Abstract
The sequence of the VP7 gene of two rotaviruses isolated from dogs in southern Italy was determined and the inferred amino acid sequence was compared with that of other rotavirus strains. There was very high nucleotide and amino acid identity between canine strain RV198/95 and other canine strains, and to the human strain HCR3A. Strain RV52/96, however, was found to have about 95% identity to the G3 serotype canine strains K9, A79-10 and CU-1 and 96% identity to strain RV198/95 and to the simian strain RRV. Therefore both of the canine strains belong to the G3 serotype. Nevertheless, detailed analysis of the VP7 variable regions revealed that RV52/96 possesses amino acid substitutions uncommon to the other canine isolates. In addition, strain RV52/96 exhibited a nucleotide divergence greater than 16% from all the other canine strains studied; however, it revealed the closest identity (90.4%) to the simian strain RRV. With only a few exceptions, phylogenetic analysis allowed clear differentiation of the G3 rotaviruses on the basis of the species of origin. The nucleotide and amino acid variations observed in strain RV52/96 could account for the existence of a canine rotavirus G3 sub-type.
Keywords: Canine rotavirus, VP7, G3 serotype
1. Introduction
Group A rotaviruses are a major cause of neonatal diarrhea in humans and animals. Rotaviruses are non-enveloped and possess a triple-layered capsid, enclosing 11 fragments of double-stranded RNA (dsRNA). The outer viral capsid proteins, VP7 and VP4, elicit neutralizing antibody responses and form the basis of the current dual classification system by G and P types. The VP7 expresses the major neutralizing antigen and is distinguishable by means of both serological and genomic techniques in 14 G types, with good correlation between the serological and genomic classifications. Also, since the VP4 expresses the minor neutralizing antigen, the serological classification of the P types is much more difficult than the genomic classification. To date, 13 P serotypes (including subtypes) and 20 P genotypes have been defined, but no precise correlation has been made between the serological and the genomic classifications. Thus a different designation has been adopted for the P serotype (open numbers) and the P genotype (numbers in square brackets) (Estes and Cohen, 1989, Estes, 1996). As regards canine rotavirus, all the strains isolated display the G3 and P5A[3] specificities (Hoshino et al., 1984, Nakagomi et al., 1989, Gouvea et al., 1994a, Gouvea et al., 1994b, Taniguchi et al., 1994).
Canine rotavirus is usually responsible for subclinical or mild forms of enteritis, associated with anorexia and vomiting, especially in pups younger than 2 weeks of age (Pollock and Carmichael, 1990). The canine infection is considered a minor disease problem in pups (Pollock and Carmichael, 1990); however, serological investigations have shown a high prevalence of antibodies to canine rotavirus in adult dogs (Mochizuki et al., 1986).
There are only a few reports on the isolation of canine rotavirus from dogs affected with gastroenteritis. To date, three strains have been isolated in the US, CU-1, A79-10 and LSU79C-36 (also referred to as K9); one isolate (RS15) has been reported from Japan (Fulton et al., 1981, Hoshino et al., 1982, Hoshino et al., 1983, Mochizuki and Hsüan, 1984). In addition, two strains have been isolated from humans in the USA, HCR3A (Li et al., 1993) and HCR3B (Santos et al., 1998), and one isolate has been reported in Israel, Ro1845 (Aboudy et al., 1988), all which have been shown to be genetically related to canine rotavirus and to share the same G3P5A[3] specificity (Gouvea et al., 1990, Nakagomi et al., 1990, Li et al., 1993, Nakagomi et al., 1993, Vonsover et al., 1993, Li et al., 1994, Taniguchi et al., 1994, Santos et al., 1998).
In the present note, the VP7 nucleotide and the corresponding inferred amino acid sequences of two canine rotaviruses isolated in Italy are reported and analyzed. A phylogenetic tree based on G3 human and animal rotaviruses was also elaborated.
2. Materials and methods
2.1. Viruses
Two crossbred pups of 2.5 and 6 months of age (#52/96 and #198/95, respectively) with acute gastroenteritis were presented, on separate occasions, to the Small Animal Clinic of the Veterinary Faculty, University of Bari, for clinical examination and treatment. Both pups recovered 1 week after the onset of clinical signs. Electron microscopy (EM) examination of fecal specimens from both pups revealed the presence of viral particles with rotaviral morphology. In addition, parvovirus- and coronavirus-like particles were observed in the feces of pup #198/95.
Two rotavirus isolates (RV52/96 and RV198/95) were made on MA-104 (foetal monkey kidney) cells in the presence of trypsin (5 μg/ml in maintenance medium). Viral growth was monitored by an indirect immunofluorescence assay. Viral dsRNA was recovered from the infected cells by standard phenol-chloroform extraction procedures and subjected to electrophoresis. Both the isolates had virtually identical mobilities of the genomic segments with an electrophoretical pattern 4:2:3:2, typical of group A rotaviruses (data not shown).
2.2. Polymerase chain reaction (PCR) amplification
For PCR amplification, rotaviral dsRNA was extracted from infected MA-104 cells, using the RNeasy Kit (Qiagen Gmbh, Hilden, Germany). Reverse transcription (RT) PCR was carried out as previously described (Gouvea et al., 1990, Isegawa et al., 1993). The pair of generic primers Beg9 and End9, which amplifies the entire VP7 gene, was used both for reverse transcription of genomic RNA and for PCR.
2.3. Sequencing and sequence analysis
The RT-PCR amplicons were purified on Ultrafree-DA Columns (Amicon, Millipore) and then sequenced directly with ABI-PRISM 377 (Perkin Elmer, Applied Biosystem Instruments) using an overlapping strategy; i.e. starting with the generic primers Beg9 and End9 and choosing additional primers on the basis of the sequence obtained, in order to completely sequence the VP7 gene in both directions. The sequence was determined twice for each virus. The primers used for sequencing are listed in Table 1 .
Table 1.
Primers used for sequencing strains RV198/95 and RV52/96a
| Primer | Sequence 5′–3′ | Sense |
|---|---|---|
| Beg9b (1–28) | GGC TTT AAA AGA GAG AAT TTC CGT CTG G | + |
| End9b (1062–1036) | GGT CAC ATC ATA CAA TTC TAA TCT AAG | − |
| 198R1 (245–226) | GCA GTA TCC ATT GAA CCA GT | − |
| 198R2 (600–583) | CCA CTT ATT AGC TTC ATC | − |
| 198F1 (361–377) | CTA TTT CTA ACT AAA GG | + |
| 198F2 (723–742) | GAT CAC TGA TGT CGT TGA TG | + |
| 198F3 (900–918) | GAT GCG TAT TAA TTG GAA G | + |
| 52R1 (278–259) | GGA AAG TCT CTT CCT GTG TG | − |
| 52R2 (672–653) | TAG ACA CCC AAT TCC AAG AG | − |
| 52F1 (357–374) | CAC AGT TGT TTT TGA CC | + |
| 52F2 (620–637) | GTA CAA TAA AAG TGT GTC | + |
The position and the sense of the primers is also reported.
Sequence analysis was performed with NCBI's and EMBL's analysis tools. The alignment of sequences was performed with CLUSTAL W (Thompson et al., 1994). Phylogenetic analysis was carried out with TREECOON package (Van De Peer and De Wachter, 1993). A neighbor-joining tree (Saitou and Nei, 1987) was generated using the gamma distribution model (Ota and Nei, 1993), with bootstrapping over 1000 replicates. For comparative analysis, selected VP7 sequences of G3 human and animal rotaviruses were used (Table 2 ). The nucleotide sequences of strains CU-1, A79-10, CAT2 and CAT97 were kindly supplied by Dr Y. Hoshino. The sequences of strains RV198/95 and RV52/96 are available in GenBank under accession numbers AF271089 and AF271090, respectively.
Table 2.
List of the G3 animal and human rotaviruses used in this studya
Multiple references are reported for viruses sequenced more times. n.r. not registered in the databases.
3. Results
Similar to most of the group A rotaviruses (Estes and Cohen, 1989, Bellamy and Both, 1990), the VP7 nucleotide sequence of strains RV198/95 and RV52/96 is 1062 nucleotides long. The ORF starts at nucleotide 49 with an AUG (ATG) start codon and ends at nucleotide 1029 with a UAG (TAG) termination codon, comprising 981 nucleotides. The inferred VP7 amino acid sequence of strains RV198/95 and RV52/96, aligned with the sequences of other G3 animal and human rotaviruses, is reported in Fig. 1 . As expected, the sequence of both the strains is 326 amino acids long and conserves the potential glycosylation site NST at residues 69–71 (Bellamy and Both, 1990).
Fig. 1.
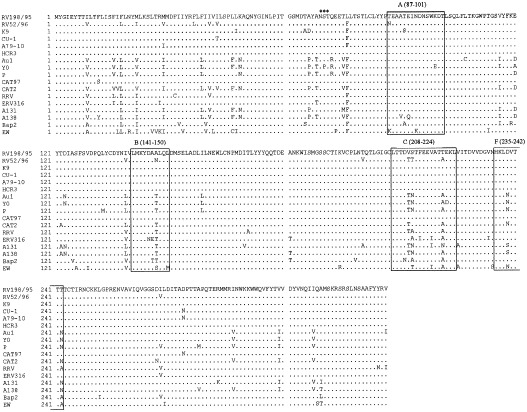
Amino acid sequence of the VP7 protein of strains RV198/95 and RV52/96 aligned with that of other G3 animal and human rotaviruses. The strains are referenced in Table 2. The variable regions A (aa 87–101), B (aa 141–150), C (aa 208–224) and F (aa 235–242) are shown. The NST glycosylation site is indicated by asterisks.
On the basis of sequence analysis, both strains RV198/95 and RV52/96 clearly belong to the G3 serotype. Rotaviruses belonging to the same G serotype generally share more than 91% VP7 amino acid similarity (Kapikian and Chanock, 1996, Palombo et al., 1997), even if strains displaying an overall identity of only 88.7% are included into the G3 serotype (Li et al., 1994). As shown in Table 3 , strain RV198/95 had the highest nucleotide identity (about 96–97%) to the canine strains K9, CU-1, A79-10 and to the human strain HCR3A. Sequence similarity of strain RV52/96 was highest (90%) to the rhesus rotavirus RRV, followed by the equine strain ERV316 (85.8%), rather than to the canine strains (83–84%). In regard to the amino acid composition, strain RV198/95 revealed the highest identity to the G3 human strain HCR3A (100%), followed by the canine strains K9, CU-1, A79-10 (97.5–99%) and the feline strain CAT97 (99.4%). As regards strain RV52/96, a high amino acid similarity was found to the simian strains RRV (96.1%) and SA11 (95.1%), whereas identity to the canine strains was about 94–95%. The two Italian isolates showed about 84.0% nucleotide and 96.2% amino acid similarity to each other.
Table 3.
VP7 nucleotide and amino acid identity among the G3 animal and human rotavirusesa
| Strain | 198/95 | 52/96 | HCR3 | CU-1 | A79-10 | CAT2 | CAT97 | SA11 | RRV | Erv316 | A-138 | YR-1 | BAP-2 | TK28 | Y0 | Au-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K9 Dog | 96.9/97.5 | 82.9/94.2 | 95.6/98.5 | 96.4/98.8 | 96.6/98.8 | 79.8/90.5 | 96.0/99.1 | 82.5/93.3 | 84.3/93.9 | 87.8/93.6 | 80.1/89.6 | 74.9/87.2 | 84.4/92.1 | 80.0/90.2 | 78.9/89.0 | 78.7/90.2 |
| RV198/95 Dog | ■ | 84.0/96.2 | 96.1/99.8 | 96.2/99.1 | 96.6/99.1 | 80.4/91.5 | 96.8/99.4 | 83.3/94.2 | 84.7/94.8 | 89.1/94.5 | 80.5/90.2 | 76.0/87.5 | 86.6/93.0 | 81.4/91.2 | 80.3/90.8 | 80.1/91.2 |
| RV52/96 Dog | ■ | 83.8/95.1 | 84.1/95.1 | 83.9/95.1 | 82.8/92.1 | 83.9/94.5 | 83.6/95.1 | 90.4/96.1 | 85.8/94.5 | 80.0/90.8 | 76.6/89.4 | 82.7/92.7 | 82.1/91.5 | 82.9/90.5 | 82.4/90.8 | |
| HCR3 Man | ■ | 97.0/99.1 | 97.2/99.1 | 80.9/91.5 | 95.1/99.4 | 83.9/94.2 | 85.0/94.8 | 88.5/94.5 | 80.9/90.2 | 76.3/87.5 | 85.0/92.3 | 81.1/91.2 | 80.5/90.8 | 80.0/91.1 | ||
| CU-1 Dog | ■ | 98.7/99.4 | 80.8/91.5 | 95.8/99.1 | 83.6/93.9 | 85.7/94.5 | 89.1/94.2 | 81.3/89.9 | 76.2/87.2 | 85.9/92.7 | 81.3/90.8 | 80.7/90.5 | 80.3/90.8 | |||
| A79-10 Dog | ■ | 80.8/91.8 | 96.2/99.1 | 83.9/94.5 | 85.3/94.5 | 89.1/94.2 | 80.7/90.5 | 76.2/87.8 | 85.6/92.7 | 81.3/91.5 | 80.5/91.2 | 80.3/91.5 | ||||
| CAT2 Cat | ■ | 80.6/90.8 | 81.1/91.8 | 82.4/91.8 | 82.3/90.5 | 87.1/95.8 | 75.3/84.1 | 78.9/89.7 | 91.2/95.4 | 91.6/96.1 | 91.1/96.7 | |||||
| CAT97 Cat | ■ | 83.2/93.6 | 85.0/94.2 | 89.9/93.9 | 80.9/89.9 | 76.4/86.9 | 86.3/82.4 | 81.6/90.5 | 81.1/90.2 | 80.7/90.5 | ||||||
| SA11 Monkey | ■ | 84.7/95.4 | 84.3/93.9 | 80.3/91.8 | 78.1/88.1 | 81.1/92.4 | 81.9/91.4 | 81.9/92.2 | 81.6/92.4 | |||||||
| RRV Monkey | ■ | 85.8/95.1 | 79.6/90.8 | 77.2/87.8 | 82.1/93.6 | 82.4/91.8 | 82.6/90.8 | 82.1/91.8 | ||||||||
| Erv316 Horse | ■ | 80.9/90.2 | 77.1/87.8 | 85.2/93.6 | 82.5/91.5 | 82.6/90.2 | 82.3/90.5 | |||||||||
| A138 Pig | ■ | 76.3/83.2 | 78.9/89.0 | 86.4/94.2 | 87.1/94.8 | 87.3/95.4 | ||||||||||
| YR-1 Mouse | ■ | 75.4/86.9 | 74.9/84.7 | 75.2/83.5 | 74.8/83.8 | |||||||||||
| BAP-2 Rabbit | ■ | 80.1/89.9 | 80.0/88.4 | 79.8/88.7 | ||||||||||||
| TK-28 Man | ■ | 96.5/96.3 | 96.4/97.0 | |||||||||||||
| Y0 Man | ■ | 97.8/98.2 |
The strains are referenced in Table 2. The nucleotide percent values are indicated on the left, the amino acid percent value on the right.
In Fig. 2 the neighbor-joining tree based on the VP7 amino acid sequence of human and animal G3 rotaviruses is presented. Human G3 rotaviruses differ from the animal G3 isolates, with exception of the human strain HCR3A, which is clustered together with the canine strains, and the feline strain CAT2, which is grouped with the human isolates. Strain RV52/96 seems to be genetically more similar to the simian strain RRV.
Fig. 2.
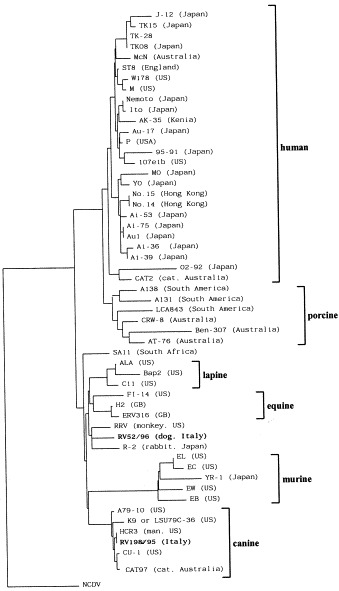
Neighbour-joining tree based on the VP7 amino acid sequences of G3 human and animal rotaviruses. Horizontal branches are drawn to scale and the tree is rooted with the G6 serotype bovine strain NCDV (accession M63266). Bootstrap values are not shown. The strains are referenced in Table 2. The geographical origin of the strains is reported in brackets. The host species of isolation of the viruses is reported in brackets only when necessary.
4. Discussion
G3 serotype rotaviruses have been found in a broad range of host species, including humans, monkeys, dogs, cats, horses, rabbits, mice, sheep and pigs (Hoshino et al., 1984, Paul et al., 1988, Fitzgerald et al., 1995).
Studies of escape mutants made with neutralizing monoclonal antibodies and sequence analysis of viruses with different serotypes have identified six variable regions in the rotaviral VP7 protein. However, only region A (residues 87–101), B (residues 141–150), C (residues 208–224) and F (residues 235–242) have been confirmed to have a major role in neutralization (Dyall-Smith et al., 1986, Green et al., 1987, Coulson and Kirkwood, 1991, Kirkwood et al., 1993, Ciarlet et al., 1997b). Comparative analysis of the VP7 protein of G3 human and animal rotaviruses showed, at the amino acid level, that sequence divergence among G3 strains from different host species occurs primarily in regions A, B and C. In contrast, those regions exhibit a high degree of conservation among G3 strains belonging to the same host species (Nishikawa et al., 1989).
The high degree of conservation of region A is believed to be very important for G3 serotype strains to maintain serotype specificity (Nishikawa et al., 1989, Ciarlet et al., 1997b). Strain RV52/96 has a substitution at amino acid 87 (asn for thr), which is considered a key contact residue of the immunodominant epitopes defining serotype G3 rotavirus strains (Ciarlet et al., 1997b). In region B, the sequence LMKYDAALQL is considered species-specific for canine and feline strains (Nishikawa et al., 1989) and residue 147 is considered to be critical for both serotype specificity (Dyall-Smith et al., 1986) and monotype specificity (Coulson and Kirkwood, 1991). In this region, strain RV52/96 shows a substitution at the critical residue 147 (asn for ala). In region C, which has been suggested to be an immunodominant antigenic site (Kirkwood et al., 1993, Ciarlet et al., 1997b), RV52/96 possesses three changes with respect to the other canine strains.
In summary, strain RV52/96 possesses amino acid variations in regions A, B and C and about 16–17% nucleotide divergence from to the other described canine isolates K9, CU-1, A79-10 and RV198/95. The differences observed demonstrate that variability exist among the canine rotaviruses. Similarly, nucleotide and amino acidic variation in the VP7 of human G3 rotaviruses has been recently described and serotype 3 viruses are currently considered to be intraserotypically more heterologous than serotype 1, 2 and 4 viruses (Wen et al., 1997, Suzuki et al., 1998). Furthermore, by cross-neutralization tests, monoclonal antibody analysis and RNA hybridization, two G3 subtypes of equine rotavirus have been established (Browning et al., 1992). Therefore, we hypothesize that the differences observed in strain RV52/96 could account for the existence of a G3-subtype of canine rotavirus. However, the analysis of additional canine isolates and serological evaluations are required to confirm this hypothesis.
The neighbor-joining tree based on the VP7 gene shows the presence of different clusters among the G3 serotype rotaviruses. With few exceptions, clustering seems to follow a species-specific pattern. The animal isolates are phylogenetically distinct from the human isolates. The porcine, murine, lapine, equine and canine strains, moreover, are clearly distinguishable from each other. In agreement with previous observations (Ciarlet et al., 1995), the porcine isolates seem to be more related to human than to animal rotaviruses. The human strain HCR3 shows a high homology with the canine strains, whereas the feline strain CAT2 is grouped into the human cluster. This is not surprising since strain HCR3A has been shown to be genetically highly related to the canine strains (Gouvea et al., 1990, Li et al., 1993, Li et al., 1994, Taniguchi et al., 1994, Santos et al., 1998). On the other hand, strain CAT2 shares the VP4 specificity, P3A[9], with human strains such as Au1 and K8 (Gouvea et al., 1994a, Taniguchi et al., 1994). As regard strain RV52/96, the homology with strain RRV is consistent with previous findings (Taniguchi et al., 1994) describing the close genetic relationship between the VP4 of canine rotaviruses and RRV. These results point out the usefulness of phylogenetic analysis for obtaining a more comprehensive understanding of the complex ecology of rotaviruses.
In conclusion, this is the first report describing the isolation of canine rotaviruses outside the USA and Japan, and confirms that the G3 is the unique serotype within the canine rotaviruses. Finally, the data provided by sequence comparison and phylogenetic analysis revealed nucleotide and amino acid variability which has not been reported previously among the canine rotavirus strains.
Acknowledgements
We thank Mr Donato Narcisi for his technical collaboration and Dr Yasutaka Hoshino for supplying the VP7 nucleotide sequences of the canine and feline strains. We thank Professor Leland E. Carmichael for revising the English and for his suggestions.
References
- Aboudy, Y., Shif, I., Ziberstein, I., Gotlieb-Stematsky, T., 1988. Use of polyclonal and monoclonal antibodies and analysis of viral RNA in the detection of unusual group A human rotaviruses. J. Med. Virol. 25, 351–359. [DOI] [PubMed]
- Bellamy A.R., Both G.W. Molecular biology of rotaviruses. Adv. Virus Res. 1990;38:1–38. doi: 10.1016/s0065-3527(08)60858-1. [DOI] [PubMed] [Google Scholar]
- Both G.W., Mattick J.S., Bellamy A.R. Serotype-specific glycoprotein of simian rotavirus: coding assignment and gene sequence. Proc. Natl. Acad. Sci. USA. 1983;80:3091–3095. doi: 10.1073/pnas.80.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning G.F., Chalmers R.M., Fitzgerald T.A., Snodgrass D.R. Evidence for two serotype G3 subtypes among equine rotaviruses. J. Clin. Microbiol. 1992;30:485–491. doi: 10.1128/jcm.30.2.485-491.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning G.F., Begg A.P. Prevalence of G and P serotypes among equine rotaviruses in the feces of diarrhoeic foals. Arch. Virol. 1996;141:1077–1089. doi: 10.1007/BF01718611. [DOI] [PubMed] [Google Scholar]
- Choi A.H., Basu M., Rae M.N., McNeal M.M., Ward R.L. Particle-bombardment-mediated DNA vaccination with rotavirus VP4 or VP7 induces high levels of serum rotavirus IgG but fails to protect mice against challenge. Virology. 1998;250:230–240. doi: 10.1006/viro.1998.9370. [DOI] [PubMed] [Google Scholar]
- Ciarlet M.L., Ludert J.E., Liprandi F. Comparative amino acid sequence analysis of the major outer capsid protein (VP7) of porcine rotaviruses with G3 and G5 serotype specifities isolated in Venezuela and Argentina. Arch. Virol. 1995;140:437–451. doi: 10.1007/BF01718422. [DOI] [PubMed] [Google Scholar]
- Ciarlet M.L., Estes M.K., Conner M.E. Comparative amino acid sequence analysis of the outer capsid protein VP4 from four lapine rotavirus strains reveal identity with P[14] human rotaviruses. Arch. Virol. 1997;142:1059–1069. doi: 10.1007/s007050050142. [DOI] [PubMed] [Google Scholar]
- Ciarlet M.L., Hoshino Y., Liprandi F. Single point mutation may affect the serotype reactivity of G11 porcine rotavirus strains: a widening spectrum? J. Virol. 1997;71:8213–8220. doi: 10.1128/jvi.71.11.8213-8220.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson B., Kirkwood C. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J. Virol. 1991;65:5968–5974. doi: 10.1128/jvi.65.11.5968-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith M.L., Lazdins I., Tregar G.W., Holmes I.H. Location of major antigenic sites involved in rotavirus serotype-specific neutralization. Proc. Natl. Acad. Sci. USA. 1986;83:3465–3468. doi: 10.1073/pnas.83.10.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S.J., Burns J.W., Cross T.L., Vo O.T., Ward R.L., Bremont M., Greenberg H.B. Comparison of VP4 and VP7 of five murine rotavirus strains. Virology. 1994;203:250–259. doi: 10.1006/viro.1994.1482. [DOI] [PubMed] [Google Scholar]
- Estes M.K., Cohen J. Rotavirus gene structure and function. Microbiol. Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M.K. Rotaviruses and their replication. In: Fields B.N., Knipe D.N., Howley P.M., Chanock R.M., Melnick J.L., Monath T.P., Roizman B., Straus S.E., editors. Vol. 2. Raven Press; New York, NY: 1996. pp. 1625–1655. (Virology). [Google Scholar]
- Fitzgerald T.A., Munoz M., Wood A.R., Snodgrass D.R. Serological and genomic characterization of group A rotaviruses from lambs. Arch. Virol. 1995;140:1541–1548. doi: 10.1007/BF01322528. [DOI] [PubMed] [Google Scholar]
- Fulton R.W., Johnson C.A., Pearson N.J., Woode G.N. Isolation of a rotavirus from newborn dog with diarrhea. Am. J. Vet. Res. 1981;42:841–843. [PubMed] [Google Scholar]
- Gouvea V., Glass R.I., Woods P., Taniguchi K., Clark H.F., Forrester B., Fang Z.Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acids from stool specimens. J. Clin. Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V., Santos N., Timenetsky M.C. VP4 typing of bovine and porcine group A rotaviruses by PCR. J. Clin. Microbiol. 1994;32:1333–1337. doi: 10.1128/jcm.32.5.1333-1337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V., Santos N., Timenetsky M.C. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 1994;32:1338–1340. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.Y., Midthun K., Gorziglia M., Hoshino Y., . Kapikian A.Z., Chanock R.M., Flores J. Comparison of the amino acid sequences of the major neutralization protein of four human rotavirus serotypes. Virology. 1987;161:153–159. doi: 10.1016/0042-6822(87)90181-4. [DOI] [PubMed] [Google Scholar]
- Hoshino Y., Wyatt R.G., Scott F.W., Appel M.J. Isolation and characterization of a canine rotavirus. Arch. Virol. 1982;72:113–125. doi: 10.1007/BF01314456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y., Wyatt R.G., Greenberg H.B., Kalica A.R., Flores J., Kapikian A.Z. Serological comparison of canine rotavirus with various simian and humans rotaviruses by plaque reduction neutralization and hemagglutination inhibition tests. Infect. Immun. 1983;41:169–173. doi: 10.1128/iai.41.1.169-173.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y., Wyatt R.G., Greenberg H.B., Flores J., Kapikian A.Z. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J. Infect. Dis. 1984;149:694–702. doi: 10.1093/infdis/149.5.694. [DOI] [PubMed] [Google Scholar]
- Huang J., Nagesha H.S., Dyall-Smith M.L., Holmes I.H. Comparative sequence analysis of VP7 genes from five Australian porcine rotaviruses. Arch. Virol. 1989;109:173–183. doi: 10.1007/BF01311079. [DOI] [PubMed] [Google Scholar]
- Isegawa Y., Nagakomi O., Nagakomi T., Ishida S., Uesugi S., Ueda S. Determination of bovine rotavirus G and P serotypes by polymerase chain reaction. Mol. Cell. Probes. 1993;7:277–284. doi: 10.1006/mcpr.1993.1041. [DOI] [PubMed] [Google Scholar]
- Kapikian A.Z., Chanock R.M. Rotaviruses. In: Fields B.N., Knipe D.M., Howley P.M., editors. 3rd edition. Vol. 2. Lippincott-Raven; Philadelphia, PA: 1996. pp. 1657–1708. (Fields Virology). [Google Scholar]
- Kirkwood C., Masendycz P.J., Coulson B.S. Characterization and location of cross-reactive and serotype-specific neutralization sites on VP7 of human G type 9. Virology. 1993;196:79–88. doi: 10.1006/viro.1993.1456. [DOI] [PubMed] [Google Scholar]
- Li B., Clark H.F., Gouvea V. Nucleotide sequence of the VP4-encoding gene of an unusual human rotavirus (HCR3) Virology. 1993;196:825–830. doi: 10.1006/viro.1993.1540. [DOI] [PubMed] [Google Scholar]
- Li B., Clark H.F., Gouvea V. Amino acid sequence similarity of the VP7 protein of human rotavirus HCR3 to that of canine and feline rotaviruses. J. Gen. Virol. 1994;75:215–219. doi: 10.1099/0022-1317-75-1-215. [DOI] [PubMed] [Google Scholar]
- Mackow E.R., Shaw R.D., Matzui S.M., Vo P.T., Benfield D.A., Greenberg H.B. Characterization of homotypic and heterotypic VP7 neutralizing sites of rhesus rotavirus. Virology. 1988;165:511–517. doi: 10.1016/0042-6822(88)90595-8. [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Hsüan S. Isolation of a rotavirus from canine diarrhoeal feces. Nippon Juigaku Zasshi. 1984;46:905–908. doi: 10.1292/jvms1939.46.905. [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Minami K., Sakamoto H. Seroepizootiologic studies on rotavirus infections of dogs and cats. Jpn. J. Vet. Sci. 1986;48:957–964. doi: 10.1292/jvms1939.48.957. [DOI] [PubMed] [Google Scholar]
- Nakagomi T., Matsuda Y., Ohshima A., Mochizucki M., Nagakomi O. Characterization of a canine rotavirus strain by neutralization and molecular hybridization assays. Arch. Virol. 1989;106:145–150. doi: 10.1007/BF01311046. [DOI] [PubMed] [Google Scholar]
- Nakagomi O., Ohshima A., Aboudy Y., Shif I., Mochizuki M., Nagakomi T., Gotlieb Stematsky T. Molecular identification by RNA–RNA hybridization of a human rotavirus that is closely related to rotaviruses of feline and canine origin. J. Clin. Microbiol. 1990;28:1198–1203. doi: 10.1128/jcm.28.6.1198-1203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagomi O., Isegawa Y., Hoshino Y., Aboudy Y., Shif I., Silberstein I., Nakagomi T., Ueda S., Sears J., Flores J. A new serotype of the outer capsid protein VP4 shared by an unusual human rotavirus strain Ro1845 and canine rotaviruses. J. Gen. Virol. 1993;74:2771–2774. doi: 10.1099/0022-1317-74-12-2771. [DOI] [PubMed] [Google Scholar]
- Nagesha H.S., Huang J., Holmes I.H. A variant serotype G3 rotavirus isolated from an unusually severe outbreak of diarrhoea in piglets. J. Med. Virol. 1992;38:79–85. doi: 10.1002/jmv.1890380202. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Hoshino Y., Taniguchi K., Green K.Y., Greenberg H.B., Kapikian A.Z., Chanock R.M., Gorziglia M. Rotavirus VP7 neutralization epitopes of serotype 3 strains. Virology. 1989;171:503–515. doi: 10.1016/0042-6822(89)90620-x. [DOI] [PubMed] [Google Scholar]
- Ota T., Nei M. Estimation of the number of amino acid substitutions per site when the substitution rate varies among different sites. J. Mol. Evol. 1993;38:642–643. [Google Scholar]
- Palombo E.A., Bugg H.C., Masendycz P.J., Coulson B.S., Barnes G.L., Bishop R.F. Sequence of the VP7 gene of an atypical human rotavirus: evidence for genetic and antigenic drift. DNA Seq. 1997;7:307–311. doi: 10.3109/10425179709034050. [DOI] [PubMed] [Google Scholar]
- Paul P.S., Lyoo Y.S., Andrews J.J., Hill H.T. Isolation of two new serotypes of porcine rotavirus from pigs with diarrhoea. Arch. Virol. 1988;100:139–143. doi: 10.1007/BF01310917. [DOI] [PubMed] [Google Scholar]
- Pollock R.V.H., Carmichael L.E. Canine viral enteritis. In: Greene C.E., editor. Infectious Diseases of the Dog and Cat. W.B. Saunders; Philadelphia: 1990. pp. 268–287. [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Santos N., Clark H.F., Hoshino Y., Gouvea V. Relationship among serotype G3P5A rotavirus strains isolated from different host species. Mol. Cell. Probes. 1998;12:379–386. doi: 10.1006/mcpr.1998.0198. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Gojobori T., Nakagomi O. Intragenic recombinations in rotaviruses. FEBS Lett. 1998;427:183–187. doi: 10.1016/s0014-5793(98)00415-3. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Urusawa T., Urasawa S. Species specificity and interspecies relatedness in VP4 genotypes demonstrated by VP4 sequence analysis of equine, feline and canine rotavirus strains. Virology. 1994;200:390–400. doi: 10.1006/viro.1994.1203. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima H., Norikawa S., Mukoyama A., Nishio O. Characterization of VP4 and VP7 of a murine rotavirus (YR-1) isolated in Japan. Jpn. J. Med. Sci. Biol. 1995;48:237–247. doi: 10.7883/yoken1952.48.237. [DOI] [PubMed] [Google Scholar]
- Van De Peer Y., De Wachter R. TREECON: a software package for the construction and drawing of evolutionary trees. Comput. Applic. Biosci. 1993;9:177–182. doi: 10.1093/bioinformatics/9.2.177. [DOI] [PubMed] [Google Scholar]
- Vonsover A., Shif I., Silberstein I., Rudich H., Aboudy Y., Mendelson E., Shulman L., Nagakomi T., Nagakomi O. Identification of feline- and canine-like rotaviruses isolated from humans by restriction fragment length polymorphism assay. J. Clin. Microbiol. 1993;31:1783–1787. doi: 10.1128/jcm.31.7.1783-1787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Nakayama M., Yamanishi Y., Nishio O., Fang Z.Y., Nagakomi O., Araki K., Nishimura S., Hasegawa A., Muller W.E., Ushijima H. Genetic variation in the VP7 gene of human rotavirus serotype 3 (G3 type) isolated in China and Japan. Arch. Virol. 1997;142:1481–1489. [PubMed] [Google Scholar]


