Graphical abstract
Phlorotannins isolated from Ecklonia cava have strong antiviral activity against PEDV via inhibiting viral entry and/or viral replication.
Keywords: Phlorotannin, Ecklonia cava, Anti-PEDV, Viral absorption, Hemagglutinin inhibition, Viral replication
Abstract
Despite the prepdominat agent causing severe entero-pathogenic diarrhea in swine, there are no effective therapeutical treatment of porcine epidemic diarrhea virus (PEDV). In this study, we evaluated the antiviral activity of five phlorotannins isolated from Ecklonia cava (E. cava) against PEDV. In vitro antiviral activity was tested using two different assay strategies: (1) blockage of the binding of virus to cells (simultaneous-treatment assay) and (2) inhibition of viral replication (post-treatment assay). In simultaneous-treatment assay, compounds 2–5 except compound 1 exhibited antiviral activities of a 50% inhibitory concentration (IC50) with the ranging from 10.8 ± 1.4 to 22.5 ± 2.2 μM against PEDV. Compounds 1–5 were completely blocked binding of viral spike protein to sialic acids at less than 36.6 μM concentrations by hemagglutination inhibition. Moreover, compounds 4 and 5 of five phlorotannins inhibited viral replication with IC50 values of 12.2 ± 2.8 and 14.6 ± 1.3 μM in the post-treatment assay, respectively. During virus replication steps, compounds 4 and 5 exhibited stronger inhibition of viral RNA and viral protein synthesis in late stages (18 and 24 h) than in early stages (6 and 12 h). Interestingly, compounds 4 and 5 inhibited both viral entry by hemagglutination inhibition and viral replication by inhibition of viral RNA and viral protein synthesis, but not viral protease. These results suggest that compounds isolated from E. cava have strong antiviral activity against PEDV, inhibiting viral entry and/or viral replication, and may be developed into natural therapeutic drugs against coronavirus infection.
1. Introduction
Coronaviruses cause acute and chronic respiratory, enteric, and central nervous system diseases in many species of humans and animals.1 Among animal pathogens, porcine epidemic diarrhea virus (PEDV) is an important agent in swine, causing severe entero-pathogenic diarrhea, dehydration, vomiting, and high mortality in nursing piglets.2 PEDV infection has become a serious issue in the swine industry, and outbreaks have led to serious economic losses in many countries.3 Unfortunately, there are currently no effective commercial vaccines or specific treatments. To date, the only measures available for controlling the disease are disinfection strategies designed to prevent the entrance of the virus to the farm.
Esculent (edible) plants are increasingly being projected as suitable alternative sources of antiviral agents, because of their minor side effects, reduced potential to cause resistance, and low cost.4 Natural sources, such as Eckolina cava (E. cava), have provided products for food preservation and fulfilled the primary healthcare needs of every known culture.5, 6 E. cava has also shown anti-viral,7, 8 anti-oxidant,9, 10, 11 anti-inflammatory,12, 13 antiplasmin-inhibitory,14 bactericidal,15 anticancer,6 anti-allergic,16 and tyrosinase-inhibitory activity.17
Phlorotannin components, which are oligomeric polyphenols of phloroglucinol units, are responsible for the pharmacological activities of E. cava. Among the phlorotannins identified in Ecklonia species are eckol (a closed-chain trimer of phloroglucinol), phlorofucofuroeckol (a pentamer), and dieckol (a hexamer).7 Although a variety of pharmacological activities associated with compounds from E. cava have been demonstrated, these studies have shed little light on antiviral activities and mechanisms of E. cava against PEDV have not been reported. Therefore, in this study, we assessed the in vitro anti-PEDV activity of an ethanol (EtOH) extract and phlorotannins isolated from E. cava and evaluated their mechanisms of antiviral activity during virus replication cycle.
2. Results and discussion
2.1. Isolation of phlorotannins from E. cava
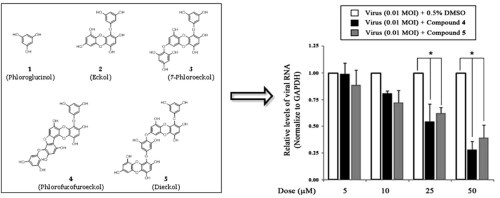
We isolated five naturally occurring compounds from the EtOH extract of E. cava, as described previously.7 An EtOH extract (157 g) of dried E. cava (2.0 kg) was solvent-fractionated into obtain n-hexane (25.4 g), ethyl acetate (EtOAc; 60.5 g), and H2O (70 g) layers. To isolate the compounds, we subjected the EtOAc layer to a succession of chromatographic procedures, including silica gel, Sephadex LH-20, and octadecyl-functionalized silica gel to yield five phlorotannins (1–5) as active principles. A spectroscopic (1H, 13C NMR, and MS) analysis and comparisons with previous studies18 identified the isolated compounds as the known species phloroglucinol (1), eckol (2), 7-phloroeckol (3), phlorofucofuroeckol (4), and dieckol (5) (Fig. 1 C).7 Also, compound 4 and 5 within EtOH extract were shown to be present in high quantities by HPLC analysis (Fig. 1A and B).
Figure 1.
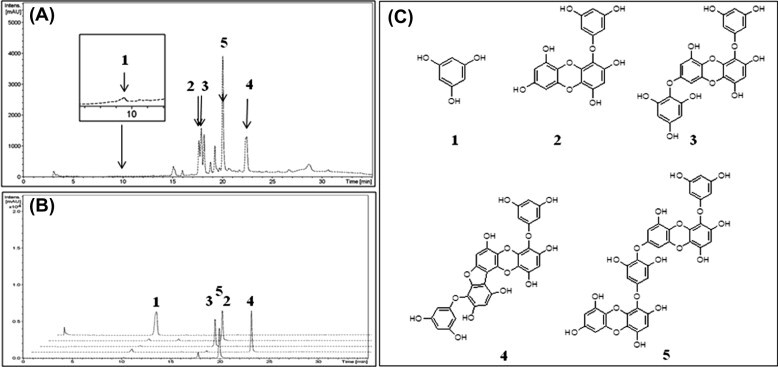
(A) Chromatogram of E. cava EtOH extract. (B) Overlapping signal peaks of five isolated phlorotannins. (C) Chemical structures of compounds 1–5 isolated from E. cava.
2.2. Cytotoxicity of the EtOH extract and phlorotannins from E. cava in Vero cells
The cytotoxicity of the EtOH extract and five isolated compounds was evaluated by determining 50% cytotoxicity concentration (CC50) values using the MTT assay. Confluent cells in α-MEM were incubated in the absence or presence of twofold diluted samples (5–600 μg/mL or μM) for 72 h, after which MTT reagents were added to the cells. The CC50 of the EtOH extract was 533.6 μg/mL and ranged from 374.4 to 579.0 μM for compounds 1–5 (Table 1 ). Subsequent experiments designed to evaluate antiviral effect were carried out at minimally toxic (>90% cell viability) concentrations of EtOH extract and compounds.
Table 1.
In vitro antiviral activity of ethanol extract and compound 1−5 isolated from E. cava against PEDV
| Extract or compounds | CC50a (μM) | Simultaneous-treatment |
Post-treatment |
||
|---|---|---|---|---|---|
| IC50b (μM) | SIc | IC50b (μM) | SIc | ||
| EtOH extract | 533.6 ± 2.6 μg/mL | 12.4 ± 2.2 μg/mL | 43.0 | 19.5 ± 3.8 μg/mL | 28.3 |
| Phloroglucinol (1) | 374.4 ± 4.0 | — | — | — | — |
| Eckol (2) | 388.3 ± 2.6 | 22.5 ± 2.2 | 17.2 | — | — |
| 7-Phloroeckol (3) | 446.2 ± 3.8 | 18.6 ± 2.3 | 23.9 | — | — |
| Phlorofucofuroeckol (4) | 579.0 ± 4.3 | 10.8 ± 1.4 | 53.6 | 12.2 ± 2.8 | 47.4 |
| Dieckol (5) | 490.6 ± 1.6 | 16.6 ± 3.0 | 29.5 | 14.6 ± 1.3 | 33.6 |
CC50: mean (50%) value of cytotoxic concentration.
IC50: mean (50%) value of inhibitory concentration.
SI: selective index, CC50/IC50.
2.3. Inhibitory activity of extract and phlorotannins from E. cava on PEDV absorption
Throughout the life cycle of coronavirus, there are several potential targets for antiviral agents: viral entry, viral penetration into cells, viral processing (viral protease), viral replication (transcription and translation), and viral release from infected cells.19 We hypothesized that the EtOH extract and compounds isolated from E. cava would exert their antiviral activity at the first two steps: (1) blockage of viral entry to the host cell, and/or (2) inhibition of viral replication after entry into the cell. We performed time-of-addition experiments to determine the stage at which the EtOH extract and isolated compounds exerted inhibitory activities, testing two distinct time points: after incubation for 1 h at 4 °C with virus prior to virus infection (simultaneous-treatment assay), and 1 h after virus inoculation (post-treatment assay).20
First, to evaluate the ability of the EtOH extract and five compounds to prevent the attachment of PEDV to Vero cells, we used a simultaneous-treatment experimental paradigm. The results showed that the EtOH extract exerted antiviral activity against the PEDV SM98 strain, exhibiting a 50% inhibitory concentration (IC50) of 12.4 ± 2.2 μg/mL. Of the five isolated phlorotannins, compounds 2–5 exhibited inhibitory activities with IC50 values ranging from 10.8 ± 1.4 to 22.5 ± 2.2 μM (Table 1). The most potent phlorotannin, phlorofucofuroeckol (4), inhibited PEDV attachment with an EC50 value of 10.8 ± 1.4 μM (SI value = 53.6). The rank-order of antiviral activities was dieckol (5) (16.6 ± 3.0 μM, SI = 29.5) > 7-phloroglucinoleckol (3) (18.6 ± 2.3 μM, SI = 23.9) > eckol (2) (22.5 ± 2.2 μM, SI = 17.2). Phloroglucinol (1) did not show significant inhibitory effects against PEDV in the simultaneous-treatment assay. From this, it may be inferred that the number of phloroglucinol moieties, and thus the number of hydroxyl groups, on the phlorotannin backbone contributes to blockage of viral entry to Vero cells. Although the structure activity relationships of phlorotannins were not thoroughly investigated, these results suggest that oligomerization and the existence of a cyclopentan ring (4) might be important for the in antiviral activity of these compounds.
2.4. Hemagglutination inhibition (HI) activity
PEDV entry into cells is a multistep process in which several interactions between viral spike (S) protein and cell surface receptors, including sialic acids (SA) and porcine aminopeptidase N (pAPN), occur.21, 22 The SA-binding activity of some coronaviruses (TGEV and IBV) helps the virus penetrate the mucus layer and proceed to the intestinal enterocytes, where it interacts with pAPN to initiate infection.22, 23 Binding of virions to SA may somehow increase viral stability and facilitate virus infection in the gastroenteric tract.23 To further explore effects on virus adsorption and determine whether the EtOH extract and five compounds can block virus adsorption and cell entry, we tested the effects of the EtOH extract and compounds 1–5 on PEDV-induced hemagglutination binding to rabbit red blood cells (rRBCs). The EtOH extract completely inhibited PEDV attachment to rRBCs at less than 7.8 μg/mL (Fig. 2 ). Among the five phlorotannins, eckol (2), phlorofucofuroeckol (4), and dieckol (5) showed particularly strong inhibition of hemagglutination, completely blocking virus attachment at 3.8–5.4 μM. But, compound 1 and 3 showed weak HI activity, requiring concentrations of 36.6 and 31.3 μM, respectively. Interestingly, the HI activity of compounds 2, 4, and 5 was similar to the antiviral activity of these compounds in simultaneous-treatment assay results (Table 1). Also, the compound 3 may be blocking the other way (such as pAPN-binding) for viral entry as well as SA-binding pathway because of exhibiting weak HI activity. Collectively, these results suggest that the effectiveness of compounds 2, 4, and 5 is mainly attributable to a strong interaction with S protein on the outer surface of PEDV, resulting in blockage of viral adsorption.
Figure 2.
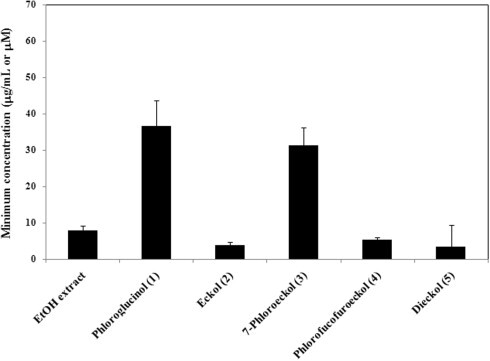
Hemagglutination inhibitory activity of the EtOH extract and compound 1–5 from E. cava. Four HAU of PEDV (SM98 strain) were incubated individually with twofold dilutions of the EtOH extract, compounds 1–5, or PBS (negative control), and rabbit RBCs, for 1 h at room temperature. The minimum concentration of each extract or compound that completely inhibited viral hemagglutination was determined.
2.5. Antiviral activity of extract and phlorotannins from E. cava on PEDV replication
To further evaluate the inhibitory effect of the EtOH extract and compounds 1–5 from E. cava on virus replication, we performed post-treatment assays. In preliminary experiments, the EtOH extract of E. cava showed antiviral activity against the PEDV SM98 strain, exhibiting an IC50 value of 19.5 ± 3.8 μg/mL. To extend these observations, we incubated Vero cells with different concentrations (1–200 μM) of the five compounds after infection with the PEDV SM98 strain for 1 h. Of the five phlorotannins, phlorofucofuroeckol (4) and dieckol (5) exhibited particularly strong inhibitory activities, with IC50 values of 12.2 ± 2.8 μM (SI = 47.4) and 14.6 ± 1.3 μM (SI = 33.6), respectively (Table 1).
To investigate the inhibitory effects of compounds 4 and 5 on viral RNA synthesis, we measured the levels of intracellular viral RNA in infected cells after treatment with drug-treated (5–50 μM) and mock-treated (0.5% DMSO) by quantitative real-time RT-PCR. Total RNA was extracted 24 h after PEDV infection and quantitative real-time RT-PCR was performed using specific primers for viral ORF3 gene. As shown in Figure 3 A, compounds 4 and 5 significantly reduced viral RNA levels in dose-dependent manner. In the presence of 50 μM compound 4 or 5, viral RNA levels were less than 30–40% of those in vehicle-treated cells (Fig. 3A). Notably, compounds 4 and 5 also inhibited the synthesis of viral proteins, such as S protein (Fig. 3B). An investigation of PEDV replication using an immunofluorescence assay revealed green fluorescence in virus-infected cells (Fig. 4 D–F), but not in mock-infected Vero cells (Fig. 4A–C). However, treatment of cells with 30 μM compound 4 or 5 reduced the number of fluorescence-positive, PEDV-infected cells (Fig. 4G–L).
Figure 3.
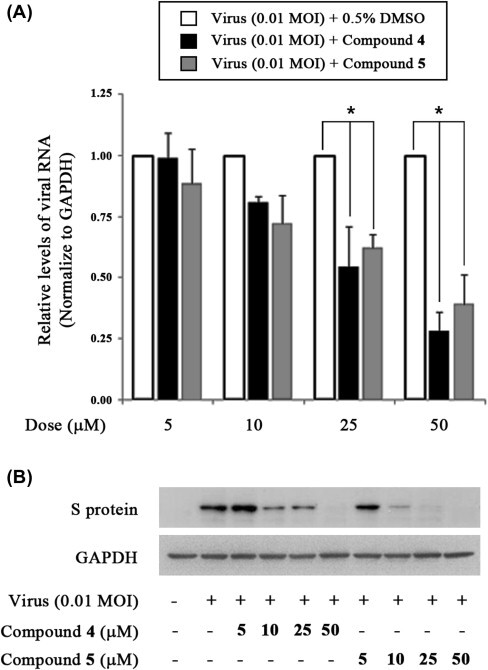
Inhibition of PEDV replication by treatment with compound 4 or 5. (A) Quantitative real-time RT-PCR of viral RNA levels (ORF3 gene) normalized to those of GAPDH. Vero cells were infected with of PEDV at a multiplicity of infection (MOI) of 0.01. After 1 h, viruses were removed and cells were treated with DMSO (0.5%) or compound 4 or 5 (5–50 μM). Total RNA was extracted 24 h after PEDV infection and the levels of intracellular viral RNA were measured. ∗P <0.05. (B) Accumulation of viral (spike) protein as determined by Western blot analysis of extracts of PEDV-infected cells treated with compound 4 or 5 (5–50 μM) at 24 h post-infection.
Figure 4.
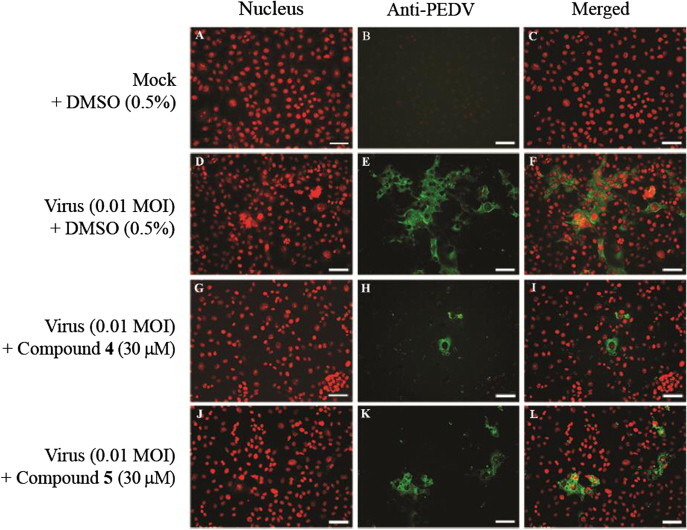
Confocal fluorescence imaging of anti-PEDV effects by compound 4 or 5. Vero cells were infected with PEDV (SM98 strain) at an MOI of 0.01 in the presence of DMSO (0.5%) (B–F), compound 4 (G–H) or compound 5 (J–L) (30 μM), or were mock infected (A–C). After 24 h, cells were fixed in 4% paraformaldehyde, blocked, and incubated with anti-PEDV antibody (green). Propidium iodide was used as nuclear counterstain (red). Scale bar = 50 μM.
Because viral RNA is synthesized in early and late stages, we evaluated the effects of compounds 4 and 5 (30 μM) on synthesis at different stages of PEDV infection in Vero cells. Total RNA was extracted 6, 12, 18, and 24 h after virus infection, and the levels of intracellular viral RNA were measured by quantitative real-time RT-PCR. As shown in Figure 5 , viral RNA levels of PEDV in cells treated with compound 4 or 5 were largely unchanged at 6 and 12 h. In contrast, viral RNA levels were markedly decreased by both compounds 4 and 5 at 18 and 24 h compared with untreated/infected cells (0.5% DMSO) (Fig. 5A). The synthesis of viral proteins was also strongly inhibited by compounds 4 and 5 at 18 and 24 h, but not at 6 and 12 h (Fig. 5B). These results indicate that compounds 4 and compound 5 exert stronger inhibitory effects on the late stage of viral replication than on the early stage.
Figure 5.
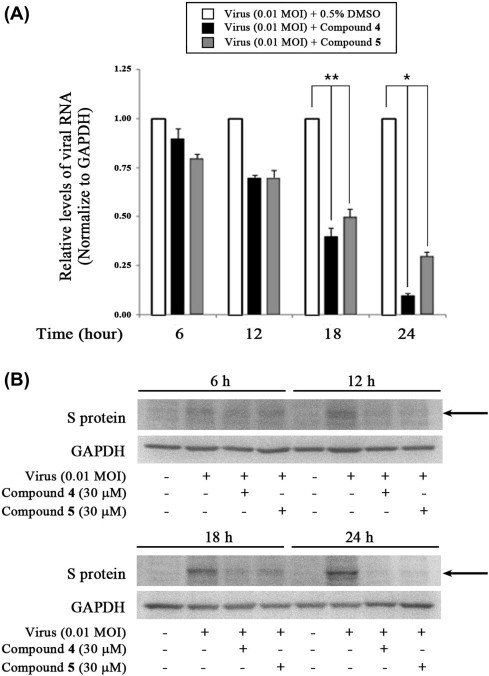
Inhibition of PEDV viral RNA and protein synthesis at a late stage of the replication cycle by treatment with compound 4 or 5. (A) Quantitative real-time RT-PCR of viral RNA levels (ORF3 gene) normalized to those of GAPDH. Vero cells were infected with PEDV at an MOI of 0.01. After 1 h, viruses were removed and cells were treated with DMSO (0.5%), or compound 4 or 5 (30 μM). Total RNA was extracted 6, 12, 18, and 24 h after virus infection and the levels of intracellular PEDV RNA were measured. ∗P <0.01 and ∗∗P <0.05. (B) Accumulation of viral S protein (arrow) as determined by Western blot analysis of extracts of PEDV-infected cell treated with compound 4 or 5 (30 μM) taken 6, 12, 18, and 24 h post-infection.
Several compounds and extracts that inhibit replication of PEDV through unknown mechanisms after viral entry have been reported, including quercetin 7-rhamnoside, WK07, and extracts of cherry fruits and Zanthoxylum species.24, 25, 26, 27 To identify antiviral mechanism of phlorotanins from E. cava against PEDV, we evaluated chymotrypsin-like cysteine proteinase (3CLpro) of PEDV inhibition assays. 3CLpro, essential for viral replication, has been recognized as a key target for anti-coronavirus drug design including SARS-CoV and PEDV. As results of 3CLpro inhibition, phlorotannins were shown to have IC50 values greater than 200 μM (data not shown). Collectively, these data indicate that compound 4 and 5 likely inhibit virus infection through two possible mechanisms: (1) blockage of viral attachment through inhibition of SA binding to host cells, and/or (2) prevention of viral replication through inhibition of viral RNA and protein synthesis in the late stage. Compounds 2 and 3, in contrast, blocked only PEDV attachment to cells.
3. Conclusion
In conclusion, this study has shown that the EtOH extract and compounds 2–5 from E. cava exert antiviral activity against the PEDV by inhibiting viral hemagglutination binding to SA receptors in the host cell. Compounds 4 and 5, which act by inhibiting both viral entry and replication, are particularly viable antiviral drug candidates. These observations further highlight the possibility that the anti-coronavirus properties of phlorotannins derived from E. cava might be harnessed for use in nutraceutical, animal feed, and pharmaceutical industries.
4. Experimental section
4.1. Extraction and isolation
Dried powder of Ecklonia cava (2.0 kg) were extracted with EtOH (20 L) for 1 week at room temperature. The EtOH extract was concentrated on a rotary evaporator, and the dried extract (157 g) was suspended in H2O and partitioned with n-hexane (25.4 g), ethyl acetate (60.5 g). The ethyl acetate soluble fraction (60 g) was chromatographed on silica gel using mixtures of CHCl3/MeOH of increasing polarity (100:0 → 20:80), yielding six fractions. Fraction 2 (3.0 g) was divided into four sub-fraction, 2-1, 2-2, 2-3, 2-4, by column chromatography on silica gel eluted with CHCl3/MeOH of increasing polarity (100:0 → 50:50). Sub-fraction 2-1 (0.23 g) was separated through chromatography on a Sephadex LH-20 column to yield compound 1 (30 mg). Compound 3(18 mg) was isolated by sub-fraction 2-3 using preparative-HPLC (CH3CN/H2O, 60/40, v/v). Fraction 4 (3.5 g) was divided into five sub-fraction, 4-1, 4-2, 4-3, 4-4, 4-5 by column chromatography on silica gel eluted with CHCl3/MeOH of increasing polarity (100:0 → 30:70). Sub-fraction 4-2 (0.5 g) was further purified by silica gel chromatography eluted with CHCl3/MeOH (70:30, v/v) and RP-C18 chromatography to yield give compound 2 (18 mg). Sub-fraction 4-3 (0.48 g) was separated through chromatography on a RP-C18 chromatography column and preparative-HPLC to yield compound 4 (23 mg), compound 5 (13 mg). The structures of isolated compounds 1–5 were confirmed by spectroscopically and compared with previously reported values.7
Compound 1: White powder; mp 218–219 °C; purity 98% (HPLC analysis conditions 70% aq acetonitrile, 1 mL/min, λ = 210 nm, R t = 3.16 min); 1H NMR (300 MHz, methanol-d 4) δ 5.80 (s, 3H); 13C NMR (75 MHz, methanol-d 4) δ 158.7, 94.0.
Compound 2: Light brown powder; mp 246–247 °C; purity 98% (HPLC analysis conditions 80% aq acetonitrile, 1 mL/min, λ = 210 nm, R t = 7.59 min); ESI-MS m/z = 371 [M−H]+; 1H NMR (500 MHz, CD3OD) δ 6.13 (s, 1H), 5.94 (d, J = 5.5 Hz, 5H); 13C NMR (125 MHz, CD3OD) δ 162.0, 160.3, 154.6, 147.3, 147.2, 144.4, 143.4, 138.6, 125.7, 124.9, 124.6, 99.9, 97.8, 95.9, 95.4.
Compound 3: Brown powder; mp 276–277 °C; purity 93% (HPLC analysis conditions 70% aq acetonitrile, 1 mL/min, λ = 210 nm, R t = 10.5 min); ESI-MS m/z = 495 [M−H]+; 1H NMR (500 MHz, CD3OD) δ 6.14 (s,1H), 6.07 (d, J = 2.45 Hz, 2H), 5.96 (m, 4H), 5.85 (s, 1H); 13C NMR (125 MHz, CD3OD) δ 160.6, 160.5, 159.0, 158.8, 155.1, 153.2, 150.8, 147.4, 145.8, 145.0, 141.7, 137.3, 125.5, 124.0, 123.4, 123.0, 98.6, 96.6, 96.5, 96.3, 94.8, 94.4, 94.2, 93.9.
Compound 4: Light brown powder; mp 292 °C (decomp); purity 95% (HPLC analysis conditions 70% aq acetonitrile, 1 mL/min, λ = 210 nm, R t = 6.02 min); ESI-MS m/z = 603 [M+H]+; 1H NMR (500 MHz, CDCl3) δ 6.62 (s, 1H), 6.39 (s, 1H), 6.25 (s, 1H), 5.95 (d, J = 2.0 Hz, 2H), 5.92 (m, 1H), 5.90 (m, 1H), 5.87 (d, J = 2.0 Hz, 2H); 13C NMR (125 MHz, CDCl3) δ 160.5, 158.8, 151.8, 150.4, 149.8, 149.3, 146.9, 146.8, 144.6, 142.6, 137.0, 133.9, 126.7, 123.6, 123.4, 104.0, 103.9, 98.6, 98.5, 96.4, 96.3, 94.8, 94.1, 94.0.
Compound 5: Dark red powder; mp 278 °C (decomp); purity 94% (HPLC analysis conditions 70% aq acetonitrile, 1 mL/min, λ = 210 nm, R t = 10.13 min); ESI-MS m/z = 743 [M+H]+; 1H NMR (300 MHz, CD3OD) δ 6.13 (s, 1H), 6.11 (s, 1H), 6.07 (s, 2H), 6.05 (d, J = 2.8 Hz, 1H), 6.03 (d, J = 2.8 Hz, 1H), 5.96 (d, J = 2.6 Hz, 1H), 5.93 (d, J = 2.8 Hz, 1H), 5.90 (m, 1H).
4.2. HPLC analysis
The profiling of isolated phlorotannins was performed on an Agilent 1200 series (Agilent Technologies, Palo Alto, CA, USA) equipped with a quaternary HPLC pump, a degasser, autosampler and VWD and a Bonus-RP C-18 column (4.6 × 160 mm, 5 μM, Agilent, USA) at 27 °C. The mobile phase, flowed of 0.5 mL/min, was consisted of distilled water (A) and acetonitrile (B) using a gradient system of 0–10 min liner increase to 30% B, 10–20 min liner increase to 60% B, 20–30 min liner increase to 90%, and 30–35 min liner increase to 100% B. The eluent was detected at 210 nm and the infection volume was 10 μL.
4.3. Viruses and cell lines
Vero (african green monkey cell line) cells were kindly provided by the American Type Culture Collection (ATCC CRL-1587; Manassas, VA, USA) and PEDV SM 98 strain was obtained from Animal, Plant and Fisheries Quarantine and Inspection Agency in Korea. Vero cells were maintained in Eagle’s minimum essential medium (EMEM) supplemented with 5% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, and 100 U/mL amphotericin B. PEDV SM98 strain was propagated onto confluent vero cells in the presence of 1 μg/mL trypsin (GIBCO Invitrogen Corporation, CA, USA).
4.4. Cytotoxicity
Vero cells were grown in 96 well plates at 1 × 105 cells/well for 48 h. The cells were replaced with media containing serially dilluted extract and compounds for 72 h. The solution was replaced with only media and 10 μL MTT (3-(4,5-dimethylthiozol-2-yl)-3,5-dipheryl tetrazolium bromide, Sigma, St. Louis, MO) solution was added to each well and incubated at 37 °C for 4 h. After removal of supernatant, 100 μL 0.04 M HCl–isopropanol was added for solubilization of formazan crystals. Absorbance was measured at 540 nm with subtraction of the background measurement at 655 nm in a microplate reader. The 50% cytotoxic concentration (CC50) was calculated by regression analysis.
4.5. Antiviral assay
The antiviral assays have been previously described,20 and the visualization of these assays was performed by neutral red method as briefly described. In the simutaneously-treatment assay: Various concentrations of EtOH extract and compounds isolated from E. cava were mixed with virus at 0.01 MOI and incubated at 4 °C for 1 h. The mixture were inoculated onto near confluent Vero cell monolayers (1 × 105 cells/well) for 1 h with occasional rocking. The solution was removed and the media was replaced. The cultures were incubated for 72 h at 37 °C under 5% CO2 atmosphere until the cells in the infected, untreated control well showed complete viral cytopathic effect (CPE) as observed by light microscopy. Each concentration of EtOH extract and compounds was assayed in triplicate.
In the post-treatment assay: PEDV SM98 strain at 0.01 MOI was inoculated onto near confluent Vero cell monolayers (1 × 105 cells/well) for 1 h with occasional rocking. The media was removed and replaced by EMEM with EtOH extract and compounds at different concentration. The cultures were incubated for 72 h at 37 °C under 5% CO2 atmosphere until the cells in the infected, untreated control well showed complete viral CPE as observed by light microscopy. Each concentration of EtOH extract and compounds was assayed for virus inhibition in triplicate.
After 72 h incubation in all antiviral assays, cells replaced with only media and 10 μL MTT solution was added to each well and incubated at 37 °C for 4 h. After removal of supernatant, 100 μL 0.04 M HCl–isopropanol was added for solubilization of formazan crystals. Absorbance was measured at 540 nm with subtraction of the background measurement at 655 nm in a microplate reader. The 50% inhibitory concentration (IC50) was calculated by regression analysis. A selective index (SI) was calculated using the formula SI = CC50/IC50.
4.6. Hemagglutination inhibition (HI) assay
The hemagglutination inhibition assay was performed to evaluate the effects of EC extract and compounds on viral adsorption to target cells. Standardized rabbit red blood cell (rRBC) solutions were prepared according to standard protocol.24 The PEDV solution (4 HAU/25 μL) was mixed with an equal volume of EC extracts and compounds (25 μL) in a twofold serial dilution in PBS (pH 7.4) for 1 h at 4 °C. 50 μL of the solution was mixed with an equal volume of a 1% rRBC suspension and incubated for 1 h at room temperature.
4.7. Reverse transcription and quantitative real-time PCR
Vero cells were grown to about 90% conflunce, infected with PEDV at 0.01 MOI, and cultured in the presence of 0.5% DMSO or compound 4 and 5 to identify antiviral activity according to the concentration (5–50 μM) of drugs, confluent Vero cells infected with PEDV at 0.01 MOI, and cultured in the presence of various concentrations of 0.5% DMSO or compounds 4 and 5. After 24 h, cells were scraped off, washed twice with PBS, and collected by centrifugation (500×g for 3 min). In order to determine the expression level of open reading frame 3 (ORF3) gene mRNA of PEDV, total RNA was isolated using Qiagen RNeasy mini kit (QIAGEN, Hilden, Germany) according to manufacturer’s instruction. The primer sequences used for quantitative real-time PCR of viral RNA were 5′-GCACTTATTGGCAGGCTTTGT-3′ (sense) and 5′-CCATTGAGAAAAGAAAGTGTCGTAG-3′ (antisense). The GAPDH was used as internal control of cellular RNAs, with primer sequences of 5′-TCAACAGCGACACCCACTC-3′ (sense) and 5′-CTTCCTCTTGTGCTCTTGCTG-3′ (antisense).
For inhibitory activity of compound 4 and 5 in early and late replication step, confluent Vero cells infected with PEDV at 0.01 MOI, cultured in the presence of compounds 4 and 5 (30 μM). After 6, 12, 18, 24 h, medium was removed and next process was performed as above mentioned.
The total RNA was reverse transcribed into cDNA using the High Capacity RNA-to-cDNA master mix (Applied Biosystems) according to the manufacturer’s protocol. Reverse transcription was performed at 42 °C for 1 h. The enzyme was inactivated at 95 °C for 5 min. The cDNA was stored at −20 °C or directly used in quantitative real-time PCR. Real-time PCR was conducted using 2 μL of cDNA and Power SYBR Green PCR 2X master mix (Applied biosystems). Cycling conditions for real-time PCR were as follows: 95 °C for 1 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 15 s. Real-time PCR was conducted using the Step One Plus Real-time PCR system, and the data were analyzed with StepOne software v2.1 (Applied Biosystems).
4.8. Western blot analysis
Vero cells were cultured about 90% confluence and infected with PEDV at 0.01 MOI for 1 h. After then, the cells were cultured in the presence of 0.5% DMSO, compound 4, and 5. Medium was removed after 6, 12, 18, 24 h. Cells were scraped off, washed twice with PBS, and collected by centrifugation (500×g for 3 min). For whole cell lysate, cells were lysed in the PhosphoSafe Extraction Reagent (Novagen, Darmstadt, Germany). Protein was separated by using NuPage 10% or 12% Bis–Tris prepacked gel (Invitrogen, CA, USA). Equal amounts of protein were then electrotransferred to nitrocellulose membranes (Invitrogen, CA, USA). Membranes were blocked by incubation for 1 h at room temperature in 5% BSA in TBS–0.1% Tween 20 solution, followed by a further 12 h at 4 °C of incubation with primary antibody against spike (S) protein (Jeno Biotech Inc, Korea) and GAPDH (Santa Cruz Biotechnology, CA, USA). Blots were washed for 15 min with 3 changes of TBS–0.1% Tween 20 solution, followed by incubation for 1 h at room temperature with the HRP-conjugated IgG antibody (Santa Cruz Biotechnology, CA, USA). Finally, they were developed in LumiGLO reagent (Cell Signaling Technology, MA, USA). GAPDH were used as a loading control.
4.9. Confocal fluorescence imaging
Vero cells were grown on 8-well chamber slides (LAB-TEK, NUNC, USA), and the monolayers were infected with PEDV at 0.01 MOI for 1 h. The virus was removed and replaced by EMEM and 30 μM of compounds 4 and 5 under test. The cells were cultured for 24 h at 37 °C in a 5% CO2 atmosphere, washed three times with PBS (pH 7.4), and fixed in 4% paraformaldehyde for 15 min at room temperature. After 3 times washed with PBS (pH 7.4), cells were incubated at 37 °C for 1 h with monoclonal antibody against S protein of PEDV (Jeno Biotech, Korea) diluted 1:50 in PBS (pH 7.4). After washing with PBS (pH 7.4), cells were incubated at 37 °C for 1 h with FITC-conjugated goat anti-mouse IgG antibody (Santa Cruz, CA, USA) diluted 1:100 in PBS (pH 7.4). Cells were washed with PBS (pH 7.4), stained with 500 nM propidium iodide (PI) solution for 10 min at room temperature, and washed 3 times with PBS (pH 8.0). Slides were mounted using antifade reagent (Invitrogen, CA, USA) and visualized under a Carl Zeiss LSM 510 META confocal microscope (Carl Zeiss Inc., Jena, Germany).
4.10. Statistical analysis
All experiments were performed three times. Data were expressed as mean ± SE. Statistical analysis was performed using Sigma Plot Statistical Analysis software. Differences between group mean values were determined by one-way analysis of variance followed by a two-tailed Student’s t-test for unpaired samples, assuming equal variances.
Acknowledgments
This research was supported by National Research Foundation Grant funded by the Korea government (Ministry of Education, Science, and Technology) (No. 2009-0081749) and KRIBB Research Initiative Program, Republic of Korea.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.2013.04.085.
Supplementary data
References and notes
- 1.Weiss S.R., Navas-Martin S. Microbiol. Mol. Biol. Rev. 2005;69:635. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debouck P., Pensaert M. Am. J. Vet. Res. 1980;41:219. [PubMed] [Google Scholar]
- 3.Hofmann M., Wyler R. Schweiz. Arch. Tierheilkd. 1987;129:437. [PubMed] [Google Scholar]
- 4.Jassim S.A., Naji M.A. J. Appl. Microbiol. 2003;95:412. doi: 10.1046/j.1365-2672.2003.02026.x. [DOI] [PubMed] [Google Scholar]
- 5.Jung W.K., Ahn Y.W., Lee S.H., Choi Y.H., Kim S.K., Yea S.S., Choi I., Park S.G., Seo S.K., Lee S.W., Choi I.W. Food Chem. Toxicol. 2009;47:410. doi: 10.1016/j.fct.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 6.Kim M.M., Ta Q.V., Mendis E., Rajapakse N., Jung W.K., Byun H.G., Jeon Y.J., Kim S.K. Life Sci. 2006;79:1436. doi: 10.1016/j.lfs.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 7.Ryu Y.B., Jeong H.J., Yoon S.Y., Park J.Y., Kim Y.M., Park S.J., Rho M.C., Kim S.J., Lee W.S. J. Agric. Food Chem. 2011;59:6467. doi: 10.1021/jf2007248. [DOI] [PubMed] [Google Scholar]
- 8.Ahn M.J., Yoon K.D., Min S.Y., Lee J.S., Kim J.H., Kim T.G., Kim S.H., Kim N.G., Huh H., Kim J.W. Biol. Pharm. Bull. 2004;27:544. doi: 10.1248/bpb.27.544. [DOI] [PubMed] [Google Scholar]
- 9.Ahn G.N., Kim K.N., Cha S.H., Song C.B., Lee J.H., Heo M.S., Yeo I.K., Lee N.H., Jee Y.H., Kim J.S., Heu M.S., Jeon Y. Eur. Food Res. Technol. 2007;226:71. [Google Scholar]
- 10.Kang H.S., Chung H.Y., Kim J.Y., Son B.W., Jung H.A., Choi J.S. Arch. Pharmacol. Res. 2004;27:194. doi: 10.1007/BF02980106. [DOI] [PubMed] [Google Scholar]
- 11.Heo S.J., Park P.J., Park E.J., Cho S.K., Kim S.K., Jeon Y.J. Food Sci. Biotechnol. 2005;14:614. [Google Scholar]
- 12.Kang K.A., Lee K.H., Park J.W., Lee N.H., Na H.K., Surh Y.J., You H.J., Chung M.H., Hyun J.W. FEBS Lett. 2000;2007:581. doi: 10.1016/j.febslet.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Crockett S.L., Wenzig E.M., Kunert O., Bauer R. Phytochem. Lett. 2008;1:37. doi: 10.1016/j.phytol.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuyama Y., Kodama M., Miura I., Kinzyo Z., Mori H., Nakayama Y., Takahashi M. Chem. Pharm. Bull. (Tokyo) 1990;38:133. doi: 10.1248/cpb.38.133. [DOI] [PubMed] [Google Scholar]
- 15.Nagayama K., Iwamura Y., Shibata T., Hirayama I., Nakamura T. J. Antimicrob. Chemother. 2002;50:889. doi: 10.1093/jac/dkf222. [DOI] [PubMed] [Google Scholar]
- 16.Le Q.T., Li Y., Qian Z.J., Kim M.M., Kim S.K. Process Biochem. 2008;44:168. [Google Scholar]
- 17.Park D.C., Ji C.I., Kim S.H., Jung K.J., Lee T.G., Kim I.S., Park Y.H., Kim S.B. J. Fish. Sci. Technol. 2000;3:195. [Google Scholar]
- 18.Li Y., Qian Z.J., Ryu B.M., Lee S.H., Kim M.M., Kim S.K. Bioorg. Med. Chem. 1963;2009:17. doi: 10.1016/j.bmc.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 19.Haagmans B.L., Osterhaus A.D. Antiviral Res. 2006;71:397. doi: 10.1016/j.antiviral.2006.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H.H., Kwon H.J., Ryu Y.B., Chang J.S., Cho K.O., Hosmillo M.D., Rho M.C., Park S.J., Lee W.S. Res. Vet. Sci. 2012;92:320. doi: 10.1016/j.rvsc.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B.X., Ge J.W., Li Y.J. Virology. 2007;365:166. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwegmann-Wessels C., Herrler G. Glycoconjugate J. 2006;23:51. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwegmann-Wessels C., Zimmer G., Schröder B., Breves G., Herrler G. J. Virol. 2003;77:11846. doi: 10.1128/JVI.77.21.11846-11848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J.H., Chae S.W., Yoon K.A., Park J.S., Choi H.J. J. Cosmatics Pub Health. 2010;6:42. [Google Scholar]
- 25.Song J.H., Shim J.K., Choi H.J. Virol. J. 2011;8:460. doi: 10.1186/1743-422X-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yook H.S., Kim K.H., Park J.E., Shin H. J. Am. J. Chin. Med. 2010;38:937. doi: 10.1142/S0192415X10008366. [DOI] [PubMed] [Google Scholar]
- 27.Choi H.J., Kim J.H., Lee C.H., Ahn Y.J., Song J.H., Baek S.H., Kwon D.H. Antiviral Res. 2009;81:77. doi: 10.1016/j.antiviral.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


