Graphical abstract
The presence of a quinone-methide moiety appears to play a relatively significant role in 3CLpro inhibition.
Abbreviations: IC50, the inhibitor concentration leading to 50% activity loss; Ki, inhibition constant; Km, Michaelis–Menten constant; Vmax, maximum velocity; NMR, nuclear magnetic resonance
Keyword: SARS-CoV, 3CLpro, Tripterygium regelii, Quinone-methide, Celastrol, Iguesterin
Abstract
Quinone-methide triterpenes, celastrol (1), pristimerin (2), tingenone (3), and iguesterin (4) were isolated from Triterygium regelii and dihydrocelastrol (5) was synthesized by hydrogenation under palladium catalyst. Isolated quinone-methide triterpenes (1–4) and 5 were evaluated for SARS-CoV 3CLpro inhibitory activities and showed potent inhibitory activities with IC50 values of 10.3, 5.5, 9.9, and 2.6 μM, respectively, whereas the corresponding 5 having phenol moiety was observed in low activity (IC50 = 21.7 μM). As a result, quinone-methide moiety in A-ring and more hydrophobic E-ring assist to exhibit potent activity. Also, all quinone-methide triterpenes 1–4 have proven to be competitive by the kinetic analysis.
The worldwide outbreak of the life-threatening disease severe acute respiratory syndrome (SARS) was caused by infection with SARS-CoV as a novel coronavirus (CoV).1, 2 The genome of SARS-CoV contains 11–14 major annotated open reading frames including those predicted to encode for known viral proteins, including the replicase polyproteins, S (spiked protein), polymerase, M (membrane protein), N (nucleocapsid protein), and E (small envelope protein).2 The replicase polyproteins ppla and pplb encoded by the SARS-CoV are extensively processed to yield the functional subunits for successful viral propagation.3 The SARS-CoV 3CLpro, which is also called main protease (Mpro) mediates the proteolytic processing of replicase polypeptides into functional proteins, plays an important role in viral replication. The active site of SARS-CoV 3CLpro has a catalytic dyad with the sulfur of Cys145 as a nucleophile and the imidazole ring of His41 as a general base.4 Therefore, the SARS-CoV 3CLpro can be considered an attractive target for developing effective drugs against SARS.
To date, a large number of inhibitors of 3CLpro have been studies, including C2-symmetric diols,5 isatin derivatives,6 anilides,7 and thiazolyl ketone-containing peptidic compounds.8 Moreover, some groups have reported that triterpenoids such as betulinic acid from Juniperus formosana 9 and glycyrrhetinic acid from Glycyrrhiza glabra and their derivatives could inhibit 3CLpro.10
During our search for novel SARS-CoV 3CLpro inhibitors from medicinal plants, we found that the MeOH (95%) extracts from the bark of Tripteryguim regelii (Celastraceae) can remarkably inhibit SARS-CoV 3CLpro activity (>70% inhibition at 30 μg/mL). Subsequent bioactivity-guided fractionation of the CHCl3 extracts led to the isolation of four quinone-methide triterpenoid derivatives11 that were identified as celastrol (1), pristimererin (2), tingenone (3), and iguesterin (4) on the basis of their spectroscopic analyses (Fig. 1 ).12 T. regelii is a woody vine native to Eastern and Southern China, Korea, Japan, and Taiwan.13 This plant known as ‘Thunder God Vine’ has been historically used in traditional Chinese medicine to treat inflammatory and autoimmune diseases, such as rheumatoid arthritis (RA), systemic lupus erythematosus, psoriatic arthritis, and Behcet’s disease.14 A number of triterpenes, diterpenes, and sesquiterpenes as a major compound have been isolated from the bark of T. regelii and are known to have a variety of biological activities with good cytotoxicity to numerous cancer cell lines and efficacy in several rodent models of arthritis and other inflammatory disease.15 However, their SARS-CoV 3CLpro inhibitory activities have not been reported. In this study, we wish to describe the SARS-CoV 3CLpro inhibitory activities of the four quinone-methide triterpene derivatives 1–4 and semi-synthetic dihydrocelastrol 5.
Figure 1.
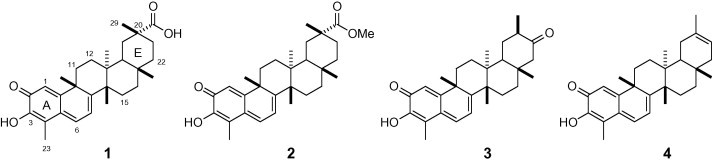
Chemical structures of isolated quinone-methide triterpenes (1–4) from T. regelii.
The enzymatic activity of SARS-CoV 3CLpro was measured by a modification of the method described by Kuo et al.16 The biological activities of 1–5 were assessed against SARS-CoV 3CLpro and confirmed by the positive control with curcumin9 which inhibited SARS-CoV 3CLpro with IC50 value of 23.5 μM. We found that the IC50 values of the 1–5 ranged from 2.6 to 21.7 μM against SARS-CoV 3CLpro (Table 1 ) and that all were dose-dependent (Fig. 2 ). Celasterol (1) having acid moiety at C-20 inhibited SARS-CoV 3CLpro with IC50 value of 10.3 μM. Pristimerin (2) substituted with methyl ester group at C-20 inhibited SARS-CoV 3CLpro with twofold greater potency (IC50 = 5.5 μM) compared to celasterol (1), whereas ketone-substituted analogue, tingenone (3), at C-21 showed an attenuated potency with IC50 value of 9.9 μM. Iguesterin (4) existing double bond in E-ring showed potent inhibitory activity with IC50 value of 2.6 μM.
Table 1.
Inhibitory effects of 1–5 on SARS-CoV 3CLpro
| Entry | IC50a (μM) | Inhibition type (Ki, μM) |
|---|---|---|
| 1 | 10.3 ± 0.2 | Competitive (4.2 ± 0.6) |
| 2 | 5.5 ± 0.7 | Competitive (3.1 ± 0.0) |
| 3 | 9.9 ± 0.1 | Competitive (4.0 ± 0.1) |
| 4 | 2.6 ± 0.3 | Competitive (0.8 ± 0.2) |
| 5 | 21.7 ± 1.9 | Not tested |
| Curcuminb | 23.5 ± 3.7 | Not tested |
All compounds were examined in a set of experiments repeated three times; IC50 values of compounds represent the concentration that caused 50% enzyme activity loss.
Curcumin was used as a positive control.
Figure 2.
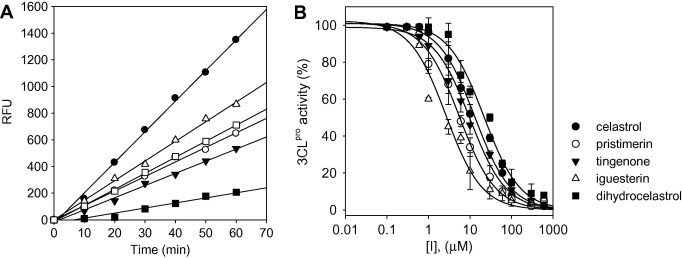
(A) Time course of hydrolysis of substrate by SARS-CoV 3CLpro in the presence of compounds 1–5 at 10 μM. Concentrations of compounds for lines from top to bottom were control (●), 5 (△), 1 (□), 3 (○), 2 (▾), and 4 (■), respectively. (B) Effects of compounds 1–5 on the activity of SARS-CoV 3CLpro.
In order to confirm whether quinone-methide moiety plays to inhibit SARS-CoV 3CLpro, celastrol (1) was reacted by dehydrogenation under Pd catalyst to give the dihydrocelastrol (5) in a quantitative yield, as shown in Scheme 1 . According to Schwalbe’s result,17 celastrol (1) possesses electrophilic sites within the A and B rings, where nucleophilic groups of cysteine thiol group of cell division cycle protein 37 (Cdc37) reacts with the quinone-methide of 1 through a Michael addition, resulting in the formation of an adduct at C6. Thus, we believe that this moiety would be valuable for enhancing the inhibitory activity of 3CLpro. In fact, our result showed that quinone-methide moiety of 1 (IC50 = 10.3 μM) increased the potency of SARS-CoV 3CLpro inhibition by twofold when compared with the reduced analogue 5 (IC50 = 21.7 μM) (Table 1). In view of this result, the presence of a quinone-methide moiety appears to play a relatively significant role in inhibition.
Scheme 1.
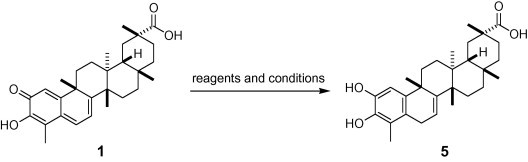
Reagents and conditions: 10% Pd/C, EtOH, H2 gas, AcOH (cat.), rt.
We then further characterized the inhibitory mechanism of the isolated quinone-methide triterpenes against SARS-CoV 3CLpro activity. This began with analysis of the mode of inhibition using both Lineweaver–Burk and Dixon plots. As shown representatively in Figure 3 , the kinetic plots show that compound 4 has a competitive inhibition mode of action. Because the Lineweaver–Burk plot of 1/V versus 1/[S] result in a family of straight lines with the same y-axis intercept, V max (44.4 ± 6.2 intensity min−1), respectively, for the SARS-CoV 3CLpro inhibitor 4 (Fig. 3A). All isolated inhibitors manifested the same inhibition mode of action. The K i value of the most potent compound 4 was determined to be 0.8 μM (Table 1), as found from the common x-axis intercept of lines on the corresponding Dixon plot (Fig. 3B).
Figure 3.
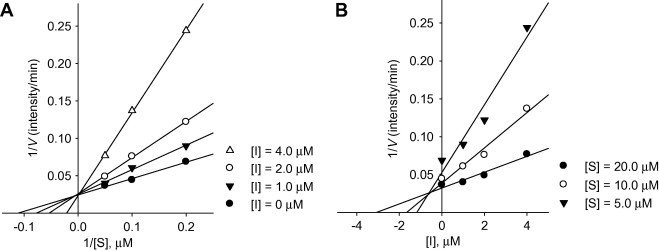
(A) Lineweaver–Burk plot for 3CLpro inhibition by compound 4. (B) Dixon plot for compound 4 determining the inhibition constant Ki.
To better investigate the interaction of these inhibitors with the enzyme, we attempted a molecular docking simulation of the binding of 4 and 5 into SARS-CoV 3CLpro active site. The X-ray structure of the SARS-CoV 3CLpro in complex with a substrate-analogue inhibitor (coded 1uk4 or 2z3e)18, 19 obtained from the Protein Data Bank (PDB; http://www.rcsb.org/pdb/) was used for modeling analysis. Computer docking analysis20 revealed that iguesterin (4) can fit well into the substrate-binding pocket of SARS-CoV 3CLpro. As shown in Figure 4 A, the hydroxyl group of C3 of 4 formed a hydrogen bond with the oxygen atom of the carbonyl group of Cys 44 and OH of Thr 25 located in domain I (residues 8–101),9 whereas dihydrocelastrol (5) did not form any intermolecular bonds with the enzyme besides the hydrophobic interaction (Fig. 4B). Iguesterin (4) binding mode has different from betulinic acid (His164 and Thr24) and savinin (Cys145, Gln166, Gln189, Gly143 and Ser144) with 1uk4. These differences of intermolecular interaction apparently are reflected by the 10-fold smaller IC50 value of iguesterin (4) compared with that of reference compounds in inhibiting the 3CLpro. Moreover, the calculated binding energy (kcal/mol)19 of all quinone-methide triterpenes are shown in Table 2 . These docking experiments support the observation in the enzymatic assay, which reveals the important roles of the quinone-methide moieties inhibition of the SARS-CoV 3CLpro. Although this preliminary molecular modeling study cannot be final proof for this critical argument, it constitutes a positive sign for the possibility of the mechanism.
Figure 4.
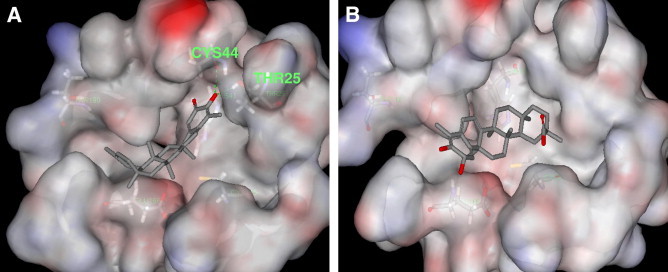
Computer modeling of compounds 4 (A) and 5 (B) binding to SARS-CoV 3CLpro (1uk4).
Table 2.
The calculated binding energy values of 1–5 on SARS-CoV 3CLpro
| Entry | Docking energy (kcal/mol) |
|---|---|
| 1 | −9.58 |
| 2 | −9.87 |
| 3 | −9.75 |
| 4 | −9.97 |
| 5 | −9.18 |
In summary, we have identified quinone-methide triterpene derivatives as novel SARS-CoV 3CLpro inhibitors. The newly identified bioactive compounds with SARS-CoV 3CLpro inhibitory activity in the μM range include celastrol (1), pristimerin (2), tingenone (3) and iguesterin (4). Although all isolated quinone-methide triterpenes has previously been known compounds, this is the first time it has been shown to possess SARS-CoV 3CLpro inhibitory activity. We are hopeful that these preliminary data may be able to open new avenues for research targeted toward reducing the threat of SARS to the global community. Further research and drug development on quinone-methide triterpenes as promising SARS-CoV 3CLpro inhibitors are ongoing in our laboratory and the results will be reported in due course.
Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF) (No. 2009-0081749) grant funded by the Korea government (MOST) and a KRIBB Research Initiative Program, Republic of Korea.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2010.01.152.
Contributor Information
Mun-Chual Rho, Email: Rho-m@kribb.re.kr.
Woo Song Lee, Email: wslee@kribb.re.kr.
Supplementary data
References and notes
- 1.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., De Risi J., Yamg J.Y., Cox N., Hughes J.M., Le Duc J.W., Bellini W.J., Anderson L.J. N. Eng. J. Med. 2003;348:1953. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., can Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Lancet. 2003;362:263. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.-H., Tong S., Tamin A., Lowe L., Frace M., De Risi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D.H.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Anand K., Ziebuhr J., Wadhwang P., Mesturs J.R., Hilgenfield R. Science. 2003;300:1763. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 5.Wu C.-Y., Jan J.-T., Ma S.-H., Kuo C.-J., Juan H.-F., Cheng E.Y.-S., Hsu H.-H., Huang H.C., Wu D., Brik A., Liang F.-S., Liu R.-S., Fang J.-M., Chen S.-T., Liang P.H., Wong C.-H. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10012. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L.-R., Wang Y.-C., Lin Y.-W., Chou S.-Y., Chen S.-F., Liu L.-T., Wu Y.-T., Kuo C.-J., Chen T.S.-S., Juang S.-H. Bioorg. Med. Chem. Lett. 2005;15:3058. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shie J.-J., Fang J.-M., Kuo T.-H., Kuo C.-J., Liang P.-H., Huang H.-J., Yang W.-B., Lin C.-H., Chen J.-L., Wu Y.-T., Wong C.-H. J. Med. Chem. 2005;48:4469. doi: 10.1021/jm050184y. [DOI] [PubMed] [Google Scholar]
- 8.Regnier T., Sarma D., Hidaka K., Bacha U., Freire E., Hayashi Y., Kiso Y. Bioorg. Med. Chem. Lett. 2009;19:2722. doi: 10.1016/j.bmcl.2009.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen C.-C., Kuo Y.-H., Jan J.-T., Liang P.-H., Wang S.-Y., Liu H.-G., Lee C.-K., Chang S.-T., Kuo C.-J., Lee S.-S., Hou C.-C., Hsiao P.-W., Chien S.-C., Shyur L.-F., Yang N.-S. J. Med. Chem. 2007;50:4087. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 10.Hoever G., Baltina L., Michaelis M., Kondratenko R., Baltina L., Tolstikov G.A., Doerr H.W., Cinatl J., Jr. J. Med. Chem. 2005;48:1256. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- 11.The stem root (5 kg) of T. regelii were air-dried, chopped and extracted three times with 95% MeOH (3 × 10 L) for 7 days at room temperature. The combined extract was concentrated, and the dark residue (453 g) was partitioned between water and chloroform (1 L:1 L). The organic layer was washed with brine, dried over anhydrous Na2SO4, and concentrated to give a dark brown residue (310 g) which was chromatographed on a silica gel column. The resultant extract (310 g) was suspended in H2O (2 × 1 L). The diluted with H2O has been partitioned with organic solvents (CHCl3 and BuOH) of the different polarities to afford CHCl3 (162 g), BuOH (125 g), and H2O (150 g) extracts, respectively. The CHCl3 and BuOH extracts were subjected to column chromatography using silica gel with hexane–acetone gradient and hexane–EtOAc gradient. The CHCl3 extract (162 g) was subjected to column chromatography (glass column 10 × 80 cm) over silica gel (1.5 kg; 70–230 mesh; Merk), eluted with gradient mixtures of hexane (4 L) and hexane–acetone, of increasing polarity (30/1→1/6), and finally with MeOH. Fifteen pooled fractions (fr.1–fr.30) were obtained after combining fractions with similar TLC profiles from this initial column chromatography. The column was eluted with solvents of increasing polarity (CHCl3–acetone) to give 30 fractions. Fraction 5 (14.3 g) was chromatographed on a silica gel column with hexane–acetone to give 45 fractions (5.1–5.30). The fractions from 5.11–5.18 and 5.21–32.27 were evaporated to give compounds 1 (280 mg) and compound 3 (100 mg), respectively. Fraction 10 (21.6 g) was chromatographed on a silica gel column and eluted with hexane–EtOAc to give 23 fractions (10.1–10.23). The fractions 10.6–10.14 were combined and evaporated to give 120 mg of compound 2 and compound 4 (78 mg).
- 12.(a) Lee B.W., Seo W.D., Gal S.W., Yang M.S., Park K.H. Agric. Chem. Biotechnol. 2004;47:77. [Google Scholar]; (b) Morita H., Hirasawa Y., Muto A., Yoshida T., Sekita S., Shirota O. Bioorg. Med. Chem. Lett. 2008;18:1050. doi: 10.1016/j.bmcl.2007.12.016. [DOI] [PubMed] [Google Scholar]; (c) Gonzalez A.G., Fraga B.M., Gonzalez C.M., Ravelo A.G., Ferro E. Phytochemistry. 1975;14:1067. [Google Scholar]
- 13.Ma J.-S., Brach A.R., Liu Q.-R. Edinburgh J. Bot. 1999;56:33. [Google Scholar]
- 14.Xie D.E. Zong Xi Yi Jie He Za Zhi. 1983;3:349. [PubMed] [Google Scholar]
- 15.(a) Kim D.H., Shin E.K., Kim Y.H., Lee B.W., Jun J.-G., Park J.H.Y., Kim J.-K. Eur. J. Clin. Invest. 2009;39:819. doi: 10.1111/j.1365-2362.2009.02186.x. [DOI] [PubMed] [Google Scholar]; (b) Byun J.-Y., Kim M.-J., Eum D.-Y., Yoon C.-H., Seo W.-D., Park K.H., Hyun J.-W., Lee Y.-S., Lee J.-S., Yoon M.-Y., Lee S.-J. Mol. Pharm. 2009;76:734. doi: 10.1124/mol.109.056259. [DOI] [PubMed] [Google Scholar]; (c) Hui B., Wu Y., Wang H., Tian X. Zhongguo Taolixue Tongbao. 2003;19:656. [Google Scholar]
- 16.(a) Kuo C.-J., Chi Y.-H., Hsu T.-A., Liang P.-H. Biochem. Biophys. Res. Commun. 2004;318:862. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Briefly, a fluorogenic peptide Dabcyl-KNSTLQSGLRKE-Edans is used as the substrate, and the enhanced fluorescence due to cleavage of this substrate catalyzed by the protease was monitored at 590/20 nm with excitation at 360/40 nm. The IC50 value of individual compound was measured in a reaction mixture containing 10 μg/mL of the 3CLpro (final concentration was 2.5 μg) and 10 μM of the fluorogenic substrate in 20 mM Bis–Tris buffer. Reactions were run at 25 °C with continuous monitoring of fluorescence for 60 min. Kinetic parameters were obtained using various concentrations of FRET peptide in the fluorescent assay. The maximal velocity (Vmax), Michaelis–Menten constant (Km) and inhibition constant (Ki) were calculated from the Lineweaver–Burk and Dixon plot.
- 17.Sreeramulu S., Gande S.L., Göbel M., Schwalbe H. Angew. Chem., Int. Ed. 2009;48:5853. doi: 10.1002/anie.200900929. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y.H., Chen A.P., Chen C.T., Wang A.H., Liang P.H. J. Biol. Chem. 2002;277:7369. doi: 10.1074/jbc.M110014200. [DOI] [PubMed] [Google Scholar]
- 19.(a) Kasam V.Z.M., Maass A., Schwichtenberg H., Wolf A., Jacq N., Breton V., Hofmann-Apitius M. J. Chem. Inf. Model. 2007;47:1818. doi: 10.1021/ci600451t. [DOI] [PubMed] [Google Scholar]; (b) Yin J., Niu C., Cherney M.M., Zhang J., Huitema C., Eltis L.D., Vederas J.C., James M.N. J. Mol. Biol. 2007;371:1060. doi: 10.1016/j.jmb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Ryu Y.B., Curtis-Long M.J., Lee J.W., Kim J.H., Kim J.Y., Kang K.Y., Lee W.S., Park K.H. Bioorg. Med. Chem. 2009;17:2744. doi: 10.1016/j.bmc.2009.02.042. [DOI] [PubMed] [Google Scholar]; (b) Wu G., Robertson D.H., Brooks C.L., Vieth M. J. Comput. Chem. 2003;24:1549. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


