Graphical abstract
Highlights
-
•
Many pathogens have resisted vaccine development efforts due to their antigenic diversity and immunodominance of non-neutralizing epitopes.
-
•
Structural characterization of conserved, functional sites targeted by neutralizing antibodies has identified bona fide vaccine targets.
-
•
Structure-based immunogen design has the potential to elicit precise antibody responses targeting neutralizing epitopes.
-
•
Structure-based approaches can tune antibody specificity and diversity tailored to distinct pathogens.
Abstract
Vaccines have been one of the most successful interventions in global health. However, traditional vaccine development has proven insufficient to deal with pathogens that elude the immune system through highly variable and non-functional epitopes. Emerging B cell technologies have yielded potent monoclonal antibodies targeting conserved epitopes, and their structural characterization has provided templates for rational immunogen design. Here, we review immunogen design strategies that leverage structural information to steer bulk immune responses towards the induction of precise antibody specificities targeting key antigenic sites. Immunogens designed to elicit well-defined antibody responses will become the basis of what we dubbed precision vaccines. Such immunogens have been used to tackle long-standing vaccine problems and have demonstrated their potential to seed the next-generation of vaccines.
Current Opinion in Structural Biology 2018, 51:163–169
This review comes from a themed issue on Engineering & design
Edited by Giovanna Ghirlanda and Ivan Korendovych
For a complete overview see the Issue and the Editorial
Available online 29th June 2018
https://doi.org/10.1016/j.sbi.2018.06.002
0959-440X/© 2018 Published by Elsevier Ltd.
Introduction
Vaccination is likely the most efficacious prophylactic approach in modern medicine and has greatly reduced the burden of infectious diseases [1]. Despite their numerous successes, classical vaccine strategies that rely on attenuated or inactivated pathogen formulations, have failed to elicit neutralizing antibodies against a number of pathogens. Some notable examples are long-standing viral threats like Human Immunodeficiency Virus 1 (HIV-1), Respiratory Syncytial Virus (RSV), human Metapneumovirus (hMPV) or Dengue as well as emerging pathogens such as Zika, amongst others. Additionally, the constant antigenic drift of Influenza requires formulation of seasonal vaccines, and has prevented the development of a universally protective vaccine [2].
Many of these pathogens have evolved strategies to evade targeted, neutralizing immune responses [3]. The immunodominance of antigenic sites that do not confer broad and potent neutralization over those that can be targeted by potent neutralizing antibodies (nAbs) is a poorly understood phenomenon, but it is well established that non-neutralizing antibodies can facilitate virus entry into host cells between different serotypes of certain pathogens (e.g. Dengue), thereby causing antibody dependent disease enhancement (ADE) [4,5].
To overcome current limitations in vaccine development, a rational vaccine strategy, known as reverse vaccinology [6], has been proposed, aiming to focus the immune response on epitopes where the pathogen is vulnerable to antibody-mediated neutralization. In essence, this strategy relies on the isolation of nAbs from human or animal repertoires, followed by the structural characterization of the nAb–antigen complex, and finally exploit the acquired atomic-level information to design novel immunogens. A key challenge for next-generation vaccines, then, is to place neutralizing epitopes in the immune system’s spotlight for efficient recognition and enhanced epitope-specific antibody elicitation. We refer to such class of vaccines as precision vaccines, given their extremely well-defined epitope-directed antibody response. An essential requisite for precision vaccines is to encode the structural information of epitopes targeted by broad and potent neutralizing antibodies in the designed immunogens.
Here, we review four structure-based immunogen design approaches that aim to elicit focused antibody responses: (I) silencing non-neutralizing epitopes; (II) conformational stabilization; (III) germline targeting; (IV) epitope scaffolding. A special emphasis is placed on linking the design strategy to the immunological outcomes for different pathogens (Figure 1 ).
Figure 1.
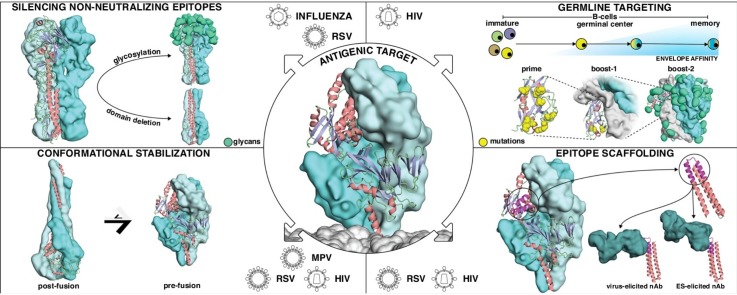
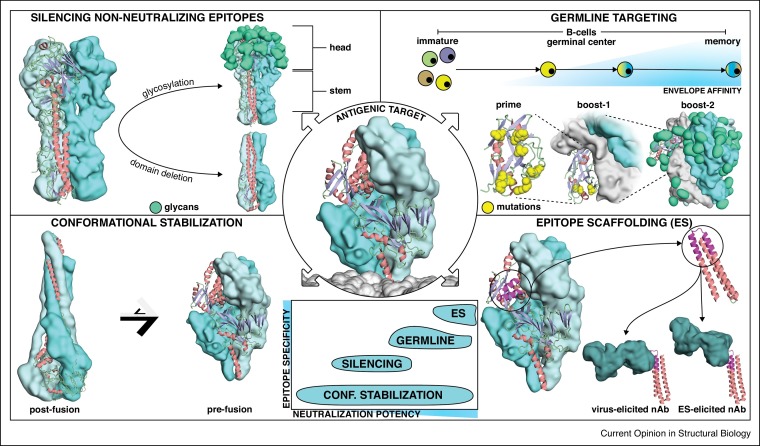
Structure-based approaches for immunogen design. Four main strategies have been used to design improved immunogens for the elicitation of neutralizing antibodies that target precise conformational states, epitopes or antibody lineages. Center: All these strategies were employed to stabilize viral fusion proteins or to enhance antibody responses towards neutralizing epitopes located in viral fusion proteins. Center-bottom: The structure-based approaches can be classified according to expected epitope specificity and neutralization potency of the elicited antibody responses. Upper left: Silencing non-neutralizing epitopes. Glycosylation or deletion of Influenza’s hemagglutinin head domain enables the focusing of antibody responses towards more conserved epitopes targeted by bnAbs. Lower left: Antigenic conformational stabilization. Respiratory Syncytial Virus fusion protein (RSVF) adopts two distinct conformations, a metastable prefusion state (right), and a stable postfusion state (left). Stabilizing the prefusion conformation boosts the induction of neutralizing antibodies. Upper right: Germline targeting. A domain of the HIV-1 Envelope protein (Env) is modified to trigger the activation of inferred germline precursors of HIV-1 bnAbs. After priming, B-cell maturation is guided through consecutive boostings with increasingly native Env versions to select for antibodies that can bind the epitope in its native context. Lower right: Epitope scaffolding. A structurally characterized neutralizing epitope is transplanted onto a heterologous, synthetic protein scaffold, which is used to elicit virus-reactive antibodies targeting the desired epitope.
Silencing non-neutralizing epitopes
The most intuitive strategies to focus antibody responses on immunologically subdominant epitopes is to remove or mask non-neutralizing epitopes. In this section, we describe two approaches to favor the immune recognition of conserved epitopes: (I) excise antigen domains mainly targeted by non-neutralizing antibodies; (II) reduce the accessibility of non-neutralizing epitopes through glycan masking.
In the Influenza virus, the hemagglutinin (HA) surface protein is the main target of humoral responses, and is one of the most notorious examples where the antibody responses mostly target a highly sequence variable region (head domain) and yield mostly strain specific responses [7]. Despite this immunodominance towards the head domain, several epitopes in the HA stem region are targeted by broadly neutralizing antibodies (bnAbs) [8,9], providing a strong rationale for vaccines that aim to induce antibody responses towards the HA stem. Two independent studies reported the design of immunogens comprising only the HA-stem domain [10,11•]. Several rounds of structure-based design were performed, including the removal of the entire head domain followed by the introduction of stabilizing core mutations and linker design. Upon vaccination with stem derived antigens, mice, ferrets and cynomolgus monkeys mounted a cross-reactive antibody response and survived a challenge with a lethal dose of a highly pathogenic viral isolate.
By contrast to HA, the major target of Respiratory Syncytial Virus (RSV) neutralizing antibodies is the head domain of the Fusion protein (RSVF) [12]. Boyington et al. designed truncated versions of RSVF comprising only the head region. This head-only immunogen elicited comparable neutralization titers to those of the full length, prefusion RSVF (see the next section). However, mice primed with RSVF and boosted with a head-only immunogen showed enhanced antibody titers towards neutralization epitopes located on the apex of the head domain, indicating the value of heterologous prime-boost immunizations to focus antibody responses [13].
A structurally less aggressive approach to mask dominant epitopes is to hyper-glycosylate non-functional epitopes or domains. Such strategy has been used by Eggink et al. to silence immunodominant epitopes in HA, generating hyper-glycosylated HA head domains (HGHD). In mice, the HGHDs yielded a heightened antibody response towards the immunosubdominant stem domain and showed improved protection upon viral challenge [14].
Masking of non-neutralizing epitopes through glycosylation has also led to novel HIV-1 Envelope (Env) immunogens with reduced antigenicity to non-neutralizing epitopes. Two main targets of non-nAbs are the flexible Env V3 loop, as well as the bottom of recombinant Env, which is inaccessible on membrane-bound Env spikes. The introduction of two glycans on V3 was shown to dampen the immune response against this site upon rabbit immunization [15]. Similarly, glycosylation of the Env base showed reduced reactivity with sera of animals immunized with non-masked Env, indicating that glycan masking can render non-neutralizing epitopes inaccessible [16•].
Overall, the domain deletion and masking of epitopes induce measurable immunological outcomes, nevertheless they do not seem to radically transform the immune responses obtained. Perhaps the most promising are the HA headless immunogens which elicited antibodies with increased reactivity across heterosubtypic Influenza strains.
Antigenic conformational stabilization
Viruses are dependent on entering host cells for replication, a process mediated by viral surface proteins. Generally, these proteins tend to adopt a metastable prefusion state on the virion’s surface (e.g. HIV, RSV, hMPV and others), undergoing large structural rearrangements to a stable postfusion conformation upon viral and host cell membrane fusion [17]. Serum studies by Magro et al. have revealed that RSV nAbs mostly target the prefusion state of the RSVF [18]. This fueled the idea of promoting the elicitation of antibodies directed against the prefusion conformation rather than postfusion. In Table 1 , we summarize the design strategies used to accomplish the antigenic conformational stabilization.
Table 1.
General design strategies for antigenic conformational stabilization of viral fusion proteins
| Design strategy | Structural stabilization effect | Examples |
|---|---|---|
| Cavity filling mutations |
|
|
| Disulfides |
|
|
| Substitution by prolines |
|
|
| Fusion of trimerization domains |
|
|
| Structural deletions |
|
The structural determination of the prefusion conformation of RSVF [19] provided a template for designing prefusion-stabilized versions of RSVF, latter achieved by introducing intra-protomer disulfide bonds and cavity-filling hydrophobic mutations (Ds-Cav1) [20]. Similarly, Krarup et al. reported a stabilization strategy for RSVF, by introducing proline residues that prevent structural rearrangements required to adopt the postfusion conformation [21]. In immunization studies, prefusion RSVF was eight times more potent in terms of neutralization than postfusion RSVF, as observed in mice, non-human primates and cotton rats. A large fraction of the elicited antibodies was directed against the apex of RSVF, a site that undergoes profound conformational changes in the postfusion state [20]. In a follow up article, Joyce and colleagues further stabilized Ds-Cav1, resulting in an immunogen with improved physical stability in the prefusion state (RSVF-DS2). In mice, DS2 variants elicited up to four-fold more potent neutralizing responses as compared to Ds-Cav1, indicating that conformational stability correlated with immunogenicity [22•]. As a more relevant model for human RSV disease, a bovine RSVF-DS2 version elicited bovine RSV neutralizing antibodies >100-fold higher than postfusion F in calves, and conferred full protection of upper and lower respiratory tract upon viral challenge [23].
The structural similarity between RSV and hMPV allowed similar design strategies to stabilize the MPVF prefusion state. Interestingly, immunization studies in mice have not yielded higher neutralizing antibody responses than those elicited by the postfusion conformation, showing that although design strategies seem to be transferable to other pathogen’s fusion proteins, the immunological outcome can differ [24].
The largest class I fusion proteins are found in human coronaviruses, and known as spike proteins (S). The structure of the spike protein of a human Coronavirus HKU1 has been solved [25], serving as template to stabilize a MERS-CoV spike protein in its prefusion conformation. Immunogenicity studies in mice revealed that prefusion MERS-CoV S elicited neutralizing antibodies with increased breadth and potency compared to postfusion S, showing that for this particular protein the antigenic conformational stabilization strategy yields a superior immunological outcome [26].
Using analogous design strategies, extensive efforts have been made to stabilize the HIV Env glycoprotein in a prefusion, closed conformation. A particular challenge for Env engineering is to maintain its native conformation upon recombinant expression, and reduce the conformational `breathing’ between the open and closed conformations that have been reported for the prefusion Env. The most widely used construct for native-like Env design is the SOSIP.664 trimer, which is stabilized by an intermolecular disulfide bond, an isoleucine-proline substitution within the fusion peptide and a truncation to remove the hydrophobic membrane proximal region. While a comprehensive analysis of the efforts to stabilize Env is beyond the scope of this article and have been the subject of several reviews [27,28], we highlight two recent studies that employed rational stabilization strategies to suppress non-neutralizing antibody responses.
The Env V3 loop is a major target of non-neutralizing Abs, and is exposed in the Env open conformation. To reduce the V3 conformational dynamics, structure-based design was employed to strengthen hydrophobic packing, preventing V3 loop to adopt the open conformation [29,30]. Similarly, Kulp et al. employed computational design to replace a network of buried hydrophilic residues by hydrophobic amino acids, thereby limiting V3 exposure [16•]. Both stabilization strategies reduced reactivity to V3 directed antibodies, and immunogenicity studies in rabbits confirmed dampened antibody responses against the V3 loop.
Another main target of non-neutralizing antibodies within Env are the CD4-induced epitopes. To prevent the CD4-induced conformational change while maintaining an epitope targeted by bnAbs, de Taeye et al. introduced two mutations that occur in HIV-1 strains unable to undergo CD4-induced conformational changes [30]. Using a computational multi-state design protocol, Kulp et al. rationally designed mutations that abrogate CD4 binding to Env, while maintaining binding to bnAbs targeting the CD4 receptor binding site (CD4bs) [16•]. While these engineered Envs showed reduced binding to antibodies targeting CD4-induced epitopes, neither study could prove a direct impact of such modifications on antibody specificity or neutralization in vivo.
In summary, the antigenic conformational stabilization is clearly one of the most promising strategies for immunogen engineering, having shown the ability to dramatically transform the immunological outcome by presenting the most relevant antigenic conformation for the elicitation of functional antibodies. In this strategy, the precision character arises from the conformational specificity rather than epitope specificity which will be much more dominant in the following design strategies.
Germline targeting
Typical bnAbs against HIV-1 carry a large number of somatic hypermutations [36, 37, 38]. Based on this observation, germline targeting has emerged as a novel strategy to elicit well defined antibody lineages. Germline targeting aims to engage the unmutated precursors of the bnAbs, and drive their maturation towards a bnAb by gradually acquiring the necessary somatic mutations to broadly neutralize HIV-1.
Using computational design and in vitro evolution, Jardine et al. developed an engineered outer domain (eOD) of the viral gp120 protein, and introduced mutations that enable binding of inferred germline precursors of the bnAb VRC01 (glVRC01) [36,39,40]. Both in a glVRC01 heavy chain knock-in mouse model, and in mice transgenic for human immunoglobulin loci, it was shown that the germline targeting immunogen (eOD-GT8) primed antibodies with characteristic features of VRC01-like antibodies [41, 42, 43]. Primed B cells were shown to be recalled upon boosting immunizations with gradually more native Env versions, and somatic mutations were driven towards those found in VRC01-class bnAbs [44•]. Similarly, Medina-Ramirez et al. have engineered a SOSIP.664 trimer to bind germline precursors of CD4bs antibodies, and this engineered native-like trimer was shown to activate glVRC01 antibodies in vitro and in vivo [45].
In order to target another bnAb class binding to a different epitope on Env (PGT121-like bnAbs), Steichen et al., employed mammalian cell surface display to engineer germline targeting Env trimers, which were then shown to activate B cells carrying a PGT121 germline receptor in vitro and to prime PGT121-like antibody responses in knock-in mice [46].
Together, these studies established a first milestone in using structure-based design together with a stepwise vaccination protocol that targets and expands specific germline precursors. Albeit an important contribution, it remains to be shown what outcome the germline targeting strategy will have in terms of eliciting broadly neutralizing antibodies in relevant animal models. Unlike the other described approaches, the precision aspect of this strategy is related to the specific antibody lineages that are deemed necessary to achieve broad HIV neutralization activity.
Epitope scaffolding
The presentation of single epitopes in the context of scaffold-proteins has gained traction in the last years with the aim of eliciting epitope-specific antibodies. Essentially, this approach uses structural information of the exact epitope conformation recognized by a nAb to design heterologous proteins that mimic the structure of the epitope and are structurally compatible with the antibody binding mode.
The first reports of epitope-scaffolds were presented in the context of HIV using as targets the neutralization epitopes 4E10 [47] and 2F5 [48,49]. In these two studies, the epitopes were grafted onto heterologous scaffold-proteins using computational design approaches [50], biochemical and structural characterization confirmed accurate epitope mimicry. Immunologically, the most remarkable result was that the epitope-scaffolds were able to induce antibodies with fine specificities similar to those of the parent antibodies [47, 48, 49], showing that the epitope-scaffolds achieved in vivo the immunological outcome for which they were primarily designed, although no serum neutralization was obtained.
A later effort by McLellan and colleagues applied the epitope scaffolding strategy to design immunogens that presented the antigenic site II of RSVF, the target of the FDA-approved monoclonal antibody Palivizumab. However, only the side chains of the epitope were grafted onto a scaffold with a similar backbone conformation as the epitope, yielding a scaffold with imperfect epitope mimicry. Much like the HIV epitope-scaffolds, in mouse immunizations, epitope-specific antibodies were elicited but no serum neutralization was achieved [51].
To improve the structural mimicry of site II in a synthetic scaffold, Correia et al. developed a new protein design algorithm, where protein folding and sequence design simulations were coupled to generate immunogens with the epitope structure stabilized in the exact native conformation. These designs showed an extremely high affinity to site II antibodies, and structural characterization revealed a perfect epitope mimicry. In mouse immunogenicity studies the second-generation epitope-scaffold still did not elicit viral neutralization titers, nevertheless when used in non-human primates (NHP) low, but consistent levels of neutralization were detected. Monoclonal antibodies isolated from the NHPs were site II-specific, and bound with high affinity to RSVF. Structural studies confirmed that the elicited antibodies recognized antigenic site II similarly to Palivizumab, and most importantly, neutralized RSV with superior potency compared to Palivizumab [52•].
In summary, the epitope-scaffolding approach is an efficient way of eliciting epitope-specific antibodies. Nevertheless, these are simplified immunogens that mostly encode the epitope binding motif surface and lack important molecular features regarding the tertiary and quaternary environments of the native viral proteins. This class of immunogens faces important challenges; despite their proven ability to induce very potent and epitope specific antibodies, it remains to be seen how to increase the fraction of functional antibodies within the overall repertoire.
Conclusion/outlook
The field of vaccinology is facing incredible challenges to develop efficacious vaccines against sophisticated pathogens with the ability to escape immune responses in many different ways. Due to those escape mechanisms, mounted antibody responses against some of those pathogens have limited breadth and do not afford protection against antigenically drifted strains. Additionally, the induced neutralization titers are often low and decay over time allowing reinfection. The reverse vaccinology strategy was envisioned as an integrated approach to design immunogens that elicit or boost preexisting antibody responses focused on bona fide neutralization epitopes. In the age of systems approaches and big-data science, our understanding of complex biological phenomena, such as an immune response, has been hugely expanded. Therefore, we are now able to define with exquisite accuracy the molecular determinants desirable for a particular vaccine development endeavor. Those determinants include, but are not limited to, antigen conformations, desired antibody lineages and key neutralization epitopes, which lead us to conclude that we have entered into a new age of precision vaccines. Structure-based approaches, as reviewed here, are essential to define the target problem and realize the full potential of precision vaccines. Despite the successes so far, none of the strategies described here have proven to be the `one-fits-all’ solution. While a prefusion stabilized RSVF was a far superior immunogen than the postfusion counterpart, the same approach for MPVF did not enhance the neutralizing responses. Similarly, the sole conformational stabilization of HIV Env is deemed unlikely to elicit bnAbs [53] and germline targeting approaches hold great promise to prime a bnAb response. Finally, while epitope-scaffolds elicited very potent and specific monoclonal antibodies in NHPs, the average neutralization titers were rather low.
The strengths and weaknesses of each of the presented strategies suggests a complex balance between the approach, the target pathogen and the expected immune response. Ultimately, a combination of the different strategies may be the best course of action to overcome their specific limitations and bring forth a new generation of rationally designed, precision vaccines (Table 2 ).
Table 2.
Overview of immunogen engineering approaches and immunological outcomes
| Approach | Immunological outcome | Examples |
|---|---|---|
| Silencing non-neutralizing epitopes | Immune response focused on HA stem region, conferring heterosubtypic protection upon challenge Boosting of RSVF-head directed antibodies |
Influenza [10,11•] RSV [13] |
| Conformational stabilization | Increased serum neutralization Antibody responses focused on sites that are inaccessible in alternative conformation |
RSV [22•] HIV [16•,30] |
| Germline targeting | Activation of germline precursors in humanized mice Sequential boosting protocol selected productive somatic mutations |
HIV [42,44•] |
| Epitope scaffolding | Elicitation of epitope specific antibodies Successful elicitation of RSV antibody with superior neutralization potency compared to clinically approved mAb |
HIV [47] RSV [52•] |
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
Acknowledgments
Bruno Correia is a grantee from the European Research Council (Starting grant: 716058), the Swiss National Science Foundation, Novartis Foundation for Medical-Biological Research and the Biltema Foundation. Fabian Sesterhenn is funded by the Swiss Systemsx.ch initiative for systems biology. Jaume Bonet is sponsored by an EPFL-Fellows grant funded by an H2020 Marie Sklodowska-Curie action.
Glossary
- Immunogen
complete antigen composed of a macromolecular carrier and one or multiple epitopes capable of inducing an immune response.
- Immunodominance
immune responses that preferentially target certain antigens or antigenic sites.
- Precision vaccine
vaccine composed of designed immunogens that elicits an antibody response directed to key antigenic sites that are sensitive to neutralization.
- Broadly neutralizing antibody (bnAb)
neutralizing antibody that is broadly effective against different strains of the same pathogen.
- Germline antibodies
unmutated antibody repertoire encoded by the variable (V), diversity (D) and joining (J) genes.
References
- 1.Greenwood B. The contribution of vaccination to global health: past, present and future. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130433. doi: 10.1098/rstb.2013.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nabel G.J. Designing tomorrow’s vaccines. N Engl J Med. 2013;368:551–560. doi: 10.1056/NEJMra1204186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finlay B.B., McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 4.Dejnirattisai W. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takada A., Kawaoka Y. Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Rev Med Virol. 2003;13:387–398. doi: 10.1002/rmv.405. [DOI] [PubMed] [Google Scholar]
- 6.Rappuoli R., Bottomley M.J., D’Oro U., Finco O., De Gregorio E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med. 2016;213:469–481. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angeletti D. Defining B cell immunodominance to viruses. Nat Immunol. 2017;18:456–463. doi: 10.1038/ni.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekiert D.C. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sui J. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Impagliazzo A. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 11•.Yassine H.M. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]; The immunodominant region of Influenza HA, the head domain, constantly undergoes antigenic drift to escape the immune response. To focus antibody responses on the immunologically subdominant HA stem region, the authors engineered a stem-only immunogen. Vaccination of mice and ferrets elicits a protective immune response against heterosubtypic virus challenge. This study is an important step towards eliciting subdomain-focused antibody responses, which are known to protect against diverse group 1 influenza strains.
- 12.Ngwuta J.O. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med. 2015;7:309ra. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyington J.C. Structure-based design of head-only fusion glycoprotein immunogens for respiratory syncytial virus. PLoS One. 2016;11:e0159709. doi: 10.1371/journal.pone.0159709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggink D., Goff P.H., Palese P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol. 2014;88:699–704. doi: 10.1128/JVI.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringe R.P. Reducing V3 antigenicity and immunogenicity on soluble, native-like HIV-1 Env SOSIP trimers. J Virol. 2017;91 doi: 10.1128/JVI.00677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Kulp D.W. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun. 2017;8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; To reduce the exposure of non-neutralizing epitopes on native-like HIV-1 Env trimers, Kulpet al. employed computational and structure-guided design to reduce exposure of epitopes from three Env antigenic sites. Additional glycosylation sites were introduced to block antibody access to the membrane proximal region, a network of buried hydrophilic residues was replaced by hydrophobic residues to lock the V3 conformation. In addition a multistate design protocol was used to abrogate CD4 receptor binding while retaining affinity for bnAbs. The rational design approach to dampen the immunogenicity of non-neutralizing epitopes could potentially be employed for other pathogens.
- 17.Harrison S.C. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magro M. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A. 2012;109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLellan J.S., Yang Y., Graham B.S., Kwong P.D. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLellan J.S. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krarup A. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Joyce M.G. Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat Struct Mol Biol. 2016;23:811–820. doi: 10.1038/nsmb.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]; A first generation of the prefusion stabilized RSVF was optimized for improved stability and immunogenicity, yielding four-fold increased neutralization titers. The design and screening strategies for over 500 variants could be transferable to engineer other fusion proteins for improved biophysical and antigenic properties.
- 23.Zhang B. Protection of calves by a prefusion-stabilized bovine RSV F vaccine. NPJ Vaccines. 2017;2:7. doi: 10.1038/s41541-017-0005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battles M.B. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun. 2017;8:1528. doi: 10.1038/s41467-017-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchdoerfer R.N. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallesen J. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders R.W., Moore J.P. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev. 2017;275:161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward A.B., Wilson I.A. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev. 2017;275:21–32. doi: 10.1111/imr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Taeye S.W. Stabilization of the gp120 V3 loop through hydrophobic interactions reduces the immunodominant V3-directed non-neutralizing response to HIV-1 envelope trimers. J Biol Chem. 2018;293:1688–1701. doi: 10.1074/jbc.RA117.000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Taeye S.W. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon Y.D. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders R.W. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blais N. Characterization of Pre-F-GCN4t, a modified human respiratory syncytial virus fusion protein stabilized in a noncleaved prefusion conformation. J Virol. 2017;91 doi: 10.1128/JVI.02437-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klasse P.J. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol. 2013;87:9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgiev I.S. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol. 2015;89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jardine J. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein F. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao X. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jardine J.G. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jardine J.G. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sok D. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science. 2016;353:1557–1560. doi: 10.1126/science.aah3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian M. Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell. 2016;166:1471–1484. doi: 10.1016/j.cell.2016.07.029. e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Briney B. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell. 2016;166:1459–1470. doi: 10.1016/j.cell.2016.08.005. e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brineyet al. showed that an heterologous prime boost immunization regimen, consisting of a germline-targeting prime to activate germline precursors of VRC01 bnAbs, followed by boosting with a `bridging’ immunogen carrying a closer to native epitope and ultimately native HIV Env, drove the desired precursors to mature towards antibodies with critical features of HIV bnAbs. This sequential immunization strategy, imposing a strong selective pressure to guide bnAb precursors to acquire defined somatic mutations found in mature HIV bnAbs is an important milestone for HIV vaccine development.
- 45.Medina-Ramirez M. Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J Exp Med. 2017;214:2573–2590. doi: 10.1084/jem.20161160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steichen J.M. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Correia B.E. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Ofek G. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guenaga J. Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS One. 2011;6:e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azoitei M.L. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 51.McLellan J.S. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J Mol Biol. 2011;409:853–866. doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Correia B.E. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a novel computational design approach, the authors transplanted a neutralizing epitope of the RSVF protein onto a small, non-viral and synthetic protein scaffold. Immunization of nonhuman primates elicited neutralization activity and potent RSV-neutralizing antibodies that targeted the transplanted epitope were isolated. This study provided a proof of concept to rationally elicited narrowly focused antibody responses towards defined epitopes.
- 53.Medina-Ramirez M., Sanders R.W., Sattentau Q.J. Stabilized HIV-1 envelope glycoprotein trimers for vaccine use. Curr Opin HIV AIDS. 2017;12:241–249. doi: 10.1097/COH.0000000000000363. [DOI] [PMC free article] [PubMed] [Google Scholar]