Dear Editor,
Most recently, Yuen and colleagues have prospected, in this journal, that the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) may be the beginning of another SARS-like pandemic and the research preparedness against this potential pandemic is an important precautionary strategy.1
The rapid identification of HCoV-EMC that caused a SARS-like disease in Saudi Arabia2 is attributed to the success in discovery of the SARS coronavirus (SARS-CoV).3 Therefore, the knowledge gained from the research on SARS-CoV and the structures of its spike (S) protein may provide a useful template for identifying receptor for HCoV-EMC and developing vaccines against HCoV-EMC.4
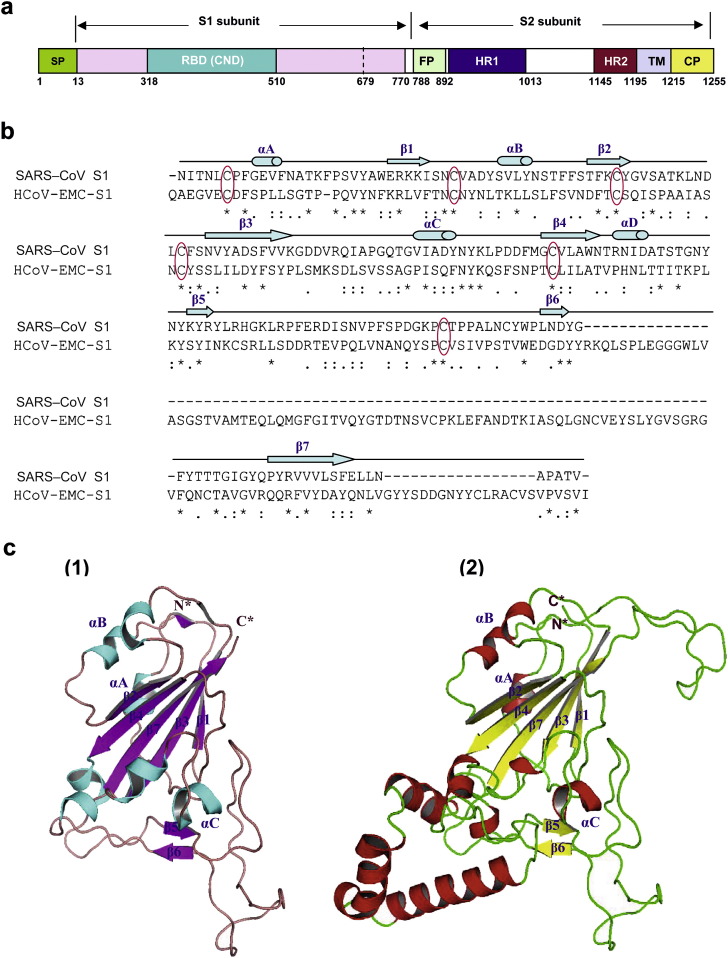
SARS-CoV S protein consists of S1 and S2 subunits (Fig. 1 a). The S1 subunit contains the receptor-binding domain (RBD, residues 318–510) responsible for its binding to the angiotensin-converting enzyme 2 (ACE2) receptor.5 We previously demonstrated that the RBD is also a critical neutralizing domain (CND), which could induce highly potent neutralizing antibody responses in the immunized animals and protect against SARS-CoV challenge.6, 7 Therefore, the immunogen containing this CND is expected to be effective SARS vaccine candidates.8
Figure 1.
Prediction of the RBD/CND in the HCoV-EMC S protein S1 subunit based on the RBD in SARS-CoV S protein. (a) Schematic representation of the SARS-CoV S protein. SP, signal peptide; RBD, receptor-binding domain; CND, critical neutralizing domain; FP, fusion peptide; HR, heptad repeat; TM, transmembrane domain; and CP, cytoplasm domain. The residue numbers of each region represent their positions in the S protein of SARS-CoV. (b) Alignment analysis of the sequence of the RBD/CND (residues 321–508) in the SARS-CoV S protein9 with the corresponding region (residues 377–662) in the HCoV-EMC S protein. The secondary structure assignments are listed above the primary sequence with β-sheets highlighted as arrows and α-helices highlighted by cylinders, respectively.9 The conserved cysteines are highlighted with red circles. (c) Crystal structures of the RBD/CND in SARS-CoV S protein S1 subunit9 (1) and predicted structure of RBD/CND in HCoV-EMC S protein S1 subunit (2). A core consists of a five-stranded anti-parallel β-sheet (β1–β4, β7) connecting with three short α-helices (αA–αC), and an extended loop contains two-stranded β-sheet (β5, β6). N* and C* stand for the N- and C-termini of RBD/CND, respectively.
Sequence alignment of the RBD/CND in SARS-CoV S with that of the corresponding region (residues 377–662) in HCoV-EMC S protein revealed that both fragments have low homology (14% identity and 38% similarity). However, the core domain consisting of β-sheets and α-helices in both fragments have higher homology (23% identity and 61% similarity). Strikingly, six cysteines are located at the same sites in both fragments (Fig. 1b), suggesting that they share conserved conformational structures.
Based on the X-ray crystal structure of the RBD/CND domain in the SARS-CoV S protein (PDB id: 2DD8),9 the structure of the corresponding region in the HCoV-EMC S protein was predicted using the Swiss-Model Workplace homology modeling server.10 The results indicate that like the RBD/CND domain in the SARS-CoV S protein,9, 11 the fragment of residues 377–662 in HCoV-EMC S protein also contains a core domain consisting of 5 β-sheets (β1–β4, β7) and 3 α-helices (αA–αC) and a long extended loop containing 2 anti-parallel β-sheets (β5–β6) (Fig. 1c). It has been demonstrated that the core in the RBD/CND domain of the SARS-CoV S protein is responsible for maintaining the overall conformation of the protein, while the extended loop is responsible for its binding with the receptor ACE2 or a neutralizing antibody.9, 11 These findings suggest that the region (residues 377–662) in HCoV-EMC S protein may also serve as a RBD/CND and can be used as a probe to identify HCoV-EMC's receptor and as an immunogen to design vaccines to prevent HCoV-EMC infection.
Potential conflicts of interest
No reported conflicts.
Acknowledgments
SJ was supported by funding from 973 Programme of China (#2012CB519001). LL was supported by “Chen Guang” Project of SMEC and SEDF (11CG03).
References
- 1.Chan J.F.W., Li K.S.M., To K.K.W., Cheng V.C.C., Chen H., Yuen K.Y. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J Infect. 2012;65:477–489. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Anderson L.J., Baric R.S. Emerging human coronaviruses: disease potential and preparedness. N Engl J Med. 2012;367:1850–1852. doi: 10.1056/NEJMe1212300. [DOI] [PubMed] [Google Scholar]
- 4.Butler D. SARS veterans tackle coronavirus. Nature. 2012;490:20. doi: 10.1038/490020a. [DOI] [PubMed] [Google Scholar]
- 5.Li W.H., Moore M.J., Vasilieva N., Sui J.H., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y.X., Zhou Y.S., Liu S.W., Kou Z.H., Li W.H., Farzan M. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y., Li J., Du L., Yan X., Hu G., Zhou Y. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: revealing the critical antigenic determinants in inactivated SARS-CoV vaccine. Vaccine. 2006;24:5498–5508. doi: 10.1016/j.vaccine.2006.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV: a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 11.Li F., Li W.H., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]