Abstract
The current frequency of Canine Parvovirus variants (CPV2a, CPV2b and CPV2c) in the Argentine dog population was investigated by PCR amplification of a 583 bp fragment in the VP2 gene. From a total of 79 rectal swab samples that have been submitted to our laboratory since 2008, 55 (69.6%) resulted positive and were further analyzed by direct DNA sequencing. Fifty positives samples (91%) were characterized as CPV2c variant, which appeared in Argentina in the year 2003 and has been the prevalent type since 2008, whereas CPV2a and CPV2b, still found in Argentine dogs, were represented in 3.6% and 5.4% of the population, respectively. Considering that CPV2c is spreading worldwide, and that this variant is also affecting vaccinated dogs, efforts should be made towards the development of new matched CPV vaccines.
Keywords: Canine Parvovirus, VP2 gene, CPV2c variants, Sequence analysis
1. Introduction
Four different families of animal viruses have been identified as the etiological agents of severe enteritis in dogs: Canine Parvovirus (CPV), Canine Coronavirus, Canine Rotavirus and Canine Distemper Virus (Mochizuki et al., 1993, Pollock and Carmichael, 1983, Decaro et al., 2009a). Because of the severity of the disease and its rapid spread throughout the canine population, CPV has arisen a great deal of public interest.
CPV is a non-enveloped virus with a single-stranded DNA genome of approximately 5000 bases which encodes two structural (VP1 and VP2) and two non-structural (NS1 and NS2) proteins (Cotmore and Tattersall, 1987).
CPV was first identified in May 1978 as a new virus infecting dogs (Parrish and Kawaoka, 2005, Kapil et al., 2007). In the late 1970s and early 1980s both live and inactivated Feline Panleukopenia (FPL) vaccines were used to protect dogs against CPV disease. However the level of protection they afforded was poor and the duration of immunity was short. These vaccines were replaced by live attenuated CPV vaccines, which provided good protection and longer lasting immunity (Spibey et al., 2008).
Some biological properties of the virus, particularly its rate of nucleotide substitution which is closer to that of RNA viruses (aprox. 10−3/10−4) than to that of double-stranded DNA viruses (Shackelton et al., 2005), determined the continuous appearance of new variants with high sanitary impact in the vaccinated and unvaccinated dog populations. Two antigenic variants, named CPV2a and CPV2b, appeared a few years after the discovery of the first characterized CPV2.
Currently, the original CPV2 strain is not circulating in dog populations, although it is still present in vaccine formulations, whereas the variants CPV2a and CPV2b are distributed worldwide (Parrish et al., 1991, Martella et al., 2005a, Decaro et al., 2006b, Mochizuki et al., 1993, Buonavoglia et al., 2000, de Ybanez et al., 1995, Greenwood et al., 1996, Sagazio et al., 1998, Steinel et al., 1998, Truyen et al., 1996, Pereira et al., 2000, Cavalli et al., 2008, Buonavoglia et al., 2001, Martella et al., 2004, Martella et al., 2005b, Truyen et al., 2000, Martella et al., 2006).
CPV2a, differs from the original type 2 strain in the presence of five amino acid (aa) changes in the VP2 coat protein, while the antigenic differences observed in CPV2b are the consequence of only one aa substitution (Asn426Asp), located in the major antigenic site of the capsid (epitope A) (Parrish et al., 1991).
A new antigenic variant, carrying the aa substitution Asp426Glu, in the major antigenic site of the viral capsid of the VP2 protein, was reported in Italy in the year 2001 (Buonavoglia et al., 2001, Desario et al., 2005). This newest variant -designated CPV2c- has already been detected in other European countries, as well as in Asia, Africa and America (Decaro et al., 2009a, Decaro et al., 2009b, Nakamura et al., 2004, Kapil et al., 2007, Martella et al., 2004, Decaro et al., 2007, Hong et al., 2007, Perez et al., 2007, Joao Vieira et al., 2008, Calderon et al., 2009, Nandi et al., 2010).
It is important to emphasize that the updating of methodologies by incorporation of the Minor Groove Binder probe technology, as well as PCR amplification followed by sequencing, allowed a precise identification of emerging CPV variants, therefore becoming important tools for the study of CPV epidemiology (Decaro et al., 2006a, Decaro et al., 2005c, Decaro et al., 2005b).
In the present study, we report an update of the identification and characterization of different CPV variants in Argentina.
2. Materials and methods
2.1. Clinical specimens
A total of 79 rectal swabs samples were obtained from domestic dogs from the Argentine cities of Buenos Aires (n = 50), Dolores (n = 2), Tandil (n = 4), Pehuajó (n = 1), Mar del Plata (n = 1), Bahía Blanca (n = 12), Córdoba (n = 2), Misiones (n = 1) and Río Negro (n = 3), located 216, 350, 370, 400, 690, 710, 1060, and 1500 Km from Buenos Aires, respectively. A sample from Paraguay was also analyzed while the origin of two other samples was unknown.
Clinical specimens were submitted to our laboratory for diagnostic purposes between the years 2008 and 2010. Information about the animals whose samples tested positive for CPV, such as clinical symptoms, age, gender, breed and vaccination status, is provided in Table 1 .
Table 1.
Age, gender, vaccination records and clinical signs of dogs infected with CPV.
| Strain | Year | Age (months) | Sex | Breed | Vaccination status | Clinical signs | CPV strain | Origin | aa at 440 |
|---|---|---|---|---|---|---|---|---|---|
| Arg19 | 2008 | 2 | M | R | V (C) | + | CPV2c | Buenos Aires | T |
| Arg20 | 2008 | 2 | NA | AD | V (C) | + | CPV2c | Bahía Blanca | T |
| Arg21 | 2008 | 2 | NA | AD | V (C) | + | CPV2c | Bahía Blanca | T |
| Arg22 | 2008 | 2 | NA | AD | V (C) | + | CPV2c | Bahía Blanca | T |
| Arg23 | 2008 | 2 | NA | AD | V (C) | + | CPV2c | Bahía Blanca | T |
| Arg24 | 2008 | 5 | M | MB | V (C) | + | CPV2c | Buenos Aires | T |
| Arg25 | 2008 | 2 | F | MS | V (C) | + | CPV2c | Tandil | T |
| Arg26 | 2008 | 2 | M | GS | V (C) | + | CPV2c | Río Negro | T |
| Arg27 | 2008 | 2 | M | GS | V (C) | + | CPV2c | Río Negro | T |
| Arg28 | 2008 | 4 | F | MP | V (C) | + | CPV2c | Buenos Aires | T |
| Arg29 | 2008 | 3 | M | GP | V (C) | + | CPV2c | Buenos Aires | T |
| Arg30 | 2008 | 19 | M | GP | V (C) | + | CPV2a | Buenos Aires | T |
| Arg31 | 2008 | 4 | M | SFT | V (C) | + | CPV2c | Bahía Blanca | T |
| Arg32 | 2008 | 1 | M | P | NV | + | CPV2c | Buenos Aires | T |
| Arg33 | 2008 | 3 | F | AD | V (C) | + | CPV2c | CórdobaT | T |
| Arg34 | 2008 | NA | NA | MS | NA | NA | CPV2c | Buenos Aires | T |
| Arg35 | 2008 | 4 | M | FT | V (C) | + | CPV2c | Tucuman | T |
| Arg36 | 2008 | NA | NA | Pi | V (NA) | NA | CPV2a | Buenos Aires | T |
| Arg37 | 2009 | 3 | F | BD | V (NA) | NA | CPV2c | Buenos Aires | T |
| Arg38 | 2009 | 3 | F | MB | NV | + | CPV2c | Buenos Aires | T |
| Arg39 | 2009 | 3 | F | MB | NV | + | CPV2c | Buenos Aires | T |
| Arg40 | 2009 | 1 | M | D | NV | + | CPV2c | Buenos Aires | A |
| Arg41 | 2009 | 2 | F | MS | V (C) | + | CPV2c | Buenos Aires | A |
| Arg42 | 2009 | 12 | M | MP | V (C) | + | CPV2c | Buenos Aires | T |
| Arg43 | 2009 | 3 | F | C | V (C) | + | CPV2c | Buenos Aires | A |
| Arg44 | 2009 | 3 | M | B | V (C) | + | CPV2c | Buenos Aires | T |
| Arg45 | 2009 | NA | F | R | NV | + | CPV2c | Buenos Aires | T |
| Arg46 | 2009 | 3 | M | BD | V (C) | + | CPV2b | Buenos Aires | T |
| Arg47 | 2009 | 4 | F | PB | NV | + | CPV2c | Buenos Aires | T |
| Arg48 | 2009 | 6 | F | LR | V (C) | + | CPV2c | Buenos Aires | A |
| Arg49 | 2009 | 3 | F | MB | NV | + | CPV2c | Buenos Aires | A |
| Arg50 | 2009 | 1 | M | G | V (C) | + | CPV2b | Dolores | T |
| Arg51 | 2009 | 1 | F | G | V (C) | + | CPV2b | Dolores | T |
| Arg52 | 2009 | 2 | M | LR | NV | + | CPV2c | Buenos Aires | A |
| Arg53 | 2009 | 2 | F | MP | V (C) | + | CPV2c | Buenos Aires | A |
| Arg54 | 2009 | 8 | M | MP | V (C) | + | CPV2c | Buenos Aires | A |
| Arg55 | 2009 | NA | NA | YT | V (NA) | NA | CPV2c | Buenos Aires | A |
| Arg56 | 2009 | 2 | M | B | V (C) | + | CPV2c | Buenos Aires | A |
| Arg57 | 2009 | 7 | M | GS | V (C) | + | CPV2c | Buenos Aires | A |
| Arg58 | 2009 | 2 | F | SFT | V (C) | + | CPV2c | Bahía Blanca | T |
| Arg59 | 2009 | 1 | M | MP | V (C) | + | CPV2c | NA | A |
| Arg60 | 2009 | 2 | F | Bea | NV | + | CPV2c | Buenos Aires | A |
| Arg61 | 2009 | 5 | M | MB | V (C) | + | CPV2c | CórdobaT | T |
| Arg62 | 2009 | 4 | M | RR | V (C) | + | CPV2c | Misiones | T |
| Py1 | 2009 | 4 | M | NM | V (NA) | + | CPV2c | Paraguay | A |
| Arg63 | 2010 | 2 | M | GR | V (C) | + | CPV2c | NA | T |
| Arg64 | 2010 | NA | NA | SFT | NA | NA | CPV2c | Bahía Blanca | T |
| Arg65 | 2010 | NA | NA | SFT | NA | NA | CPV2c | Bahía Blanca | T |
| Arg 66 | 2010 | 5 | F | Bea | V (C) | + | CPV2c | Buenos Aires | A |
| Arg67 | 2010 | NA | F | MS | V (C) | + | CPV2c | Buenos Aires | A |
| Arg 68 | 2010 | 3 | M | MS | V (C) | + | CPV2c | Mar del Plata | T |
| Arg 69 | 2010 | 2 | M | Pug | V (C) | + | CPV2c | Buenos Aires | A |
| Arg 70 | 2010 | 2 | F | GR | V (C) | + | CPV2c | Buenos Aires | A |
| Arg 71 | 2010 | 2 | M | P | V (C) | + | CPV2c | Buenos Aires | A |
| Arg 72 | 2010 | 4 | M | SFT | V (C) | + | CPV2c | Bahía Blanca | T |
F: female; M: male; MB: Mixed Breed; GR: Golden Retriever; LR, Labrador Retriever; GS: German Shepherd; R: Rottweiler; D: Doberman; AD: Argentine Dogo; MS: Miniature Schnauzer; FT: Fox Terrier; YT: Yorkshire Terrier; SFT: Smooth Fox Terrier; C: Cocker; P: Poodle; MP: Miniature Poodle; B: Boxer; Bea: Beagle; BD: Bulldog; Pi: Pinscher; PB: Pitbull; G: Greyhound; NM: Neapolitan Mastiff; RR: Rhodesian Ridgeback; GP: Giant Poodle. V: vaccinated; NV: no vaccinated; NA: no information available. C: complete vaccination according to its age.
2.2. Preparation of samples for PCR analysis
CPV genomic DNA was extracted directly from rectal swabs and from a commercial vaccine (Vanguard® Plus CPV, Pfizer), in a lysis buffer containing 50 mM Tris–HCl pH 8, 100 mM NaCl, 25 mM sucrose, 10 mM EDTA and 1% SDS. After lysis, the extracts were digested with proteinase K (Invitrogen®, USA) at 56 °C for 30 min and the DNAs were extracted with phenol–chloroform.
2.3. Primers and PCR amplification
For the amplification of a large fragment of the VP2 capsid protein-encoding gene of CPV, the 555for/555rev primer set (5′-CAGGAAGATATCCAGAAGGA-3′/5′-GGTGCTAGTTGATATGTAATAAACA-3′), was used. The resulting product (583 bp), comprises at least six or seven informative aa responsible for important biological properties of the virus (Buonavoglia et al., 2001).
PCR amplification was performed using Taq recombinant polymerase (Invitrogen, USA) in an MJ Research cycler (PTC-100, Sierra Point, CA), with an initial denaturation step at 94 °C for 10 min, followed by 40 cycles of PCR (94 °C for 30 s, 50 °C for 60 s, and 72 °C for 60 s). The final concentration of MgCl2 was 1.5 mM.
2.4. Sequencing of the amplified DNA
The DNA fragments amplified from clinical samples were directly sequenced by the dideoxy-mediated chain-termination method (Macrogen Inc., Korea). Multiple sequence alignment analysis was performed using ClustalW.
The GenBank accession numbers of the aa and nucleotide sequences of reference CPV strains are: CPV-b strain (CPV2, M38245); CPV-15 strain (CPV2a, M24003); CPV-39 strain (CPV2b, M74849) and 56/00 (CPV2c, FJ222821).
The GenBank accession numbers of the nucleotide sequences of the DNA fragments amplified from local variants are: HQ589341, HQ589342, HQ445626, HQ450025, HQ450027, HQ45002, and HQ450029 (sample from Paraguay). For practical purposes, identical sequences were grouped, and only one of them was registered in the database.
3. Results
3.1. CPV detection by PCR
During this study, 79 rectal swabs samples from animals suspected of CPV infection were tested. CPV specific DNA was detected by PCR amplification of a 583 bp fragment, in 55 out of 79 (69.6%) analyzed rectal swabs. The DNA extracted from a commercial vaccine was used as positive control. In all cases, the obtained PCR products were analyzed electrophoretically, and no differences were found in the size of the amplicons (data not shown).
Negative samples were assayed twice to confirm the results. Forty-nine out of 55 positive animals (96%) showed clinical symptoms compatible with CPV, while no records were available for the other six dogs. Interestingly, most of the positive samples (78.2%) were recovered from 1 to 5 months old puppies (Table 1). Forty-three of them (78%) were obtained from puppies which had been vaccinated against CPV at least once, whereas eleven (20%) were obtained from unvaccinated dogs, and the vaccination status of the remaining dog was unknown.
No relationship could be established regarding sex or breed (Table 1).
3.2. Identification of CPV variants by DNA sequencing
In order to identify and differentiate CPV variants in local clinical samples, the PCR products from all the positive samples were sequenced and the deduced aa sequences were aligned.
Fig. 1 shows the results of the sequence alignment from the position 407–456 of VP2, derived from seven representative local field samples identified as CPV2a, CPV2b and CPV2c, along with the sequences of the vaccine strain (CPV2) and three international reference strains. At the 426 position CPV2a variants showed an Asn (N), CPV2b an Asp (D) whereas the CPV2c variants showed a Glu (E) residue. Similar analysis performed with all the CPV positive samples, revealed that 50 out of 55 (91%) carried the Asp426Glu mutation, characteristic of the CPV2c variant, whereas two samples were characterized as CPV2a, and the remaining three, were defined as CPV2b (data not shown). In addition, the aa alignments performed with the complete 583 bp, amplification products, showed a low degree of variation (≤1%) among the Argentine CPV2c variants. This observation is in accordance with previous reports from other authors (Decaro et al., 2009b, Kapil et al., 2007, Pereira et al., 2007).
Fig. 1.
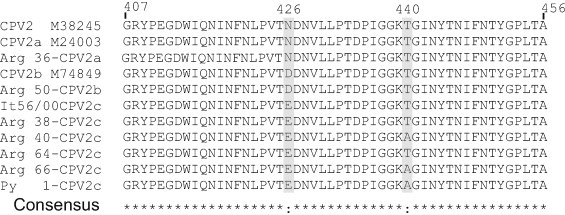
Alignment of the deduced amino acid sequences of a fragment of the 583 bp amplicon of the VP2 gene amplified by PCR from clinical samples (aa 407–456). The aligned sequences correspond to a vaccine strain (CPV2, M38245), a CPV2a reference strain (CPV2a, M24003), a CPV2b reference strain (CPV2b M74849), an Italian CPV2c strain (FJ222821.1), and Argentine CPV2a, CPV2b and CPV2c variants. The positions 426 and 440 are highlighted.
From the results shown in Fig. 2 and Table 1, it is clear that all CPV2 variants are at present affecting the Argentine dog population. However since the year 2008 CPV2c has become the predominant variant considering that 91% of the positive samples belonged to this variant, whereas CPV2a and CPV2b were represented only in 3.6% and 5.4% of the clinical samples, respectively.
Fig. 2.
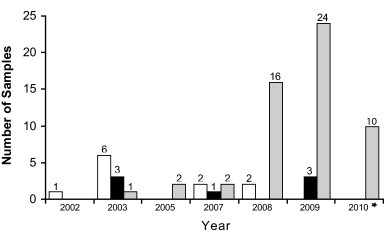
Updating of the evolution of CPV variants in Argentina. The number of cases detected in the years 2002–2010 belonging to each CPV variant are shown on top of the bars (previous study, Calderon et al., 2009). *Up to October 2010. White bars, CPV2a variants; black bars, CPV2b variants; grey bars, CPV2c variants.
3.3. A potentially relevant aminoacidic change
Besides the aa substitution at the 426 position, analysis of the sequences of Argentine CPV variants, showed the appearance of a new mutation, T440A, which is restricted to the CPV2c local variants, and whose worldwide presence has been recently reported (Decaro et al., 2009b, Kapil et al., 2007, Hong et al., 2007, Chinchkar et al., 2006, Kang et al., 2008).
The analysis performed, clearly demonstrated that 100% of the samples characterized as CPV2c obtained in 2008, presented the typical aa T at the 440 position, whereas 58% and 50% of the CPV2c samples obtained in 2009 and 2010, respectively, showed the presence of a new aa substitution (T440A), indicating that the variant CPV2cA440 has been introduced recently in the Argentine dog population.
3.4. Age distribution of CPV2c variant
As can be seen in Fig. 3 , the distribution of the CPV2c variant according to age clearly shows that the majority of the puppies (86%) affected by this variant were between 1 and 5-month-old, with most of the samples belonging to 2 month-old puppies.
Fig. 3.
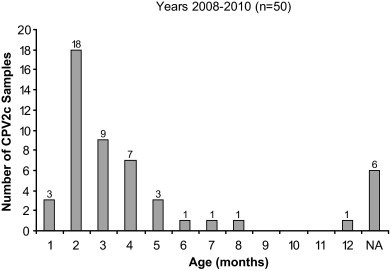
Distribution of the CPV2c variants in Argentine dogs, according to age.
4. Discussion
The present work represents an updating of the incidence of different CPV variants in Argentina, including the cases characterized in a previous report (Calderon et al., 2009). Although CPV2c was first detected in 2003, the results presented herein clearly show that CPV2c has become the predominant variant since the year 2008, as 91% of the samples analyzed were found to carry the typical aa substitution Glu426 (Table 1 and Fig. 2). An additional mutation located at an exposed domain of the VP2 capsid protein (Glu426-Ala440) was also found in some of the CPV2c samples (Fig. 1), which is in agreement with previous reports (Decaro et al., 2009b, Kapil et al., 2007, Hong et al., 2007, Chinchkar et al., 2006, Kang et al., 2008). It is interesting to point out that this mutation was found exclusively in samples collected during the years 2009 and 2010, indicating its recent appearance in the Argentine dog population.
The correlation of the T440A substitution with biological properties of the virus is still unknown, but since it is located at an important VP2 domain, exposed to selective pressure of vaccine or colostral antibodies, epidemiological surveillance is required to investigate the evolution, behavior and eventual sanitary impact of the viruses carrying this mutation (Hoelzer and Parrish, 2010, Hoskins, 2009).
Except for the two relevant aa substitutions mentioned, a low degree of variability (≤1%) was found in the VP2 sequences of CPV2c Argentine field variants. This observation is in agreement with previous reports from other regions of the world (Decaro et al., 2009b).
It has been reported that dogs infected with the newest CPV2c variant showed frequently atypical clinical symptoms, thus making the diagnosis of CPV disease difficult (Decaro et al., 2005a). In addition, classical diagnostic procedures are not suitable for precise CPV strain identification. The traditional PCR amplification of a 583 bp fragment showed to be effective in the detection of CPV DNA, but as the size PCR products derived from CPV2, CPV2a, CPV2b and CPV2c are indistinguishable from each other; sequencing is mandatory to allow the precise identification of each variant.
The clinical samples analyzed in this work were obtained from a population composed predominantly by young puppies, and were submitted to our laboratory by associated dog breeder's veterinarians. Our results clearly showed that the higher incidence of CPV2c was in 1–5-month-old puppies, with a peak in 2-month-old animals (Fig. 3). The vaccination status of this particular population was variable, with animals who had received only one or up to three doses of vaccine.
Although these findings need further confirmation by analyzing a larger number of animals, it may be speculated that at an early age there is a weakness or even a gap in the protective status of the puppies, probably due to low levels of maternal antibodies, or to the presence of an immature immune system, unable to respond properly to vaccination. Similar results were reported by our laboratory in the case of calf neonatal diarrhea caused by rotavirus (Bellinzoni et al., 1989). A strong increase in the level of colostral antibodies was demonstrated in this case through immunization of pregnant dams 40 days prior to delivery or by feeding exposed calves with hyperimmune colostrum. Therefore, vaccination of pregnant bitches might also increase the specific colostral and milk antibody levels in dogs, which can help dealing with the CPV2c infections in young puppies. Another possibility might be the development of improved vaccines or the updating of the vaccination schedules, in order to induce effective early protective responses.
Canine Parvovirus evolution and its rapid spreading through dog populations, raises concerns among breeders, owners and veterinary practitioners about the need of updating current CPV vaccines (Truyen, 2006). In addition, complementary sanitary measures, such as increasing the biosecurity levels of the breeding premises and the implementation of stronger sanitary regulations in the trade or exhibitions of live dogs, could also be relevant.
Acknowledgements
This work was supported with funds from FEVAN, CONICET and Tecnovax S.A. We thank Drs. Marenda, Leonardo Mauro and Ruben Somoza for providing the clinical specimens. We also thank Drs. Maximiliano Wilda and Blanca Robiolo for the critical revision and helpful discussion of the manuscript and Miss Silvia Rojana for her technical help.
References
- Bellinzoni R.C., Blackhall J., Baro N., Auza N., Mattion N., Casaro A., La Torre J.L., Scodeller E.A. Efficacy of an inactivated oil-adjuvanted rotavirus vaccine in the control of calf diarrhoea in beef herds in Argentina. Vaccine. 1989;7:263–268. doi: 10.1016/0264-410X(89)90241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonavoglia C., Martella V., Pratelli A., Tempesta M., Cavalli A., Buonavoglia D., Bozzo G., Elia G., Decaro N., Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 2001;82:3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- Buonavoglia D., Cavalli A., Pratelli A., Martella V., Greco G., Tempesta M., Buonavoglia C. Antigenic analysis of canine parvovirus strains isolated in Italy. New Microbiol. 2000;23:93–96. [PubMed] [Google Scholar]
- Calderon M.G., Mattion N., Bucafusco D., Fogel F., Remorini P., La Torre J. Molecular characterization of canine parvovirus strains in Argentina: detection of the pathogenic variant CPV2c in vaccinated dogs. J. Virol. Methods. 2009;159:141–145. doi: 10.1016/j.jviromet.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Cavalli A., Martella V., Desario C., Camero M., Bellacicco A.L., De Palo P., Decaro N., Elia G., Buonavoglia C. Evaluation of the antigenic relationships among canine parvovirus type 2 variants. Clin. Vaccine Immunol. 2008;15:534–539. doi: 10.1128/CVI.00444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S.F., Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- Chinchkar S.R., Mohana Subramanian B., Hanumantha Rao N., Rangarajan P.N., Thiagarajan D., Srinivasan V.A. Analysis of VP2 gene sequences of canine parvovirus isolates in India. Arch. Virol. 2006;151:1881–1887. doi: 10.1007/s00705-006-0753-8. [DOI] [PubMed] [Google Scholar]
- de Ybanez R.R., Vela C., Cortes E., Simarro I., Casal J.I. Identification of types of canine parvovirus circulating in Spain. Vet. Rec. 1995;136:174–175. doi: 10.1136/vr.136.7.174. [DOI] [PubMed] [Google Scholar]
- Decaro N., Desario C., Billi M., Mari V., Elia G., Cavalli A., Martella V., Buonavoglia C. Western European epidemiological survey for parvovirus and coronavirus infections in dogs. Vet. J. 2009 doi: 10.1016/j.tvjl.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Desario C., Campolo M., Elia G., Martella V., Ricci D., Lorusso E., Buonavoglia C. Clinical and virological findings in pups naturally infected by canine parvovirus type 2 Glu-426 mutant. J. Vet. Diagn. Invest. 2005;17:133–138. doi: 10.1177/104063870501700206. [DOI] [PubMed] [Google Scholar]
- Decaro N., Desario C., Elia G., Campolo M., Lorusso A., Mari V., Martella V., Buonavoglia C. Occurrence of severe gastroenteritis in pups after canine parvovirus vaccine administration: a clinical and laboratory diagnostic dilemma. Vaccine. 2007;25:1161–1166. doi: 10.1016/j.vaccine.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Desario C., Parisi A., Martella V., Lorusso A., Miccolupo A., Mari V., Colaianni M.L., Cavalli A., Di Trani L., Buonavoglia C. Genetic analysis of canine parvovirus type 2c. Virology. 2009;385:5–10. doi: 10.1016/j.virol.2008.12.016. [DOI] [PubMed] [Google Scholar]
- Decaro N., Elia G., Campolo M., Desario C., Lucente M.S., Bellacicco A.L., Buonavoglia C. New approaches for the molecular characterization of canine parvovirus type 2 strains. J. Vet. Med. B: Infect. Dis. Vet. Public Health. 2005;52:316–319. doi: 10.1111/j.1439-0450.2005.00869.x. [DOI] [PubMed] [Google Scholar]
- Decaro N., Elia G., Desario C., Roperto S., Martella V., Campolo M., Lorusso A., Cavalli A., Buonavoglia C. A minor groove binder probe real-time PCR assay for discrimination between type 2-based vaccines and field strains of canine parvovirus. J. Virol. Methods. 2006;136:65–70. doi: 10.1016/j.jviromet.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Elia G., Martella V., Desario C., Campolo M., Trani L.D., Tarsitano E., Tempesta M., Buonavoglia C. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet. Microbiol. 2005;105:19–28. doi: 10.1016/j.vetmic.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Decaro N., Martella V., Desario C., Bellacicco A.L., Camero M., Manna L., d’Aloja D., Buonavoglia C. First detection of canine parvovirus type 2c in pups with haemorrhagic enteritis in Spain. J. Vet. Med. B: Infect. Dis. Vet. Public Health. 2006;53:468–472. doi: 10.1111/j.1439-0450.2006.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desario C., Decaro N., Campolo M., Cavalli A., Cirone F., Elia G., Martella V., Lorusso E., Camero M., Buonavoglia C. Canine parvovirus infection: which diagnostic test for virus? J. Virol. Methods. 2005;126:179–185. doi: 10.1016/j.jviromet.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Greenwood N.M., Chalmers W.S., Baxendale W., Thompson H. Comparison of isolates of canine parvovirus by monoclonal antibody and restriction enzyme analysis. Vet. Rec. 1996;138:495–496. doi: 10.1136/vr.138.20.495. [DOI] [PubMed] [Google Scholar]
- Hoelzer K., Parrish C.R. The emergence of parvoviruses of carnivores. Vet. Res. 2010;41:39. doi: 10.1051/vetres/2010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C., Decaro N., Desario C., Tanner P., Pardo M.C., Sanchez S., Buonavoglia C., Saliki J.T. Occurrence of canine parvovirus type 2c in the United States. J. Vet. Diagn. Invest. 2007;19:535–539. doi: 10.1177/104063870701900512. [DOI] [PubMed] [Google Scholar]
- Hoskins J. DVM news Magazine; 2009. Canine parvovirus: An update on variants. A review of what's happening with canine parvovirus in the United States. 3. [Google Scholar]
- Joao Vieira M., Silva E., Oliveira J., Luisa Vieira A., Decaro N., Desario C., Muller A., Carvalheira J., Buonavoglia C., Thompson G. Canine parvovirus 2c infection in central Portugal. J. Vet. Diagn. Invest. 2008;20:488–491. doi: 10.1177/104063870802000412. [DOI] [PubMed] [Google Scholar]
- Kang B.K., Song D.S., Lee C.S., Jung K.I., Park S.J., Kim E.M., Park B.K. Prevalence and genetic characterization of canine parvoviruses in Korea. Virus Genes. 2008;36:127–133. doi: 10.1007/s11262-007-0189-6. [DOI] [PubMed] [Google Scholar]
- Kapil S., Cooper E., Lamm C., Murray B., Rezabek G., Johnston L., III, Campbell G., Johnson B. Canine parvovirus types 2c and 2b circulating in North American dogs in 2006 and 2007. J. Clin. Microbiol. 2007;45:4044–4047. doi: 10.1128/JCM.01300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Cavalli A., Decaro N., Elia G., Desario C., Campolo M., Bozzo G., Tarsitano E., Buonavoglia C. Immunogenicity of an intranasally administered modified live canine parvovirus type 2b vaccine in pups with maternally derived antibodies. Clin. Diagn. Lab Immunol. 2005;12:1243–1245. doi: 10.1128/CDLI.12.10.1243-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Cavalli A., Pratelli A., Bozzo G., Camero M., Buonavoglia D., Narcisi D., Tempesta M., Buonavoglia C. A canine parvovirus mutant is spreading in Italy. J. Clin. Microbiol. 2004;42:1333–1336. doi: 10.1128/JCM.42.3.1333-1336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Decaro N., Buonavoglia C. Evolution of CPV-2 and implication for antigenic/genetic characterization. Virus Genes. 2006;33:11–13. doi: 10.1007/s11262-005-0034-8. [DOI] [PubMed] [Google Scholar]
- Martella V., Decaro N., Elia G., Buonavoglia C. Surveillance activity for canine parvovirus in Italy. J. Vet. Med. B: Infect. Dis. Vet. Public Health. 2005;52:312–315. doi: 10.1111/j.1439-0450.2005.00875.x. [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Harasawa R., Nakatani H. Antigenic and genomic variabilities among recently prevalent parvoviruses of canine and feline origin in Japan. Vet. Microbiol. 1993;38:1–10. doi: 10.1016/0378-1135(93)90070-n. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Tohya Y., Miyazawa T., Mochizuki M., Phung H.T., Nguyen N.H., Huynh L.M., Nguyen L.T., Nguyen P.N., Nguyen P.V., Nguyen N.P., Akashi H. A novel antigenic variant of Canine parvovirus from a Vietnamese dog. Arch. Virol. 2004;149:2261–2269. doi: 10.1007/s00705-004-0367-y. [DOI] [PubMed] [Google Scholar]
- Nandi S., Chidri S., Kumar M., Chauhan R.S. Occurrence of canine parvovirus type 2c in the dogs with haemorrhagic enteritis in India. Res. Vet. Sci. 2010;88:169–171. doi: 10.1016/j.rvsc.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Parrish C.R., Aquadro C.F., Strassheim M.L., Evermann J.F., Sgro J.Y., Mohammed H.O. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 1991;65:6544–6552. doi: 10.1128/jvi.65.12.6544-6552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish C.R., Kawaoka Y. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 2005;59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- Pereira C.A., Leal E.S., Durigon E.L. Selective regimen shift and demographic growth increase associated with the emergence of high-fitness variants of canine parvovirus. Infect. Genet. Evol. 2007;7:399–409. doi: 10.1016/j.meegid.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Pereira C.A., Monezi T.A., Mehnert D.U., D’Angelo M., Durigon E.L. Molecular characterization of canine parvovirus in Brazil by polymerase chain reaction assay. Vet. Microbiol. 2000;75:127–133. doi: 10.1016/s0378-1135(00)00214-5. [DOI] [PubMed] [Google Scholar]
- Perez R., Francia L., Romero V., Maya L., Lopez I., Hernandez M. First detection of canine parvovirus type 2c in South America. Vet. Microbiol. 2007;124:147–152. doi: 10.1016/j.vetmic.2007.04.028. [DOI] [PubMed] [Google Scholar]
- Pollock R.V., Carmichael L.E. Canine viral enteritis. Vet. Clin. North. Am. Small Anim. Pract. 1983;13:551–566. doi: 10.1016/s0195-5616(83)50059-4. [DOI] [PubMed] [Google Scholar]
- Sagazio P., Tempesta M., Buonavoglia D., Cirone F., Buonavoglia C. Antigenic characterization of canine parvovirus strains isolated in Italy. J. Virol. Methods. 1998;73:197–200. doi: 10.1016/s0166-0934(98)00055-x. [DOI] [PubMed] [Google Scholar]
- Shackelton L.A., Parrish C.R., Truyen U., Holmes E.C. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. U.S.A. 2005;102:379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spibey N., Greenwood N.M., Sutton D., Chalmers W.S., Tarpey I. Canine parvovirus type 2 vaccine protects against virulent challenge with type 2c virus. Vet. Microbiol. 2008;128:48–55. doi: 10.1016/j.vetmic.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Steinel A., Venter E.H., Van Vuuren M., Parrish C.R., Truyen U. Antigenic and genetic analysis of canine parvoviruses in southern Africa. Onderstepoort J. Vet. Res. 1998;65:239–242. [PubMed] [Google Scholar]
- Truyen U. Evolution of canine parvovirus—a need for new vaccines? Vet. Microbiol. 2006;117:9–13. doi: 10.1016/j.vetmic.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Truyen U., Platzer G., Parrish C.R. Antigenic type distribution among canine parvoviruses in dogs and cats in Germany. Vet. Rec. 1996;138:365–366. doi: 10.1136/vr.138.15.365. [DOI] [PubMed] [Google Scholar]
- Truyen U., Steinel A., Bruckner L., Lutz H., Mostl K. Distribution of antigen types of canine parvovirus in Switzerland, Austria and Germany. Schweiz Arch Tierheilkd. 2000;142:115–119. [PubMed] [Google Scholar]


