Abstract
Protein complexes composed of many subunits carry out most essential processes in cells and, therefore, have become the focus of intense research. However, deciphering the structure and function of these multiprotein assemblies imposes the challenging task of producing them in sufficient quality and quantity. To overcome this bottleneck, powerful recombinant expression technologies are being developed. In this review, we describe the use of one of these technologies, MultiBac, a baculovirus expression vector system that is particularly tailored for the production of eukaryotic multiprotein complexes. Among other applications, MultiBac has been used to produce many important proteins and their complexes for their structural characterization, revealing fundamental cellular mechanisms.
Multiprotein complexes catalyze cellular function
Our understanding of cellular function has remarkably expanded in recent years, brought about by technological advances in DNA and protein analysis [1]. The sequencing of complete genomes, such as the human genome, has set the stage to address proteome-wide interaction studies, which have revealed that proteins do not typically exist as isolated entities 2, 3, 4. Rather, they are assembled in complex molecular machines consisting of numerous proteins and, often, other biomolecules (such as nucleic acids, sugars, lipids and small molecules), arranged into functional modules that catalyze essential cellular processes. This molecular organization has been recently termed ‘protein sociology’ [5].
Understanding cellular processes requires detailed knowledge of the 3D structure of the molecules involved, and the parameters and architectural features that dictate their interaction. Structural genomics efforts strive to analyze comprehensively single proteins and protein domain structures on a whole-genome scale, and have provided atomic structures of thousands of protein structures and folds. Furthermore, the architectures of essential macromolecular complexes, such as ribosomes, nucleosomes and RNA polymerases, have been revealed at near atomic resolution, providing a wealth of structural detail and crucial insight into the functions of these multicomponent machines 6, 7, 8. Integrated approaches combining structural, functional and computational data are emerging, and provide views of protein organization in space and time in entire organisms 5, 9.
Notwithstanding, our molecular understanding of the very large number of protein complexes in the cell remains limited to a handful of examples for which detailed near-atomic structures are known. This is mostly because of the difficulty in obtaining samples in sufficient quality and quantity for molecular studies. Many essential complexes remain intractable because they exist in very low amounts in their endogenous host, which hinders their purification from native source material. Recombinant overproduction can resolve this bottleneck, and numerous expression systems, mostly for heterologous protein expression in Escherichia coli, have been developed and refined over the past few decades. More recently, E. coli expression systems have been designed for coexpression of several proteins by using polycistronic mRNA transcripts, or two or more plasmids that coexist in the same cell 10, 11 (Box 1 ). However, many eukaryotic protein complexes cannot be produced efficiently in E. coli. They may contain subunits that are too large for the E. coli transcription and translation machinery, or may require either specific chaperone systems for proper folding or protein modifications (such as phosphorylation or acetylation) that E. coli cannot provide. Thus, the successful overproduction of these complexes, which is required to decipher their structure and function, depends on the availability of powerful eukaryotic expression technologies. In this review, we describe MultiBac, a recent eukaryotic expression system that is specifically designed to tackle and overcome this crucial production bottleneck 12, 13. We summarize the technology concepts underlying MultiBac and review its wide range of applications.
Box 1. Coexpression toolbox for production of protein complexes.
Expression systems for production of protein complexes in E. coli frequently make use of polycistronic expression cassettes with several genes of interest, spaced apart by ribosome binding sites (Shine–Dalgarno sequences), placed under the control of a single promoter, and typically followed by a sequence for efficient termination of mRNA transcription 10, 11, 21, 60, 61, 62 (Figure I a). In eukaryotic hosts, an analogous design can involve internal ribosomal entry sites (IRESs), which are inserted between gene coding regions under control of a single promoter 24, 25, 26, 27 (Figure Ib). IRESs exist in the 5′-untranslated regions of many viral and cellular mRNAs [24], and facilitate cap-independent translation by recruiting ribosomes for efficient protein production. IRES sequences differ greatly and exhibit species specificity [26]. For example, IRES elements from encephalomyocarditis virus (EMCV) work well in mammalian cells, whereas IRESs from Perina nuda virus (PhV) and Rhopalosiphum padi virus (RhPV) have been successfully used for protein complex production in insect cells [27].
Figure I.
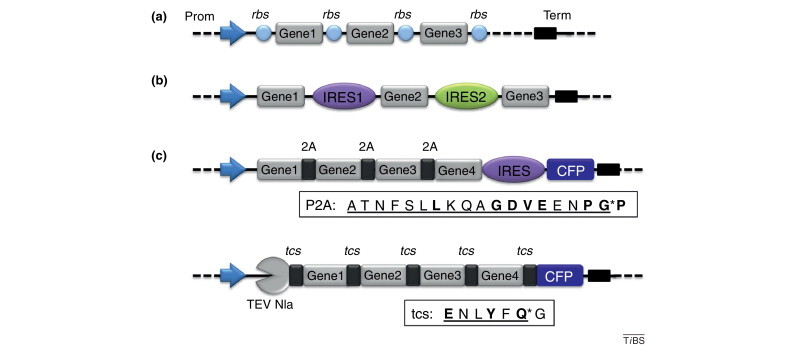
Tools for expression of protein complexes. (a) Protein complexes can be produced efficiently in E. coli by using polycistrons. In polycistrons, genes of interest are spaced apart by ribosome binding sites (rbs) and placed under control of a single promoter (Prom). A sequence (Term) ensures termination of mRNA transcription. One or several polycistrons can be provided on one or more plasmids simultaneously in one cell. (b) The equivalent construction for eukaryotic expression provides IRESs instead of rbs, resulting also in polycistronic mRNAs. (c) Polyproteins are particularly useful to balance the stoichiometry of expressed proteins. Polyprotein constructions can provide self-cleaving peptides derived from picornavirus (2A) in between the individual proteins (top). A fluorescent protein (here CFP) can be included to facilitate tracking of heterologous expression from the respective promoter by fluorescence measurement. As an alternative, polyproteins can be used that contain a highly specific protease [here tobacco etch virus (TEV) Nla], which processes the polyprotein by cleaving corresponding cleavage sites (tcs), thus liberating the individual proteins including itself (bottom). Both polyprotein strategies have the drawback of decorating the C terminus of the preceding protein with overhanging residues that are part of the cleavage sequence (boxed). Asterisks denote sites of cleavage. Conserved residues defining the site are in bold. C-terminal overhangs are underlined.
Polyproteins are long polypeptides containing individual proteins spaced by specific proteolytic cleavage sites. Certain viruses, such as coronavirus, produce their entire proteome by proteolytic processing of polyproteins encoded by a few open reading frames. Polyprotein approaches have proven to be particularly powerful for balancing the stoichiometry of coexpressed proteins 22, 28, 29, 30 (Figure Ic). Polyprotein constructions can involve self-cleaving peptides found in picornavirus (called P2A peptides) or variants thereof 28, 29. Alternatively, constructions can be used that mimic open reading frames in positive-sense single-stranded RNA viruses and provide a highly specific protease gene together with genes of interest arranged in a single large open reading frame 22, 30. Individual proteins are then liberated from the encoded polyprotein by the protease, which cleaves the proteolytic sites placed between the protein subunits (Figure Ic).
The elucidation of protein–protein interactions in complexes is of crucial importance for many applications including structural biology. A novel approach, CoESPRIT, utilizes library-based construct screening for the identification and expression of soluble protein complexes in E. coli [31]. In CoESPRIT, a subunit of a (putative) protein complex is provided as bait. The interacting partner is provided in the form of a random incremental library generated by exonuclease digestion of the full-length gene. Prior input from bioinformatics analyses such as homology alignments or domain identification is not required for this approach. Coexpression of the library with the bait protein allows identification of soluble complexes by immunofluorescence-assisted colony screening using labeled antibody markers against affinity tags present on the proteins screened. Production of protein complex from high-expressor colonies thus identified can then be scaled up to milligram amounts for high-resolution studies by NMR or X-ray crystallography [31].
The MultiBac system
Yeast, mammalian cells and insect cells have been successfully used for recombinant production of eukaryotic proteins [14]. In particular, the use of insect cell cultures infected by a recombinant baculovirus, carrying the eukaryotic gene of interest, was first demonstrated many years ago [15]. The exciting evolution of baculovirus from a pest control agent to a powerful recombinant protein production tool has recently been reviewed [16], and baculovirus expression systems have become increasingly popular for many applications 17, 18, 19.
MultiBac is a baculovirus expression system particularly designed for the production of eukaryotic multiprotein complexes with many subunits (Figure 1 ). It consists of an array of small synthetic DNA plasmids, an engineered baculovirus genome derived from the Autographa californica nuclear polyhedrosis virus (AcNPV; see Glossary) that is used to infect cells of the caterpillar Spodoptera frugiperda, and a set of protocols detailing every step from gene insertion into the plasmids to production of protein complexes in cultured insect cells 19, 20. The presence of many subunits in a protein complex requires the assembly of many encoding genes and their integration into the baculovirus in a multicomponent co-production experiment. This process is laborious and technically challenging using conventional methods that typically involve serial insertion of genes into increasingly large and difficult-to-handle DNA plasmids. MultiBac applies a different concept for multigene assembly that relies on recombination of small, custom-designed, synthetic DNA plasmid molecules (< 3 kb) that are called ‘acceptors’ and ‘donors’ (Figure 1a).
Figure 1.
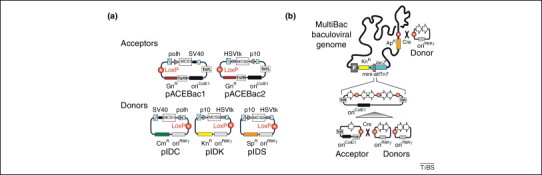
The MultiBac system. MultiBac is a baculovirus expression vector system particularly tailored for high-quality multiprotein complex production in insect cells. (a) MultiBac consists of an array of small (3 kb), synthetic DNA plasmids called acceptors and donors. Acceptors have a regular origin of replication (oriColE1), whereas donors have a conditional origin derived from R6Kγ phage (oriR6kγ) that requires special bacterial strains for their propagation. Donors and acceptors contain expression cassettes controlled by late baculoviral promoters (polh or p10) as well as strong eukaryotic polyadenylation signals (from SV40 or HSVtk). All plasmids contain the LoxP sequence (circles filled in red) for fusing donors to an acceptor by the Cre recombinase. Each plasmid has a different resistant marker: gentamycin resistance (GnR) for acceptors, and either chloramphenicol (CmR), kanamycin (KnR) or spectinomycin (SpR) resistance for donors. In addition, a ‘multiplication’ module is present to facilitate the assembly of several expression cassettes on a donor or acceptor based on specifically designed restriction sites (e.g. homing endonucleases and BstXI restriction endonuclease, shown as blue boxes flanking promoter and terminator, respectively) 18, 19, 21. Acceptors contain the DNA sequences (Tn7L and Tn7R) required for transposition by the Tn7 transposase. (b) The assembly of a composite MultiBac baculoviral genome for multigene expression. Genes are inserted into donors and acceptors by using either restriction endonucleases and ligase, or sequence-independent and ligation-independent cloning methods (bottom). Cre-mediated fusion produces a multigene construction that is inserted into the baculoviral genome by Tn7 transposition in specifically tailored E. coli cells that contain the viral DNA (also known as a bacmid). The DNA located between the Tn7R and Tn7L sequences in the multigene fusion construct is inserted by the transposase enzyme into the Tn7 attachment site (mini-attTn7). The MultiBac baculovirus was engineered for improved protein production and delayed host cell lysis by deleting specific genes [12]. The combination of sequence and ligation independent cloning (SLIC) for gene insertion and Cre–LoxP fusion for multigene construct generation is called tandem recombineering and can be performed in a parallelized mode on microtiter plates, optionally on a robot [21]. Further heterologous genes can be inserted into an additional LoxP site present on the bacmid. Composite baculoviral DNA is then purified from the bacteria and used to transfect insect cells. Virus is expanded by infecting small-volume (50–100 ml) insect cell cultures and harvesting the budded virus particles released into the culture media. This virus is then used for protein complex expression in larger (typically one to several liters) insect cell cultures 13, 20.
Acceptors and donors can be easily loaded with one or several genes each, and recombined in a single-step reaction into a multigene construct. Acceptors contain an origin of replication that allows propagation in standard cloning strains of E. coli, whereas donors harbor a conditional origin of replication (derived from phage R6Kγ) that requires the presence of a specific protein (known as π) for replication. This protein is expressed from the pir gene inserted into the chromosome of specifically tailored E. coli strains that are used to propagate donor plasmids 13, 20. Donors and acceptors contain a resistance marker, a short imperfect inverted repeat (LoxP), an expression cassette consisting of a baculoviral promoter (p10 or polh), a DNA segment for inserting heterologous genes, and an efficient signal for eukaryotic polyadenylation (Figure 1a). The expression cassettes are flanked by a homing endonuclease site and a compatible BstXI site, which allow for iterative assembly of several expression cassettes on each plasmid [21].
Importantly, donors can survive in a pir-negative background only if they are conjoined with an acceptor that provides a nonconditional origin of replication, and this is the central feature that enables flexible and efficient generation of multigene constructs. These are achieved in vitro by Cre recombinase, which fuses one or several donors (each with one or several inserted genes) to a single acceptor in a one-step reaction by conjoining via the LoxP sites; this results in plasmid dimers, trimers and tetramers. The resulting multigene expression constructs are characterized by the precise combinations of resistance markers present on the fusions, and this can be exploited for combinatorial assembly strategies based on multiple antibiotic selection [21]. The process of inserting genes into acceptors and donors, which can be optionally done by ligation independent methods followed by Cre–LoxP fusion, is termed ‘tandem recombineering’ [22].
Gene insertions into the MultiBac genome take place in bacterial strains that contain the MultiBac viral genome as an artificial chromosome together with a plasmid encoding the Tn7 transposon enzyme complex. Multigene acceptor–donor fusion constructs are transformed into these bacterial cells, and the Tn7 transposon enzyme inserts the collection of expression cassettes present on the acceptor–donor fusion in a single-step reaction into the Tn7 attachment site engineered into the baculoviral genome (Figure 1b). Productive transposition disrupts a LacZα-encoding gene, which enables blue/white screening of colonies. The MultiBac baculoviral genome has been engineered for improved protein complex production by removing genes encoding viral protease and apoptotic activities, thereby reducing proteolytic breakdown of the heterologous gene products and delaying lysis of the infected cells 12, 13. As a second site of entry in addition to the Tn7 attachment site, the MultiBac genome contains also a distal LoxP site for adding further functionalities. For example, a gene encoding λ-phosphatase was inserted into this site to remove phosphates from a coexpressed protein complex [23]. The composite MultiBac virus is then purified and used to transfect insect cells for protein production in Petri dishes containing cell monolayers or, for larger volumes, shaker flasks containing suspension cultures 13, 19, 20. MultiBac resolves several challenges encountered in protein complex production. These include constraints on handling many and often very large encoding DNAs and the necessity to revise expression experiments rapidly and flexibly if a subunit or purification tag needs to be altered or exchanged, by replacing the respective donor or acceptor with a modified version in the tandem recombineering pipeline.
In addition to MultiBac, a growing number of technologies are currently being used for expressing protein complexes (Box 1). Examples of these approaches are the use of internal ribosome entry sites (IRESs) for multigene expression, and the use of polyprotein constructions involving self-cleaving peptide sequences or proteolytic processing of large precursor polypeptides by specific proteases 22, 24, 25, 26, 27, 28, 29, 30. Particularly promising for structural biology, where often a large number of constructs need to be scrutinized in parallel, are new methods coupling gene libraries to coexpression (Box 1) [31]. These approaches are compatible with the MultiBac technology concept; for example, the polyprotein approach has been successfully used to produce a human transcription factor core complex by MultiBac, with improved production efficiency and yield [22].
MultiBac expression of protein complexes for structural studies
The MultiBac system, originally designed by X-ray crystallographers interested in studying multiprotein complexes [12], has been well received and put to good use by the structural biology community (Figure 2 , Box 2 ). Many proteins and their complexes have been produced by MultiBac, often for the first time, for structure elucidation, providing important insight into their biological function (Figure 2). In some cases, for example for producing intact full-length protein kinase CβII, an existing transfer plasmid was combined with the optimized MultiBac baculovirus genome for protein production [32]. The modular concept of gene assembly by tandem recombineering has turned out to be instrumental for accelerating the process of obtaining high-quality samples for structural biology of numerous complexes 33, 34, 35, 36.
Figure 2.
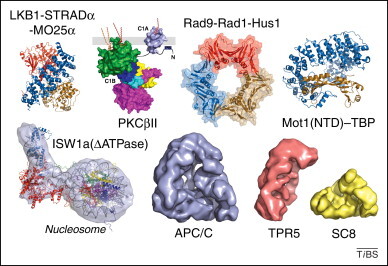
MultiBac expression for structural biology. The MultiBac system has been successfully used to produce many proteins and their complexes in high-quality for structural analysis. The structures of LKB1–STRADα–MO25α (PDB 2WTK) [55], a tumor suppressor kinase complex, the PKCβII kinase (PDB 3PFQ) [32], and the Rad9–Rad1–Hus1 complex, a DNA damage checkpoint complex (PDB 3G65) [56], were determined by X-ray crystallography providing crucial insight into the function of these important proteins. The crystal structure of the N-terminal domain (NTD) of the chromatin remodeler Mot1 bound to the TATA-box binding protein (TBP) showed a molecular bottle-opener for TBP (PDB 3G65) 57, 58. In a hybrid approach involving EM and X-ray crystallography, a structure of a different chromatin remodeling enzyme, ISW1 (lacking the ATPase domain), bound to a nucleosomal template was elucidated (PDB 2Y9Z and EMD-1877) [33]. Strikingly, the entire 13 subunit yeast APC/C, an E3 ubiquitin ligase essential for the cell cycle, was assembled by MultiBac (Box 3) as well as two multisubunit APC/C subcomplexes (TPR5 and SC8), leading to structures revealing many details of APC/C assembly by EM (EMD-1844, 1841 and 1845) [34]. Molecular illustrations were prepared with PyMOL (http://www.pymol.org/).
Box 2. Crystal structure of the Mediator head module.
Mediator is a large multiprotein complex that is central to gene expression in eukaryotes and is organized into three modules termed head, middle or arm, and tail [63]. The size and complexity of Mediator has long posed a major challenge for structural studies. Mediator head (seven subunits, 223 kDa) was first successfully produced recombinantly by infecting insect cells with single baculoviruses, each encoding one subunit, opening the door for structural studies [36]. However, this procedure required lengthy optimization to indentify suitable, highly producing recombinant baculoviruses by repeated rounds of laborious screening – a process of 8 weeks or longer for each baculovirus. The demanding procedures constrained logistics and severely compromised attempts at structure determination of Mediator head by X-ray crystallography, which typically necessitates flexible, stream-lined and efficient production protocols for generating many variants of the protein specimen studied. The MultiBac system elegantly resolved this fundamental impediment [35]. All genes encoding the subunits of the Mediator head module were inserted into a single MultiBac baculovirus by using tandem recombineering 36, 64. Combinatorial assembly of the genes lead to a series of MultiBac baculoviruses encoding either full Mediator head (Med17, Med6, Med18, Med8, Med20, Med11 and Med22) or subcomplexes including core head (Med17, Med6, Med8, Med11 and Med22) and mini head (Med17, Med11 and Med22). Investigation of these complexes by EM allowed initial assignment of subunit positions [64]. Modification of Med17 and Med18 by eliminating flexible regions was crucial to obtain diffraction-quality head module crystals, and this was easily accomplished by simply exchanging the respective unmodified genes in the original MultiBac baculovirus expressing the full head [35]. The modular acceptor–donor construction principle of MultiBac is crucial to facilitate this alteration, alleviating a need to assemble the entire multigene construct every time again de novo. The recombinant head module has been purified and crystallized, and the architecture of this 223-kDa complex has been determined by X-ray crystallography (Figure I ). The crystal structure of the Mediator head reveals how this essential complex is built from its components to provide stability as well as flexibility for transcription regulation, resulting in a platform for other transcription factors [35]. Notably, a portion of the head named ‘neck domain’ confers stability and integrity of the complex by forming a striking, novel multihelical bundle, engaging five of the seven subunits of Mediator head.
Figure I.

Architecture of Mediator head. The seven-subunit, 223-kDa complex was produced by MultiBac, crystallized and the X-ray structure determined [35]. (a) A representative gel section from SDS-PAGE showing the quality of sample prepared. (b) The crystal structure in a schematic view (top) and in an identically color-coded ribbon representation superimposed on the experimental electron density (bottom). Density is displayed at 4 Å resolution (contoured at 1.5 σ).
Two outstanding examples of the utility of MultiBac are the elegant production of the entire anaphase promoting complex, APC/C – a large (1.1 MDa) 13-subunit multiprotein assembly that regulates defined cell cycle transitions [34] – and the recent crystal structure elucidation of the Mediator head module – a transcription factor complex that is essential for the expression of class II genes in eukaryotes [35]. The gene assembly encoding the complete APC/C was inserted into two MultiBac baculoviruses, encoding eight and five subunits, respectively. Co-infection of insect cells with the two viruses allowed the purification of the entire 1.1-MDa complex and its structural determination by electron microscopy (EM), revealing the structural basis for subunit assembly (Figure 2) [34]. The production of the Mediator head module from yeast (seven subunits, 223 kDa), a paradigmatic case study for the successful application of the MultiBac system, is detailed in Box 2 [35].
Clearly, MultiBac is only one of many tools that bring such studies to fruition. Nonetheless, quality sample preparation is a crucial component of the structural determination pipeline. The studies already available exemplify the aptitude of the system, and hint at recurring challenges encountered when using MultiBac in multiprotein complex research (Box 3 ). The list of illuminating structures of complexes produced by using MultiBac can be expected to grow rapidly in the future.
Box 3. MultiBac debugged: challenges and solutions.
A multitude of factors can compromise the production of multiprotein complexes. Among the major impediments are:
Insufficient knowledge of the composition of the complex. Recombinant production of such a complex by coexpression will invariably fail if a subunit crucial for complex integrity has not been properly identified in original preparations. Although proteomics and genomics technologies increasingly contribute to improve and validate the data available (reviewed in [1]), protein complex coexpression can already reveal important information about interacting partners even if the (presumed) complete complex cannot be obtained.
Post-translational modifications (PTMs). PTMs such as phosphorylation and acetylation can be essential for proper assembly of a complex or for its activity. Thus, the modification activity needs to be either coexpressed or supplied during purification. An additional problem is that insect cells may decorate proteins with nonphysiological PTMs (frequently phosphorylation), which compromise complex formation or activity. Therefore, mutations that abolish the modification activity must be introduced in the host genome, or enzymes (such as λ-phosphatase) that remove the modification have to be coexpressed or supplied [23]. Alternatively, inhibitors of the particular modifying enzyme (if known) can be added to the growth media if compatible with cell growth and protein production.
Complex instability. Coexpression of an entire complex from a single virus may not be the ultimate solution. For example, a complex may dissociate during purification in a salt gradient. It may be worth identifying exactly what components dissociate during purification; stable subcomplexes may then be produced separately to reconstitute the complex in vitro by choosing proper conditions.
Effort required to place all subunit genes on a single baculovirus. For very large complexes, this may be challenging, and applying co-infection with a small number (two or three) of multigene viruses instead may be a viable option.
Virus instability. This is a drawback of every baculovirus expression system, because baculoviruses deteriorate during amplification and passaging. Gene deletions, notably affecting heterologous DNA inserts, can potentially abolish complex production. To minimize such deleterious events, specific protocols have been delineated 13, 20, and new approaches to overcome viral in stability by genome engineering are being developed [65].
Complexes come in many forms: the coatomer
Proteins and their complexes in their native tissue can exist as several isoforms, which may complicate their biochemical and structural characterization when extracted and purified from endogenous source material by immunoprecipitation or classical biochemical approaches. An example is the coatomer, the central protein component that covers certain vesicles, known as coat protein I (COPI)-coated vesicles, which are thought to play an important role in the early secretory pathway. Coatomer consists of seven subunits and exists in four isoforms of different subunit composition. The four isoforms co-purify when extracted from animal tissue, which hampers their structural analysis and impedes attempts to understand the potentially different functions of the individual isoforms. For example, it remains unclear whether or not COPI-coated vesicles are uniformly coated with individual isoforms, although these have been found to be unequally distributed in the Golgi stack [37].
Recently, all four coatomer isoforms have been successfully produced using MultiBac, setting the stage to individually address their structure and function [38]. This has been achieved by integrating the genes encoding the five subunits that are common to all isoforms into the Tn7 attachment site of the MultiBac viral genome. The genes for the other two subunits, which are variable and define the four isoforms, were inserted by using a donor plasmid into the LoxP site that is distal to the Tn7 attachment site on the MultiBac viral genome. Coatomer complex isoforms can then be purified successfully from insect cell cultures infected with the respective MultiBac virus [38].
A MultiBac silkworm bioreactor
Silkworms, the larvae of the moth Bombyx mori, have been used for thousands of years to produce silk for textile production, but they have gained a new role as protein production bioreactors after the successful expression of human α-interferon in these insects. A growing number of proteins, including interleukins and antibodies, have since been produced using this system 39, 40. Interestingly, it seems that silkworms can sometimes produce much higher yields of recombinant proteins than can be obtained from liquid cell cultures. For example, the activity of interleukin-3 produced in silkworms is 10 000-fold higher than that produced in cultures of immortalized African green monkey kidney (COS) cells, and 30-fold higher than that produced in insect cell suspension culture. Notably, 1 mg of purified human macrophage colony-stimulating factor can be extracted from 10 silkworms [39].
These impressive results have prompted the development of a version of the MultiBac system adjusted for protein complex production in a silkworm bioreactor [41]. In this system, the original baculovirus genome was replaced with that of a B. mori nuclear polyhedrosis virus (BmNPV), likewise provided as an artificial chromosome in bacterial cells. A Tn7 attachment site was introduced into this BmNPV genome, and a plasmid expressing the Tn7 transposon complex was co-transformed. Thus, the silkworm MultiBac system retains the ability to accept multigene vector constructions prepared by tandem recombineering or by conventional restriction and ligation cloning. A distinct feature of the B. mori MultiBac system is the method of infecting the silkworms, which occurs via injection of virus solution with a needle. Protein complexes are then purified from the hemolymph of infected silkworms after several days.
Recombinant human DNA polymerase δ has been produced using silkworm MultiBac [41]. Although the enzyme is thought to contain four subunits, the extraction from animal tissue usually leads to loss of two subunits during purification. Four milligrams of active enzyme containing all four subunits can be purified from 350 silkworms infected with a silkworm MultiBac virus carrying four genes, providing a simple, fast, reliable and economic platform to produce human DNA polymerase δ. Therefore, the silkworm MultiBac system might be useful for the production of other protein complexes, especially if an economic option for protein production is required.
Extending the concept to mammalian cells
The concept of multigene delivery of fused acceptor and donor plasmids, each carrying one or several genes, is not restricted to baculovirus expression systems. Originally, tandem recombineering was developed for generating multiexpression plasmids for production of protein complexes in E. coli to alleviate imbalances in expression levels observed with multiplasmid co-transformation [21]. Expression of protein complexes in mammalian cells can be achieved by transfection with multiple plasmids, in analogy to multiplasmid co-transformation in E. coli. However, this approach can lead to imbalanced production from the plasmids and to heterogeneous cell populations with differences in the ratio of plasmids incorporated. This impediment can be efficiently resolved by using a single plasmid that contains all the heterologous genes. This multigene plasmid can be conveniently generated by tandem recombineering of properly modified acceptors and donors, in which the baculoviral promoters are exchanged for mammalian promoters 22, 42. The resulting multigene plasmid is introduced into mammalian cells by using a transfection reagent [42].
The efficacy of this approach was demonstrated in a study where an assortment of proteins, systematically tagged with fluorescent markers, was expressed transiently or stably in animal tissue cells [42]. This MultiBac-derived system, called MultiLabel, can be used for simultaneously visualizing proteins and their actions in homogeneous cell populations by tracking the fluorescent signals. Importantly, the ratio of the heterologous proteins is constant at the single-cell level. MultiLabel has been recently implemented to dissect the interactions of epidermal growth factor receptor (EGFR), Rab GTPases and phosphoinositol phosphates, which are components of membrane trafficking processes 42, 43.
Gene therapy for obesity?
Gene therapy involves either the targeted delivery of one or several wild-type genes that replace or substitute in for disease-causing genes (which bear aberrant mutations) and produce the desired gene products. or the delivery of genes encoding therapeutic proteins that prevent, modulate or correct the disease [44]. Baculoviruses have been intensively studied as potential delivery vectors for gene therapy because they transduce mammalian cells efficiently, do not seem to be toxic, and are nonhazardous (for the target cells and for those carrying out the experiment) as they do not replicate in mammalian cells 17, 45, 46. Further advantages are the ease of producing baculoviruses and the large tolerance for heterologous gene insertions into the viral genome without compromising virion integrity. However, there are formidable obstacles to overcome, such as the observed inactivation of baculoviruses by the human complement system and the lack of specificity with respect to cell types transduced. These problems have not satisfactorily been resolved, therefore, baculoviruses have not yet played a major role as gene therapy vectors.
By contrast, recombinant adeno-associated virus (rAAV) does not have the drawbacks of baculovirus and is currently the best choice for efficient gene delivery for gene therapy [47]. However, clinical grade production of rAAVs, which are necessarily rendered replication incompetent, remains a major impediment for the field. A highly efficient scale-up protocol for rAAV production utilizes recombinant baculoviruses to produce gene-therapy-competent rAAV particles in insect cell cultures 48, 49, 50, 51. This protocol involves three recombinant baculoviruses that carry genes encoding different components of rAAV and the transgene to be delivered (Figure 3 ). Co-transfection of insect cells with the three baculoviruses results in the assembly of intact rAAV particles, which can be further processed and purified to achieve clinical grade.
Figure 3.
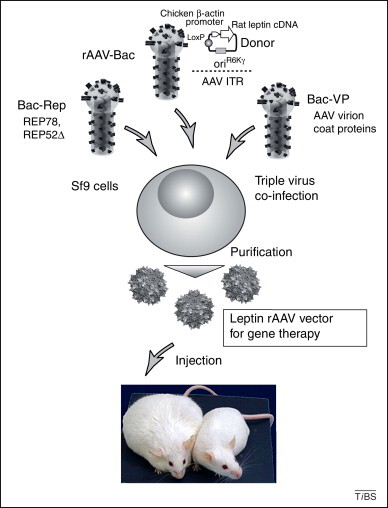
Application of the MultiBac system in gene therapy. rAAV particles are produced by co-infection with three different baculoviruses 48, 49. Bac-Rep harbors two expression cassettes that contain genes for the major AAV replication enzyme, Rep78, and an N-terminal truncation of Rep78, Rep52Δ. rAAV-Bac contains AAV inverted terminal repeat (ITR) elements that are required for rescue, replication and packaging of transgene sequences, together with rat leptin cDNA under the control of a chicken β-actin promoter, which is inserted into the rAAV-Bac baculoviral genome by Cre–LoxP-mediated fusion of a specifically tailored donor plasmid [52]. Leptin is a hormone that acts in the brain to reduce food intake and stimulate energy expenditure. Bac-VP produces the AAV virion coat proteins. Complete rAAV virions containing the leptin gene are produced in triply co-infected insect cells and purified 50, 52, and then administered to diet-induced obese rats. An obese rat is shown compared to a normal rat for illustration (bottom). Diet-induced obesity renders laboratory rats (and presumably other species) resistant to leptin treatment. Therefore, it is close to impossible to curtail diet-induced weight gain. This could be overcome by circumventing leptin resistance or restoring leptin actions in obese animals. The surprising outcome of the study involving baculovirus-produced leptin rAAV as a gene therapy vector was that exercise, in this study wheel-running, was required to prevent completely weight gain when combined with the leptin gene therapy intervention, leading to the conclusion that work-out, in tandem with leptin gene delivery, may actually develop into a potential antiobesity treatment [52]. The baculovirus schematic drawing is adapted from an image kindly supplied by K. J. Airenne (University of Kuopio, Finland). The rAAV particles shown are based on PDB entry 1LP3 [59].
A particularly noteworthy recent study has involved the production of rAAV particles for gene therapy of obesity in laboratory rodents (Figure 3) [52]. The rAAV administered to laboratory rats fed on a high-fat diet contained leptin cDNA as the transgene, introduced into the MultiBac baculoviral genome via Cre–LoxP fusion of a donor containing the leptin gene under control of a promoter that is active in mammalian cells (Figure 3).
Concluding remarks
In this review, we have presented approaches, notably the MultiBac technology, for producing important eukaryotic protein complexes that were hitherto not amenable to structural and functional analysis. Molecules as complex as the APC/C have been successfully produced using MultiBac, enabling structure determination and architectural dissection. More recently, the technology concepts underlying MultiBac have been extended to mammalian gene delivery and even gene therapy, and we anticipate that many more applications will benefit from this approach.
In addition, optimized multigene expression technologies that involve polyproteins and library approaches have become available, and the polyprotein technology has been integrated into MultiBac, increasing the prowess of the system. A beneficial next step could be to incorporate also library approaches such as CoESPRIT [31] (Box 1). This could conceivably accelerate the discovery of protein–protein interactions for complexes that rely on a eukaryotic expression host. Protein–protein interactions are intensively studied in the pharmaceutical industry, with the aim to identify intermolecular surfaces that can be targeted for drug design [53]. It will be interesting to see whether this field of discovery will also benefit from the MultiBac technology in the future.
Considerable effort has been devoted to generating custom-designed insect and mammalian host cells, which provide specialized functionalities such as humanized glycosylation [54]. A logical extension of the MultiBac system will be to make use of these host cells, for example to produce antibodies with a close-to-human glycosylation pattern for therapeutic applications.
The MultiBac system continues to catalyze progress in many fields of research. An entirely open question is where the limits of the MultiBac production technology may be. For instance, the transcriptional machinery producing eukaryotic mRNAs contains, in addition to the genetic DNA template, a stunning 100 proteins organized in multisubunit complexes. It may seem frivolous to set out with MultiBac to address this complexity structurally and functionally in a defined, fully recombinant setup. Nonetheless, the first glimpses of the molecular architectures involved are emerging: the Mediator head, produced by MultiBac, has been crystallized and the structure resolved. The stage is set. Exciting times are ahead of us.
Conflict of interest
The authors declare no competing financial interest.
Acknowledgments
We thank Christiane Schaffitzel, Darren Hart, Sergei Zolotukhin, Francisco Asturias and all members of our laboratories for helpful discussions, and Tim Richmond and Roger Kornberg for support. CB is recipient of a Swiss National Science Foundation (SNSF) Advanced Researcher grant. TI is a fellow of the Human Frontier of Science Program (HSFP). YT is supported by the US National Science Foundation (grant MCB 0843026) and the American Heart Association (grant 073595N). IB acknowledges support from the SNSF, the Agence Nationale de la Recherche (ANR), the Centre National de la Recherche Scientifique (CNRS), the EMBL and the European Commission (EC) through the joint EIPOD program, and the EC projects SPINE2-Complexes and 3D-Repertoire (Framework Program 6 (FP6)), as well as INSTRUCT, PCUBE, BioSTRUCT-X and ComplexINC (EC FP7).
Glossary
- Cre recombinase
Enzyme that recognizes, binds to, cuts and religates specific DNA sequences (called LoxP sequences). Four copies of Cre recombinase bind to two LoxP DNA sequences, and then Cre recombinase exchanges one strand each from the two LoxP sequences. If the two LoxP sequences are present on two different DNA molecules, then these will be conjoined as a result, resulting in one DNA molecule with two LoxP sites.
- Homing endonucleases
Restriction enzymes with particularly long (20–30 base pairs) recognition sequences. After cutting the DNA, they often leave four-nucleotide overhangs that can be compatible with overhangs generated by another restriction enzyme, BstXI; then, DNAs processed with homing endonucleases can be efficiently ligated to DNAs tailored by BstXI. Both recognition sequences are destroyed in the process, and the ligation product cannot be cut by either enzyme.
- Ligation independent cloning (LIC)
A method for inserting DNA sequences (i.e. genes) into a plasmid DNA molecule without using DNA ligase. DNAs to be conjoined are treated by exonucleases that trim back one strand of the DNA double helix, exposing the complementary strand as a long sticky end. If two DNA molecules have complementary sticky ends, they can be conjoined simply by mixing. LIC methods have become popular in recent years because they do not require preselected DNA sequences that serve as recognition sites for restriction enzymes. A version of LIC that is entirely independent of any DNA sequence is called SLIC (sequence and ligation independent cloning).
- LoxP sequence
Short imperfect inverted repeat of 34 base pairs which serves as a recognition site, cleavage site and religation site for Cre recombinase. The LoxP sequence is not symmetric, therefore, the conjoining of two LoxP sequences by Cre recombinase is directional.
- Nuclear polyhedrosis virus (NPV)
NPV attacks caterpillars, such as the larvae of the alfalfa looper or the silkworm. NPVs are highly specific for their host and thus useful as pest control agents.
- Polyprotein
Long polypeptide which contains several individual proteins that are connected by linker amino acids which either are capable of self-cleavage, or are recognition and cleavage sites of highly specific proteases.
- Tn7 transposon
Originally discovered as a large (14 kb) DNA segment from Tn7 phage, which can insert into a specific location in the E. coli genome. The transposon encodes the Tn7 transposase, a four-subunit protein complex that carries out this insertion reaction. Three DNA elements are minimally required for this reaction: the Tn7L and Tn7R DNA sequences which flank the DNA to be transposed, and a sequence called Tn7 attachment site (attTn7) into which the transposed DNA is inserted. This insertion mechanism can be efficiently exploited to conjoin recombinant DNA by providing the Tn7L and Tn7R sequences on a plasmid at each end of a DNA sequence of interest, and the attTn7 sequence on another DNA which will serve as recipient. The reaction is started by addition of the transposase to these DNAs.
References
- 1.Nie Y., et al. Getting a grip on complexes. Curr. Genomics. 2009;10:558–572. doi: 10.2174/138920209789503923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puig O., et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 3.Gavin A.C., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Gavin A.C., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 5.Robinson C.V., et al. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 6.Korostelev A., Noller H.F. The ribosome in focus: new structures bring new insights. Trends Biochem. Sci. 2007;32:434–441. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Tan S., Davey C.A. Nucleosome structural studies. Curr. Opin. Struct. Biol. 2010;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer P., et al. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 9.Kühner S., et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 10.Busso D., et al. Expression of protein complexes using multiple Escherichia coli protein co-expression systems: A benchmarking study. J. Struct. Biol. 2011;175:159–170. doi: 10.1016/j.jsb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Diebold M.N., et al. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 2011;175:178–188. doi: 10.1016/j.jsb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Berger I., et al. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald D.J., et al. Protein complex expression by using multigene baculoviral vectors. Nat. Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 14.Nettleship J.E., et al. Recent advances in the production of proteins in insect and mammalian cells for structural biology. J. Struct. Biol. 2010;172:55–65. doi: 10.1016/j.jsb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Smith G.E., et al. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol. 1983;3:2156–2165. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summers M.D. Milestones leading to the genetic engineering of baculoviruses as expression vector systems and viral pesticides. Adv. Virus Res. 2006;68:3–73. doi: 10.1016/S0065-3527(06)68001-9. [DOI] [PubMed] [Google Scholar]
- 17.Kost T.A., et al. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 2005;23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kost T.A., et al. Baculovirus gene delivery: a flexible assay development tool. Curr. Gene Ther. 2010;10:168–173. doi: 10.2174/156652310791321224. [DOI] [PubMed] [Google Scholar]
- 19.Trowitsch S., et al. New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 2010;172:45–54. doi: 10.1016/j.jsb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Bieniossek C., et al. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr. Protoc. Protein Sci. 2008 doi: 10.1002/0471140864.ps0520s51. Chapter 5, Unit 5.20. [DOI] [PubMed] [Google Scholar]
- 21.Bieniossek C., et al. Automated unrestricted multigene recombineering for multiprotein complex production. Nat. Methods. 2009;6:447–450. doi: 10.1038/nmeth.1326. [DOI] [PubMed] [Google Scholar]
- 22.Vijayachandran L.S., et al. Robots, pipelines, polyproteins: enabling multiprotein expression in prokaryotic and eukaryotic cells. J. Struct. Biol. 2011;175:198–208. doi: 10.1016/j.jsb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzgerald D.J., et al. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure. 2007;15:275–279. doi: 10.1016/j.str.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Kieft J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komar A.A., Hatzoglou M. Cellular IRES-mediated translation. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semler B.L., Waterman M.L. IRES mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1–5. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Chen W.S., et al. Development of a prokaryotic-like polycistronic baculovirus expression vector by the linkage of two internal ribosome entry sites. J. Virol. Methods. 2009;159:152–159. doi: 10.1016/j.jviromet.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Szymczak A.L., et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 29.Kim J.H., et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., et al. TEV protease-facilitated stoichiometric delivery of multiple genes using a single expression vector. Protein Sci. 2010;19:2379–2388. doi: 10.1002/pro.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An Y., et al. CoESPRIT: a library-based construct screening method for identification and expression of soluble protein complexes. PLoS ONE. 2011;6:e16261. doi: 10.1371/journal.pone.0016261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leonard T.A., et al. Crystal structure and allosteric activation of protein kinase C βII. Cell. 2011;144:55–66. doi: 10.1016/j.cell.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada K., et al. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber A., et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 35.Imasaki T., et al. Architecture of the Mediator head module. Nature. 2011;475:240–243. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takagi Y., et al. Head module control of mediator interactions. Mol. Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Moelleken, et al. Differential localization of coatomer complex isoforms within the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4425–4430. doi: 10.1073/pnas.0611360104. [DOI] [PubMed] [Google Scholar]
- 38.Sahlmüller M.C., et al. Recombinant heptameric coatomer complexes: novel tools to study isoform-specific functions. Traffic. 2011;12:682–692. doi: 10.1111/j.1600-0854.2011.01177.x. [DOI] [PubMed] [Google Scholar]
- 39.Kato T., et al. Silkworm expression system as a platform technology in life science. Appl. Microbiol. Biotechnol. 2010;85:459–470. doi: 10.1007/s00253-009-2267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadono-Okuda K., et al. Baculovirus-mediated production of the human growth hormone in larvae of the silkworm, Bombyx mori. Biochem. Biophys. Res. Commun. 1995;213:389–396. doi: 10.1006/bbrc.1995.2144. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y., et al. Production of recombinant human DNA polymerase delta in a Bombyx mori bioreactor. PLoS ONE. 2011;6:e22224. doi: 10.1371/journal.pone.0022224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kriz A., et al. A plasmid-based multigene expression system for mammalian cells. Nat. Commun. 2010;1:120–124. doi: 10.1038/ncomms1120. [DOI] [PubMed] [Google Scholar]
- 43.Ballmer-Hofer K., et al. Neuropilin-1 promotes VEGFR-2 trafficking through Rab11 vesicles thereby specifying signal output. Blood. 2011;118:816–826. doi: 10.1182/blood-2011-01-328773. [DOI] [PubMed] [Google Scholar]
- 44.Cavazzana-Calvo M., et al. The future of gene therapy. Nature. 2004;427:779–781. doi: 10.1038/427779a. [DOI] [PubMed] [Google Scholar]
- 45.Wang S., Balasundaram G. Potential cancer gene therapy by baculoviral transduction. Curr. Gene Ther. 2010;10:214–225. doi: 10.2174/156652310791321251. [DOI] [PubMed] [Google Scholar]
- 46.Rivera-Gonzalez G.C., et al. Baculoviruses as gene therapy vectors for human prostate cancer. J. Invert. Pathol. 2011;107:S59–S70. doi: 10.1016/j.jip.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Mingozzi F., High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 48.Urabe M., et al. Scalable generation of high-titer recombinant adeno-associated virus type 5 in insect cells. J. Virol. 2006;80:1874–1885. doi: 10.1128/JVI.80.4.1874-1885.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohlbrenner E., et al. Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol. Ther. 2005;12:1217–1225. doi: 10.1016/j.ymthe.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith R.H., et al. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol. Ther. 2009;17:1888–1896. doi: 10.1038/mt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aslanidi G., et al. An inducible system for highly efficient production of recombinant adeno-associated virus (rAAV) vectors in insect Sf9 cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5059–5064. doi: 10.1073/pnas.0810614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shapiro A., et al. Synergy between leptin therapy and a seemingly negligible amount of voluntary wheel running prevents progression of dietary obesity in leptin-resistant rats. Diabetes. 2008;57:614–622. doi: 10.2337/db07-0863. [DOI] [PubMed] [Google Scholar]
- 53.Dev K.K. Making protein interactions druggable: targeting PDZ domains. Nat. Rev. Drug Discov. 2004;3:1047–1056. doi: 10.1038/nrd1578. [DOI] [PubMed] [Google Scholar]
- 54.Shi X., Jarvis D.L. Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets. 2007;8:1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeqiraj E., et al. Structure of the LKB1–STRAD–MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326:1707–1711. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doré A.S., et al. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex–implications for clamp loading and regulation. Mol. Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 57.Wollmann P., et al. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature. 2011;475:403–407. doi: 10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lall S. A bottle opener for TBP. Nat. Struct. Mol. Biol. 2011;18:865. doi: 10.1038/nsmb0811-865. [DOI] [PubMed] [Google Scholar]
- 59.Xie Q., et al. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tolia N.H., Joshua-Tor L. Strategies for protein coexpression in Escherichia coli. Nat. Methods. 2006;3:55–64. doi: 10.1038/nmeth0106-55. [DOI] [PubMed] [Google Scholar]
- 61.Tan S., et al. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Romier C., et al. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr. D: Biol. Crystallogr. 2006;62:1232–1242. doi: 10.1107/S0907444906031003. [DOI] [PubMed] [Google Scholar]
- 63.Chadick J.Z., Asturias F.J. Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 2005;30:264–271. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 64.Cai G., et al. Mediator head module structure and functional interactions. Nat. Struct. Mol. Biol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pijlman G.P., et al. Stabilized baculovirus vector expressing a heterologous gene and GP64 from a single bicistronic transcript. J. Biotechnol. 2006;123:13–22. doi: 10.1016/j.jbiotec.2005.10.022. [DOI] [PubMed] [Google Scholar]


