805–816. doi: 10.1089/jir.1998.18.805. [DOI] [PubMed] [Google Scholar]
- Rodriguez M., Martinez V., Alazo K. The bovine IFN-omega 1 is biologically active and secreted at high levels in the yeast Pichia pastoris [J] J. Biotechnol. 1998;60(1–2):3–14. doi: 10.1016/s0168-1656(97)00152-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez M., Martinez V., Alazo K. The bovine IFN-omega 1 is biologically active and secreted at high levels in the yeast Pichia pastoris [J] J. Biotechnol. 1998;60(1–2):3–14. doi: 10.1016/s0168-1656(97)00152-1. [DOI] [PubMed] [Google Scholar]
- Rubinstein S., Familletti P.C., Pestka S. Convenient assay for interferons [J] J. Virol. 1981;37(2):755–758. doi: 10.1128/jvi.37.2.755-758.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan A.M., Womack J.E. Type I interferon genes in cattle: restriction fragment length polymorphisms, gene numbers and physical organization on bovine chromosome 8 [J] Anim. Genet. 1993;24(1):9–16. doi: 10.1111/j.1365-2052.1993.tb00912.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Green M.R. Cold Spring Harbor Laboratory Press; 2012. Molecular Cloning: a Laboratory Manual [Z] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses [J] Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W. Interferon-stimulated genes: roles in viral pathogenesis [J] Curr. Opin. Virol. 2014;6:40–46. doi: 10.1016/j.coviro.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.C. Viruses and interferons [J] Annu. Rev. Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- Shao J., Cao C., Bao J. Characterization of the biological activities and physicochemical characteristics of recombinant bovine interferon-alpha 14 [J] Mol. Immunol. 2015;64(1):163–169. doi: 10.1016/j.molimm.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Shao J., Cao C., Bao J. Characterization of bovine interferon alpha1: expression in yeast Pichia pastoris, biological activities, and physicochemical characteristics [J] J. Interf. Cytokine Res. 2015;35(3):168–175. doi: 10.1089/jir.2013.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeheli P. Interferon-induced proteins and the antiviral state [J] Adv. Virus Res. 1990;38:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr I.M., Williams B.R. How cells respond to interferons [J] Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Terenzi F., Hui D.J., Merrick W.C. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56 [J] J. Biol. Chem. 2006;281(45):34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- Tiefenthaler M., Geisen F., Schirmer M. A comparison of the antiproliferative properties of recombinant human IFN-alpha 2 and IFN-omega in human bone marrow culture [J] J. Interf. Cytokine Res. 1997;17(6):327–329. doi: 10.1089/jir.1997.17.327. [DOI] [PubMed] [Google Scholar]
- Velan B., Cohen S., Grosfeld H. Bovine interferon alpha genes. Structure and expression [J] J. Biol. Chem. 1985;260(9):5498–5504. [PubMed] [Google Scholar]
- Wacher C., Muller M., Hofer M.J. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses [J] J. Virol. 2007;81(2):860–871. doi: 10.1128/JVI.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel R. Assignment of the disulphide bonds of leukocyte interferon [J] Nature. 1981;289(5798):606–607. doi: 10.1038/289606a0. [DOI] [PubMed] [Google Scholar]
- Yang L.M., Xue Q.H., Sun L. Cloning and characterization of a novel feline IFN-omega [J] J. Interf. Cytokine Res. 2007;27(2):119–127. doi: 10.1089/jir.2006.0094. [DOI] [PubMed] [Google Scholar]
- Zhao X., Cheng G., Yan W. Characterization and virus-induced expression profiles of the porcine interferon-omega multigene family [J] J. Interf. Cytokine Res. 2009;29(10):687–693. doi: 10.1089/jir.2008.0060. [DOI] [PubMed] [Google Scholar]