Highlights
-
•
Only 2/27 kidneys and 1/81 urine samples were FeMV PCR positive.
-
•
All cats with chronic kidney disease were FeMV PCR negative on urine.
-
•
Only 1/14 cats with proteinuria was FeMV positive on urine.
-
•
The 2 FeMV PCR positive kidneys were from cats affected by FIP and adenocarcinoma.
-
•
Italian FeMV strains form a sub-cluster with strains isolated from other countries.
Keywords: Feline morbillivirus, Chronic kidney disease, Clinical pathology, Urinalysis
Abstract
Feline morbillivirus (FeMV) is an emerging virus that was first described in Hong Kong in 2012. Several reports suggested the epidemiological association of FeMV infection with chronic kidney disease (CKD) in cats. The aim of this study was to investigate the presence and the genetic diversity of FeMV as well as the relationship between FeMV infection and CKD in cats from Northern Italy. Urine (n = 81) and kidney samples (n = 27) from 92 cats admitted to the Veterinary Teaching Hospital of the University of Milan between 2014 and 2017 were investigated for FeMV infection. FeMV RNA was detected in one urine sample (1.23%; 95% CI: 0.03–6.68%) and in two kidneys (7.40%; 95% CI: 0.91–24.28%). FeMV RNA was revealed only in urine or kidneys of cats without evidence of CKD. Phylogenetic analysis showed that the three strains clustered with FeMV strains retrieved from public database, forming a distinct sub-cluster of FeMV. The presence of distinct genotypes of FeMV found in this study is in accordance with previous studies demonstrating that FeMV strains are genetically diverse. A clear relationship between the presence of FeMV infection and CKD in the cats from Northern Italy was not observed, confirming recent reports that do not support the hypothesis that FeMV infection is associated with the development of CKD.
1. Introduction
Morbilliviruses belong to the subfamily Paramyxovirinae, family Paramoxyviridae, which is composed of enveloped, single-stranded, negative-sense RNA viruses (de Vries et al., 2015). Morbilliviruses infect humans (e.g. Measles morbillivirus) and animals (e.g. Canine distemper virus, Peste des petits ruminants virus) (Nambulli et al., 2016). Feline morbillivirus (FeMV) was firstly isolated in Hong Kong between 2009 and 2011, mainly in urine, and a role of this virus in the pathogenesis of feline tubular interstitial nephritis (TIN) and chronic kidney disease (CKD) was postulated (Woo et al., 2012). Two years later, FeMV was isolated in Japan with a prevalence of 6.1% in urine, 10% in blood and 40% in kidneys with nephritis (Furuya et al., 2014). Up to date, FeMV has been identified also in USA, South America, Turkey and Europe, with sequences closely related to previously identified FeMV strains (Lorusso et al., 2015; Sieg et al., 2015; Sharp et al., 2016; Darold et al., 2017; Yilmaz et al., 2017; McCallum et la., 2018). Sieg et al (2015) found in cats from Germany viruses with sequences different from any other morbillivirus and with a nucleotide homology close to bat and rodent paramyxoviruses, thereby named feline paramyxoviruses (FPaV). The same authors found a correlation between FeMV and FPaV presence in urine and CKD (Sieg et al., 2015). Recently, a high prevalence of FeMV was found, but without a strong correlation between TIN and infection (Park et al., 2016). Moreover, Yilmaz et al (2017) did not find any significant difference in both histopathological and clinico-pathological findings between FeMV infected and not infected cats (Yilmaz et al., 2017). In addition, no relationship between the shedding of the virus in urine and laboratory alterations imputable to CKD were found in recent studies (Darold et al., 2017; McCallum et al., 2018).
In 2015, FeMV was identified in Italy from the urine of a 15-years old stray cat with symptoms and laboratory alterations imputable to chronic kidney disease (CKD). Even if a kidney biopsy was not performed, the fact that the cat continued to shed the virus in the urine for 14 days, led the Authors to suggest a possible role of this virus in the development of renal failure (Lorusso et al., 2015). FeMV is now cited as a possible cause of CKD, which is a very common feline disease, with a multifactorial and still unclear pathogenesis (Brown et al., 2016). However, only few studies on FeMV provided information about cats signalment and anamnesis, as well as on clinicopathological changes related to FeMV infection. Moreover, data regarding the correlation between CKD and FeMV infection are often discordant and they have not been investigated in Italy yet. Thus, the aims of this study were to investigate the presence of feline paramyxoviruses in Italy with molecular methods, regardless of the presence of clinical signs consistent with CKD, to perform molecular characterization and phylogenetic analysis and to evaluate the existence of a correlation between paramyxoviruses infection and clinicopathological as well as histopathological changes imputable to renal damage.
2. Material and methods
2.1. Sample selection and processing
Cats routinely subjected to urine and hematological analyses for diagnostic purposes at the Veterinary Teaching Hospital of Milan in 2017 were involved. Signalment (e.g. age, breed, sex, living environment), clinical history and health status, including results of Feline Immunodeficiency virus (FIV) and Feline Leukemia virus (FeLV) serology, were recorded for each cat. Either cats with a diagnosis or a clinical suspicion of CKD, clinically healthy cats, and cats affected by other diseases were enrolled, regardless of age and breed. Cats were examined and received additional diagnostic investigations (e.g. X-Rays, ultrasound or other instrumental diagnostic approaches) depending on the clinical presentation. Urine and blood samples were collected in the context of clinicopathological investigations requested from the referring veterinarians. Additionally, 27 RNA samples obtained from kidneys of euthanized or spontaneously deceased cats were also collected during necropsies performed for diagnostic purposes between 2014 and 2017.
All of the above procedures were performed within routine diagnostic workouts and therefore, according to the decisions of the Ethical Committee of the university of Milan, residual aliquots of samples or tissues collected under informed consent of the owners can be used for research purposes without any additional formal request of authorization to the Ethical Committee.
Cats were excluded from the study in the absence of enough urine volume or if signalment, clinical history and diagnosis as well as histological description of the kidney were not available.
2.2. Urine samples
Urine samples (8–10 mL) collected by spontaneous micturition or by ultrasound-guided cystocentesis were sent to the laboratory and immediately subjected to urinalysis. In particular, specific gravity (SG) by refractometry, urine dipstick (Combur 10 test, Roche diagnostics, Risch-Rotkreuz, Switzerland) and sediment analysis were performed. After centrifugation (1250 rpm × 5 min), supernatants were used to determine the urine protein:creatinine ratio (UPC) and to measure the activity of urinary gamma-glutamyltransferase (uGGT) with a spectrophotometer (Daytona, Randox Laboratories, Crumlin, UK). Residual aliquots of supernatants were then frozen at −80 °C upon molecular analyses, as described below.
On blood samples collected in EDTA tubes, when available, a complete blood count (CBC) along with blood smear examination were performed. On serum, creatinine, urea, sodium potassium and chloride, cholesterol and albumin concentration as well as other analytes if requested by the clinical presentation, were measured with a spectrophotometer (Daytona, Randox Laboratories, Crumlin, UK).
Cats were categorized as healthy in the absence of clinical signs or of relevant laboratory changes, and as diseased if history, clinical signs, diagnostic imaging, or laboratory changes were consistent with a specific disease. Sick cats were further classified in specific disease groups according to their diagnosis: CKD alone; CKD along with other diseases; urinary tract disorders without CKD; neoplastic, infectious, endocrine or miscellaneous diseases. Based on the International Renal Interest Society (IRIS) guidelines (http://www.iris-kidney.com), the concentration of serum creatinine and the UPC values were used to stage and substage cats for CKD, respectively.
2.3. Kidney samples
Kidneys were collected during necropsies performed either within studies on feline infectious peritonitis (FIP) or during routine diagnostic activities of the Pathology unit of the Veterinary teaching hospital. During necropsies, two sections approximately 1 cm thick each including, when present, macroscopically apparent lesions were collected. One section was immediately frozen at −80 °C and subjected to RNA extraction within two weeks from sampling, and one section was put in 10% neutral-buffered formalin for histology. Sections used for molecular biology were always collected adjacent to the sections used for histology. Histology was performed on formalin-fixed paraffin-embedded (FFPE) samples using microtomic sections (3 μm) stained with haematoxylin-eosin.
2.4. PCR for FeMV and phylogenetic analysis
Frozen-thawed urine samples were centrifuged (3000 rpm × 5 min) to remove debris. Then, 150 μL of the obtained supernatant were used for RNA extraction using the NucleoSpin RNA Virus commercial kit (Macherey-Nagle, Bethlehem, PA), following manufacturer’s instructions. RNA extraction from renal tissues was performed using the NucleoSpin RNA kit (Macherey-Nagel, Bethlehem, PA) according to the manufacturer’s instruction. RNA samples obtained from urine and kidneys were then frozen at −80 °C or immediately investigated for the presence of FeMV by molecular analysis.
A FeMV specific nested RT-PCR (nRT-PCR) was performed using primers and methods amplifying a 401 bp fragment of the L gene, according to Furuya et al (2014). PCR product were visualized under UV transilluminator on a 2% agarose gel stained with ethidium bromide. FeMV RNA extracted from a naturally infected cat was used as positive control and RNase-free water as negative control. The amplicons of the expected size were purified and sent for sequencing using the forward and reverse inner primers used for the nRT-PCR to a commercial service (GATCBiotech, Konstanz, Germany). The sequence data were assembled and manually corrected using BioEdit software version 7.0 (freely available at http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The sequences were then compared with those available in GenBank using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
For phylogenetic analysis, the sequences were aligned with representative FeMV strains retrieved from GenBank using Clustal X; manual editing was performed with Bioedit software version 7.0 (Kumar et al., 2016). Phylogeny was estimated by the neighbor-joining algorithm (NJ) and the maximum likelihood (ML) method, with 1000 bootstrap replicates (Felsenstein, 1981; Kimura, 1980).
After sequences comparison, the percentage of nucleotide similarity of pairwise evolutionary distances was calculated using MEGA version 7.
3. Results
3.1. Caseload
Eighty-one urine samples were collected from 65 cats. Six cats (9.2%) were stray cats and all other cats were privately-owned. Fifty-six cats (86.1%) were European Shorthair, the remaining cats were Persian (4 cats), Norwegian Forest (2 cats), one Exotic, one Ragdoll and one Siamese cat. Thirty-nine cats were male (36 neutered, 3 tomcats) and 26 were spayed female. Cat’s age ranged from 1 to 17 years (mean age: 9,7 years; median age: 10 years; 95% CI = 8,5–10,9). Data regarding the healthy and disease groups and the number of cats for each group are reported in Table 1 .
Table 1.
Number of cats and urine samples involved in this study. Cats are divided in groups based on their diagnosis. Mixed: CKD together with other diseases; NPL: neoplastic.
| Groups | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| CKD | Other urinary tract disorders | Mixed | NPL | Infectious | Endocrine | Miscellaneous | Healthy | Tot Cats | |
| N° of cats | 20 | 8 | 4 | 2 | 2 | 8 | 13 | 8 | 65 |
| Tot Urine samples |
|||||||||
| N° of urine samples | 28 | 11 | 7 | 2 | 2 | 9 | 14 | 8 | 81 |
Thirty-two (32/65; 49.2%) cats were affected by urinary tract disorders. In particular, 20/65 were affected by CKD only, 4/65 by CKD along with other diseases (tumors in 3 cats and pancreatitis in 1 cat) and eight (8/65; 12.3%) cats had other urinary tract disorders (crystalluria in 4 cats, urinary obstruction, hematuria, pyelonephritis and nephromegaly in single cats).
Serum was submitted along with urine for 14/20 CKD cats. Based on creatinine measurement, 3/14 were in stage 1, 8/14 cats in stage 2, 2/14 in stage 3 and 1/14 in stage 4 of the International Renal Interest Society (IRIS) guidelines and all their urine samples had the UPC ratios below 0.4, which is the cut-off to consider a cat proteinuric. Only four samples had values compatible with the IRIS borderline substage (UPC: 0.2 to 0.4). In the remaining 6/20 cats, CKD was diagnosed by previous tests on serum, and therefore only urine samples were submitted to the laboratory, to substage the patients according to the UPC ratio. Two of them were proteinuric (UPC: 0.77 and 1.05) while the others had UPC values below 0.2. UPC values above 0.4 were recorded also in 3 of the cats affected by CKD along with other disorders and in 2/8 cats with other urinary tract disorders.
More than one urine sample were collected during the study from 11 cats belonging to the CKD, CKD with other diseases and other urinary tract disorders groups, to determine changes in urine specific gravity and proteinuria as well as to monitor the efficacy of therapies administered for CKD.
Cats belonging to the remaining groups were affected by tumors (2/65; 3.1%) such as one lymphoma and one oral carcinoma, infectious diseases (2/65; 3.1%) such as one case of feline immunodeficiency virus (FIV) infection and one case of feline infectious peritonitis (FIP), endocrinopathies (8/65; 12.3%) such as hyperthyroidism (n = 4), diabetes mellitus (n = 3), Addison’s disease (n = 1) or miscellaneous diseases (13/65; 20.0%) such as gastrointestinal (10 cats), traumatic (1 cat), parasitic (1 cat) and dermatological disorders (1 cat). A smaller group of cats (8/65; 12.3%) was represented by clinically healthy cats without laboratory abnormalities, sampled during wellness visits or during the follow up in the case of positive responses to treatments.
UPC values above 0.4 were recorded in two cats from the miscellaneous group, 2 from the metabolic group and in one of each remaining group.
Kidney samples were collected from twenty-seven cats (Table 2 ). Sixteen cats (16/27; 59.3%) were affected by infectious diseases (15 FIP, confirmed by immunohistochemistry for feline coronavirus, FCoV and 1 feline panleukopenia virus, FPV). Three cats (3/27; 11.1%) had died for severe traumatic events. The remaining cats were affected by tumors (2/27; 7.4%), i.e. one pulmonary adenocarcinoma and one thymic carcinoma, or by miscellaneous diseases (6/27; 22.2%) such as end stage cardiac insufficiency (n = 3), rodenticide poisoning (n = 1), chronic pneumonia (n = 1) and diabetes (n = 1). Of the 15 cats affected by FIP, 8 were IHC positive on kidneys. The other FIP-affected cats were IHC positive on one or more of the other tested organs (e.g. brain, liver, intestine, lungs). Conventional nRT-PCR for FCoV was performed on all the kidney samples for research purposes and it resulted positive on 13/15 FIP cats’ kidneys. One additional cat not affected by FIP (IHC negative on all organs) tested positive on nRT-PCR for FCoV. Indeed, viral systemic spread is not uncommon in FCoV-infected cats not affected by FIP (Porter et al., 2014; Barker et al., 2017).
Table 2.
Number of cats from which kidney samples were collected. Cats are divided in groups based on their diagnosis. FIP: feline infectious peritonitis; FPV: feline panleukopenia virus; NPL: neoplastic.
| Groups |
||||||
|---|---|---|---|---|---|---|
| Infectious |
Trauma | NPL | Others | Tot cats | ||
| FIP | FPV | |||||
| Number of cats | 15 | 1 | 3 | 2 | 6 | 27 |
Twenty-three of the necropsied cats were European shorthair, 2 Maine Coon, 1 Exotic and 1 Ragdoll. Thirteen were female (5 spayed) and fourteen were male (10 neutered, 4 tomcats). The cats age ranged from 1 month to 15 years (mean age: 3.5 years; median: 1 year; 95% CI = 1,6–4,8).
Among the FIP group, histological examination of the kidneys revealed lymphoplasmacytic interstitial nephritis in 9/15 cats, along with granulomatous lesions in 2/9 and vasculitis and necrosis in 3/9 cats. Two kidneys from cats with FIP showed only granulomatous lesions, while 4/15 FIP cats did not have histological relevant lesions in the kidney (although typical FIP lesions were found in other organs). Histological signs of lymphoplasmacytic interstitial nephritis were found in four cats belonging to the other groups. Specifically, these lesions were observed in cats affected by diabetes, cardiac insufficiency, chronic pneumonia (along with amyloidosis) and traumatic injuries. The kidney belonging to the cat euthanized for pulmonary adenocarcinoma showed granulomatous lesions and necrosis. No histological lesions were found in the kidneys of the remaining 7 cats included in this study.
3.2. FeMV RT-PCR results
In total, 3 out of 108 (2.77%; 95% CI: 0.94–7.85%) samples examined in this study tested positive for FeMV RNA and were obtained from one cat sampled in 2014 and from two cats sampled in 2017. In particular, one urine (1/81, 1.23%; 95% CI: 0.03–6.68%) collected from a stray cat and two kidney samples (2/27, 7.40%; 95% CI: 0.91–24.28%) resulted positive.
One out of fourteen cats (7.14%; 95% CI: 0.18–33.86%) which showed UPC values above reference values or urinary sediment alterations suggestive of tubular damage (e.g. urinary casts) had FeMV RNA in the urine. At the moment of sample collection, the only FeMV positive cat with urine diagnostic results suggestive of renal disorder had ultrasound and laboratory alterations suggestive of cholangiohepatitis. In this cat, urinalysis showed UPC value compatible with proteinuria (UPC: 0.56) and hyaline and granular casts were observed in the urinary sediment. After appropriate anti-inflammatory, antibiotic and supportive therapy the cat had fully recovered, and another urine sample was taken 8 months after the first sampling. In the second sample, no laboratory alterations, including urinalysis, were found and the PCR for FeMV in the second urine sample resulted negative.
The two positive kidneys were collected from one cat affected by non-effusive FIP and from another cat affected by pulmonary adenocarcinoma. Both the FeMV positive cats showed granulomatous lesions at the histological examination of the kidneys, in one case in the framework of typical FIP lesions and in the other case associated with severe necrosis. One of the FeMV positive kidneys was also nRT-PCR for FCoV positive.
3.3. Phylogenetic analysis
FeMV PCR positive samples were sequenced and named as follows:
-
•
Urine sample, Italy 2017 1073U
-
•
Kidney sample, Italy2014 434K
-
•
Kidney sample, Italy2017 1568K
BLAST analysis identified the three sequences as FeMV, showing L gene sequence homologies of 92-89% for samples Italy2014 434 K and Italy2017 1568 K and of 91%-89% for the sample Italy2017 1073U compared with FeMV sequences deposited in the GenBank database.
The nucleotide percentages of similarity of pairwise evolutionary distances between sequences isolated in this work and a representative selection of sequences deposited in the GenBank database are reported in Table 3 . The sequences isolated in this study showed 99.2–99.6% nucleotide similarity, while the homology between the sequences of this work and the selected FeMV sequences deposited in the GenBank database was between 91.3 and 83.7%.
Table 3.
Percentages of nucleotide similarity between the L gene sequences obtained in this study and selected FeMV sequences deposited in the GenBank database. GenBank access numbers are reported for each strain in the first column.
| Percentages of similarity |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | 434 K | 1073U | 1568 K | Piuma/ 2015 |
90604 | Lisa | Mickie | Schmusi | FeMoV/ TR/Moj |
US1 | Juninha | OtJP001 | MiJP003 | N076U | SS2 | SS1 | M252A | 776U |
| Italy 2014 434 K | ||||||||||||||||||
| Italy 2017 1073U | 99.2 | |||||||||||||||||
| Italy 2017 1568 K | 99.6 | 99.6 | ||||||||||||||||
| KT825132 Italy 2015 Piuma/2015 | 88.6 | 88.6 | 89 | |||||||||||||||
|
KP159803 Germany 2013 90604 |
83.7 | 84.4 | 84 | 84.8 | ||||||||||||||
|
KR269599 Germany 2015 93436 Lisa |
84 | 84.8 | 84.4 | 85.2 | 99.6 | |||||||||||||
|
KR269600 Germany 2015 98993 Mickie |
90.9 | 90.9 | 91.3 | 89.4 | 82.9 | 83.3 | ||||||||||||
|
KR269601 Germany 2015 66883 Schmusi |
90.9 | 90.9 | 91.3 | 90.1 | 85.9 | 86.3 | 93.9 | |||||||||||
|
KT808322 Turkey 2015 FeMoV/TR/Moj |
90.5 | 90.5 | 90.9 | 89.7 | 83.3 | 83.7 | 98.9 | 94.3 | ||||||||||
|
KR014147 USA 2013 US1 |
90.1 | 90.1 | 90.5 | 95.4 | 84.4 | 84.8 | 88.6 | 89.4 | 88.2 | |||||||||
|
KX452077 Brazil 2016 Juninha |
89.7 | 89.7 | 90.1 | 88.2 | 81.7 | 82.1 | 98.1 | 92 | 97 | 88.2 | ||||||||
| NC 025264 Japan 2013 OtJP001 |
90.9 | 90.9 | 91.3 | 89.4 | 82.9 | 83.3 | 100 | 93.9 | 98.9 | 88.6 | 98.1 | |||||||
|
AB924121 Japan 2013 MiJP003 |
89.7 | 89.7 | 90.1 | 95.1 | 85.6 | 85.9 | 89.7 | 91.3 | 90.1 | 92.8 | 88.6 | 89.7 | ||||||
|
LC080850 Japan 2013 N076U |
88.6 | 88.6 | 89 | 95.8 | 83.7 | 84 | 87.8 | 89.4 | 88.2 | 97.3 | 86.7 | 87.8 | 93.2 | |||||
|
LC036586 Japan 2014 SS2 |
89.4 | 89.4 | 89.7 | 93.9 | 83.7 | 84 | 90.1 | 87.8 | 89.7 | 92.4 | 89.7 | 90.1 | 95.1 | 91.3 | ||||
|
AB910309 Japan 2014 SS1 |
90.1 | 90.1 | 90.5 | 88.6 | 82.9 | 83.3 | 99.2 | 93.2 | 98.1 | 87.8 | 97.3 | 99.2 | 89.7 | 87.1 | 90.1 | |||
|
JQ411016 China 2010 M252A |
89.4 | 89.4 | 89.7 | 93.9 | 85.9 | 85.6 | 89.4 | 90.9 | 89.7 | 92 | 88.2 | 89.4 | 98.9 | 92.4 | 93.9 | 89.4 | ||
| JQ411015 Hong Kong 2009 776U | 88.6 | 88.6 | 89 | 95.8 | 85.2 | 85.6 | 87.8 | 89.4 | 88.2 | 97.3 | 86.7 | 87.8 | 93.9 | 98.5 | 92.8 | 87.8 | 93.2 | |
|
KU646850 Malaysia 2015 UPM53 |
89.4 | 89.4 | 89.7 | 94.7 | 85.2 | 85.6 | 89.4 | 90.9 | 89.7 | 92.4 | 88.2 | 89.4 | 99.6 | 92.8 | 94.7 | 89.4 | 98.5 | 93.5 |
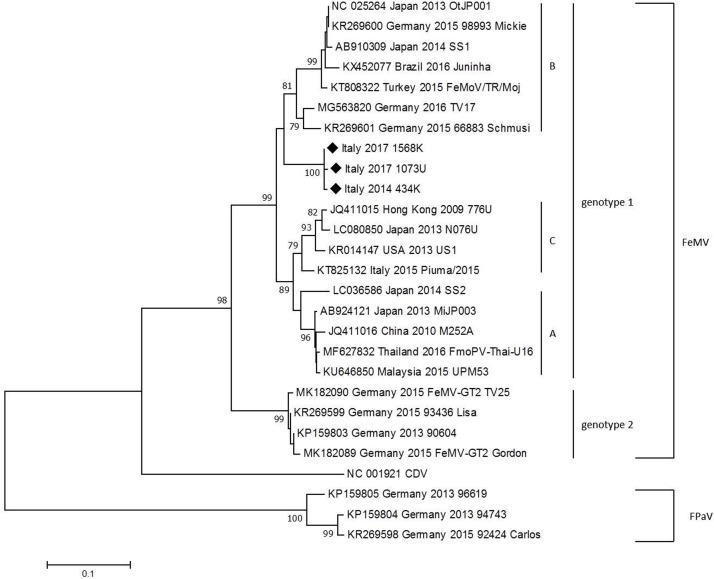
The FeMV L gene phylogenetic tree based on Neighbour-Joining method is reported in Fig. 1 . Phylogenetic analysis of the three FeMV-positive cats showed that the three strains formed a significant sub-clade that clustered with the majority of FeMV strains retrieved from public database and did not cluster with the newly described feline paramyxoviruses identified in Germany and proposed as new paramyxoviruses (FpaV) (Sieg et al., 2015). Two highly significant clusters of FeMV, one with the majority of FeMV sequences and the other with sequences reported in Germany and proposed as two different FeMV genotypes were observed, as previously reported (Sieg et al., 2015, 2019). Phylogenetic analysis also showed, beside the significative sub-clade of the three strains isolated in this study, three sub-clades A, B and C previously described. The only other strain previously reported in Italy (GenBank accession number KT825132) clusters with the sub-clade C (Marcacci et al., 2016; Park et al., 2016). Results of the phylogenetic analysis are similar using Maximum Likelihood analyses (data not shown).
Fig. 1.
Phylogenetic tree generated with NJ analysis.
A 401 bp region of the L gene obtained from FeMV, CDV and feline paramyxoviruses sequences retrieved from GenBank databases and the three sequences obtained in this study was used. Sequences are indicated by GenBank accession number (available at http://www.ncbi.nlm.nih.gov/pubmed/), country, year of origin and name of the strain. Distances were computed using the Kimura 2-parameter model. Bootstrap values above 70% are given. The symbol ◆ indicates sequences obtained in this study. A, B, C: FeMV sub-clusters described by Park et al (2016); genotype 1 and genotype 2: FeMV genotypes proposed by Sieg et al (2019).
4. Discussion
The 89–92% nucleotide identity of the FeMV strains obtained in this study with other FeMV sequences deposited in the GenBank database supports their classification as FeMVs. A nucleotide identity above 84% is consistent with the classification within the same viral species (Kuno et al., 1998).
Data of this study showed an overall prevalence of FeMV similar to previously reported prevalences, but the prevalence in urine was slightly lower compared with other studies (Furuya et al., 2014; Yilmaz et al., 2017; Park et al., 2016). This finding could be explained by the population of cats included in the current study, since it is believed that stray cats are more easily infected, and most of the cats included in the current study were client-owned (Furuya et al., 2014). In fact, when considering the FeMV prevalence on the population of stray cats included in this study, which should also be considered when evaluating the circulation of a virus in a given geographic area, the prevalence increases to 16.7% (1/6: 95% CI: 0.42–64.12%).
Another explanation to the lower prevalence recorded in the current study compared with published data could be the presence of viral RNA in low titres and the low analytical sensitivity of standard nRT-PCR compared to real time RT-PCR. It was demonstrated that samples with low titres of FeMV RNA can result positive at quantitative PCR (qPCR) but negative at RT-PCR (De Luca et al., 2018; Furuya et al., 2015). However, this hypothesis seems unlikely since previous studies reporting slightly higher prevalences were done with conventional PCR. Moreover, the overall prevalence recently obtained with qPCR was only slightly higher compared whit the prevalence registered with conventional PCR (De Luca et al., 2018). Unfortunately, we were not able to perform serological tests on the cats used in this study, therefore no data about the seroprevalence are available. Nevertheless, based on other studies reporting a higher rate of seropositivities in FeMV RNA positive cats, it can be assumed that a low seroprevalence would have been recorded, but this needs to be investigated in further studies (Woo et al., 2012; McCallum et al., 2018). Further studies should also evaluate the presence of other unrelated feline paramyxoviruses (FPaV) previously described in cats (Sieg et al., 2015), that were not investigated in this study.
No association between CKD was apparently found in this study. This finding is in contrast with previous studies reporting a correlation between FeMV and CKD, but in line with more recent studies demonstrating no clear relationships between FeMV infection and CKD (Furuya et al., 2014; Sieg et al., 2015; Yilmaz et al., 2017; McCallum et al., 2018).
In particular, none of the cats with a diagnosis of CKD or clinicopathological signs consistent with CKD was FeMV positive in urine. Similarly, none of the cats with histological signs of tubulointerstitial nephritis was FeMV positive in the kidney.
The cat with FeMV in the urine was affected by cholangiohepatitis. Long-term shedding of FeMV through the urine has been reported, showing similarity between FeMV in cats and canine distemper virus in dogs (Elia et al., 2015). The analysis of a follow up sample demonstrates that the cat either stopped to shed the virus or intermittently shed the virus, as described in a recent study evaluating the viral RNA shedding within a 110 days’ time span; although a previous study demonstrated the persistent shedding of FeMV RNA within a two-years period (Sharp et al., 2016; De Luca et al., 2018). However, the above-mentioned study evaluated the type of shedding in one cat only, thus more information on this issue are needed. The detection of FeMV in a cat with cholangitis suggests designing future studies to investigate whether the virus may be involved in the pathogenesis of hepatic disorders. The presence of FeMV in the liver of infected cats, most of which were affected by cholangiohepatitis, was already demonstrated through immunohistochemistry (Yilmaz et al., 2017). Moreover, studies in vitro demonstrated that FeMV may have tropism for several cellular lines therefore being potentially able to infect different organs (Sakaguchi et al., 2015). The cat with FeMV-positive urine had no medical requirements to perform a kidney biopsy. Therefore, it was not possible to rule out histological signs of tubular damage and to correlate FeMV infection with kidney damage. Similarly, the rapid recovery of symptoms associated with liver diseases hampered the possibility to further investigate through biopsies or other techniques the possible presence of the virus within the hepatic tissues.
The absence of tubulointerstitial nephritis in the kidneys positive for FeMV nRT-PCR suggests, in accordance with recent studies, that FeMV should not be considered as a specific cause of TIN. On the other hand, the presence of FeMV viral RNA in the two positive kidneys could suggest that FeMV is possibly able to cause lesions other than tubulointerstitial nephritis.
In fact, while granulomatous lesions are known to be typical of FIP, and therefore FeMV and FCoV infection may coexist, it is difficult to find a correlation between granulomatous lesions and pulmonary adenocarcinoma in the other FeMV positive cat. The design of this study does not allow to find a correlation between FeMV infection and granulomatous inflammation, both because of the scarce number of kidneys examined and because it was not possible to perform FeMV immunohistochemistry to investigate the presence of viral antigen inside the inflammatory lesions. However, in other studies based on immunohistochemistry, the FeMV antigen has been observed in the cytoplasm of tubular cells. It would be interesting to study whether the morbillivirus may be observed in other cellular lines of infected kidneys, as it is known that FeMV is also capable of infecting monocytic/macrophagic line cells, which are abundant within granulomatous lesions (Woo et al., 2012).
It is also interesting to notice that the only two FeMV positive cats did not show the typical pattern of interstitial nephritis thought to be associated with FeMV infection according to Woo et al (2012), while many cats (13/27) had histological patterns consistent with interstitial nephritis but were FeMV negative. Unfortunately, urinary bladders were not collected, but it would have been interesting to evaluate FeMV presence also in this organ.
Regarding the FeMV positive FIP-cat, the fact that all the other cats affected by FIP were FeMV PCR negative supported the hypothesis that the association between FeMV infection and FIP was likely a casual finding.
Although the amount of sequences publicly available is relatively low, it is interesting to note that the first FeMV identified in Italy was from a sample collected in 2014, only few years after the first identification of the virus in Hong Kong and one year before the first descriptions of FeMV in Italy (Woo et al., 2012; Lorusso et al., 2015). Phylogeographical analyses are suggested to help understand the origin and epidemiological distribution pattern of the virus.
Phylogeny showed that the three strains of this study were grouped in a separated and specific sub-cluster, within the same FeMV proposed genotype (Fig. 1). Therefore, it appears that there are at least two defined sub-clusters of FeMV in Italy: one with the first FeMV strain isolated from Lorusso et al (2015), and the second containing the three sequences obtained in this study. This finding is in accordance with the presence of different sub-clusters previously reported in the same geographic area (Sieg et al., 2015; Park et al., 2016).
It would be interesting in the future to investigate whether different FeMV sub-clusters of the same geographic area also differ in their pathogenetic behavior. For example, it should be determined if there are differences in the tropism of the virus or in the timespan of shedding related to the geographic location. However, these studies would be possible only with experimental infections of cats.
5. Conclusion
Based on our results, feline morbillivirus is scarcely present in client-owned cats of Northern Italy, while the prevalence in stray cats seems higher but needs to be confirmed on a higher number of cats. The recorded data suggests the presence of a FeMV sub-cluster well distinct from the strain previously isolated in southern Italy. Possible differences in the behavior of these strains may be suggested and need to be investigated in the future. As our findings did not correlate FeMV infection with renal disorders, the possible role of the virus as one triggering factor of a disease with such a multifactorial pathogenesis as CKD, remains uncertain.
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
All the procedures reported on this study were performed within routine diagnostic workouts. According to the decisions of the Ethical Committee of the University of Milan, residual aliquots of samples or tissues collected under informed consent of the owners can be used for research purposes without any additional formal request of authorization to the Ethical Committee.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or
not-for-profit sectors.
Acknowledgements
We thank Dr. Valentina Gualdi, Parco Tecnologico Padano, Lodi, for her support in the sequencing of FeMV L gene PCR products.
Contributor Information
Angelica Stranieri, Email: angelica.stranieri@unimi.it.
Stefania Lauzi, Email: stefania.lauzi@unimi.it.
Annachiara Dallari, Email: annachiara.dallari@outlook.it.
Maria Elena Gelain, Email: mariaelena.gelain@unipd.it.
Federico Bonsembiante, Email: federico.bonsembiante@unipd.it.
Silvia Ferro, Email: silvia.ferro@unipd.it.
Saverio Paltrinieri, Email: saverio.paltrinieri@unimi.it.
References
- Barker E.N., Stranieri A., Helps C.R., Porter E.L., Davidson A.D., Day M.J., Knowles T., Kipar A., Tasker S. Limitations of using feline coronavirus spike protein gene mutations to diagnose feline infectious peritonitis. Vet. Res. 2017;48:60–74. doi: 10.1186/s13567-017-0467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.A., Elliott J., Schmiedt C.W., Brown S.A. Chronic kidney disease in aged cats: clinical features, morphology, and proposed pathogeneses. Vet. Pathol. 2016;53:309–326. doi: 10.1177/0300985815622975. [DOI] [PubMed] [Google Scholar]
- Darold G.M., Alfieri A.A., Muraro L.S., Amude A.M., Zanatta R., Yamauchi K.C.I., Alfieri A.F., Lunardi M. First report of feline morbillivirus in South America. Arch. Virol. 2017;162:469–475. doi: 10.1007/s00705-016-3124-0. [DOI] [PubMed] [Google Scholar]
- De Luca E., Crisi P.E., Di Domenico M., Malatesta D., Vincifori G., Di Tommaso M., Di Guardo G., Di Francesco G., Petrini A., Savini G., Boari A., Lorusso A. A real-time RT-PCR assay for molecular identification and quantitation of feline morbillivirus RNA from biological specimens. J. Virol. Methods. 2018;258:24–28. doi: 10.1016/j.jviromet.2018.05.002. [DOI] [PubMed] [Google Scholar]
- de Vries R.D., Duprex W.P., de Swart R.L. Morbillivirus infections: an introduction. Viruses. 2015;7:699–706. doi: 10.3390/v7020699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia G., Camero M., Losurdo M., Lucente M.S., Larocca V., Martella V., Decaro N., Buonavoglia C. Virological and serological findings in dogs with naturally occurring distemper. J. Virol. Methods. 2015;213:127–130. doi: 10.1016/j.jviromet.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Furuya T., Sassa Y., Omatsu T., Nagai M., Fukushima R., Shibutani M., Yamaguchi T., Uematsu Y., Mizutani T. Existence of feline morbillivirus infection in Japanese cat populations. Arch. Virol. 2014;159:371–373. doi: 10.1007/s00705-013-1813-5. [DOI] [PubMed] [Google Scholar]
- Furuya T., Wachi A., Sassa Y., Omatsu T., Nagai M., Fukushima R., Shibutani M., Yamaguchi T., Uematsu Y., Shirota K., Mizutani T. Quantitative PCR detection of feline morbillivirus in cat urine samples. J. Vet. Medl. Sci. 2015;77:1701–1703. doi: 10.1292/jvms.15-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G., Chang G.J.J., Tsuchiya K.R., Karabatsos N., Cropp C.B. Phylogeny of the genus flavivirus. J. Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Di Tommaso M., Di Felice E., Zaccaria G., Luciani A., Marcacci M., Aste G., Boari A., Savini G. First report of feline morbillivirus in Europe. Vet. Ital. 2015;51:235–237. doi: 10.12834/VetIt.833.4136.2. [DOI] [PubMed] [Google Scholar]
- Marcacci M., De Luca E., Zaccaria G., Di Tommaso M., Mangone I., Aste G., Savini G., Boari A., Lorusso A. Genome characterization of feline morbillivirus from Italy. J. Virol. Methods. 2016;234:160–163. doi: 10.1016/j.jviromet.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum K.E., Stubbs S., Hope N., Mickleburgh I., Dight D., Tiley L., Williams T.L. Detection and seroprevalence of morbillivirus and other paramyxoviruses in geriatric cats with and without evidence of azotemic chronic kidney disease. J. Vet. Intern. Med. 2018;32:1100–1108. doi: 10.1111/jvim.15097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambulli S., Sharp C.R., Acciardo A.S., Drexler J.F., Duprex W.P. Mapping the evolutionary trajectories of morbilliviruses: what, where and whither. Curr. Opin. Virol. 2016;16:95–105. doi: 10.1016/j.coviro.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.S., Suzuki M., Kimura M., Mizutani H., Saito R., Kubota N., Hasuike Y., Okajima J., Kasai H., Sato Y., Nakajima N., Maruyama K., Imaoka K., Morikawa S. Epidemiological and pathological study of feline morbillivirus infection in domestic cats in Japan. BMC Vet. Res. 2016;12:228–239. doi: 10.1186/s12917-016-0853-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter E., Tasker S., Day M.J., Harley R., Kipar A., Siddell S.G., Helps C.R. Amino acid changes in the spike protein of feline coronavirus correlate with systemic spread of virus from the intestine and not with feline infectious peritonitis. Vet. Res. 2014;45:49–60. doi: 10.1186/1297-9716-45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Koide R., Miyazawa T. In vitro host range of feline morbillivirus. J. Vet. Med. Sci. 2015;77:1485–1487. doi: 10.1292/jvms.15-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp C.R., Nambulli S., Acciardo A.S., Rennick L.J., Drexler J.F., Rima B.K., Williams T., Duprex W.P. Chronic infection of domestic cats with feline morbillivirus, United States. Emerg. Infect. Dis. 2016;22:760. doi: 10.3201/eid2204.151921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieg M., Heenemann K., Rückner A., Burgener I., Oechtering G., Vahlenkamp T.W. Discovery of new feline paramyxoviruses in domestic cats with chronic kidney disease. Virus Genes. 2015;51:294–297. doi: 10.1007/s11262-015-1232-7. [DOI] [PubMed] [Google Scholar]
- Sieg M., Busch J., Eschke M., Böttcher D., Heenemann K., Vahlenkamp A., Reinert A., Seeger J., Heilmann R., Scheffler K., Vahlenkamp T.W. A new genotype of feline morbillivirus infects primary cells of the lung, kidney, brain and peripheral blood. Viruses. 2019;11:E146. doi: 10.3390/v11020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wong B.H., Fan R.Y.Y., Wong A.Y.P., Zhang A.J.X., Wu Y., Choi G.K.Y., Li K.S.M., Huie J., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5435–5440. doi: 10.1073/pnas.1119972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz H., Tekelioglu B.K., Gurel A., Bamac O.E., Ozturk G.Y., Cizmecigil U.Y., Tarakci E.A., Aydin O., Yilmaz A., Berriatua E., Helps C.R., Richt J.A., Turan N. Frequency, clinicopathological features and phylogenetic analysis of feline morbillivirus in cats in Istanbul, Turkey. J. Fel. Med. Surg. 2017;19:1206–1214. doi: 10.1177/1098612X16686728. [DOI] [PMC free article] [PubMed] [Google Scholar]