Abstract
Angiotensin-converting enzyme (ACE) is a metallopeptidase that converts angiotensin I into angiotensin II. ACE is crucial in the control of cardiovascular and renal homeostasis and fertility in mammals. In vertebrates, both transmembrane and soluble ACE, containing one or two active sites, have been characterized. So far, only soluble, single domain ACEs from invertebrates have been cloned, and these have been implicated in reproduction in insects. Furthermore, an ACE-related carboxypeptidase was recently characterized in Leishmania, a unicellular eukaryote, suggesting the existence of ACE in more distant organisms.
Interestingly, in silico databank analysis revealed that bacterial DNA sequences could encode putative ACE-like proteins, strikingly similar to vertebrates' enzymes. To gain more insight into the bacterial enzymes, we cloned the putative ACE from the phytopathogenic bacterium, Xanthomonas axonopodis pv. citri, named XcACE. The 2 kb open reading frame encodes a 672-amino-acid soluble protein containing a single active site. In vitro expression and biochemical characterization revealed that XcACE is a functional 72 kDa dipeptidyl-carboxypeptidase. As in mammals, this metalloprotease hydrolyses angiotensin I into angiotensin II. XcACE is sensitive to ACE inhibitors and chloride ions concentration. Variations in the active site residues, highlighted by structural modelling, can account for the different substrate selectivity and inhibition profile compared to human ACE. XcACE characterization demonstrates that ACE is an ancestral enzyme, provoking questions about its appearance and structure/activity specialisation during the course of evolution.
Abbreviations: Ac, acetyl; ACE, Angiotensin-converting enzyme; ACE, gene encoding ACE; Ang, Angiotensin; bp, base pair(s); C-, carboxy; cDNA, DNA complementary to RNA; CHO, chinese hamster ovary; DMSO, dimethylsulfoxide; DNase, desoxyribonuclease; dNTP, deoxyribonucleoside triphosphate; DpaOH, N3(2,4-dinitrophenyl)l-2,3-diaminopropionyl-OH; enk, enkephalin; enk-NH2, enkephalinamide; FPLC, fast protein liquid chromatography; HHL, Hippuryl-Histidyl-Leucine; HPLC, high-performance liquid chromatography; kb, kilobase(s); kDa, kilodalton; Mca, 7-methoxycoumarin-4-yl)-diacetyl; N-, amino; ng, nanogram(s); nXcACE, native Xanthomonas axonopodis pv citri Angiotensin-converting enzyme; ORF, open reading frame; PAGE, polyacrylamide gel electrophoresis; pv, pathovar RNase: ribonuclease; rXcACE, recombinant Xanthomonas axonopodis pv citri Angiotensin-converting enzyme; SDS, sodium dodecyl sulfate; Xc, Xanthomonas axonopodis pathovar citri.
Keywords: Evolution, Cloning, Biochemistry, Structure, Conservation, Protease
1. Introduction
Angiotensin-converting enzyme (ACE, dipeptidyl-peptidase A, kininase II, E.C. 3.4.15.1, DCP1) is a dipeptidyl-carboxypeptidase that belongs to the M2-metalloprotease family. In mammals, this zinc-dependent metalloprotease participates in blood pressure elevation (review in (Corvol and Eyries, 2004)), generating the vasopressor angiotensin II (Ang II) from the inactive angiotensin I (Ang I), and by inactivating the vasodilator bradykinin. In vertebrates, somatic (sACE) and testicular (germinal) ACE (tACE) have been characterized. These two separate isoforms are transcribed from two alternate promoters within a single gene (Howard et al., 1990), which arose from duplication of an ancestral gene (Hubert et al., 1991). Consequently, sACE possesses two domains (termed N-domain and C-domain, respectively) bearing the highly conserved gluzincin motif HExxH, whereas tACE possesses only one, corresponding to the C-terminal domain of sACE. Crystal structures of both the human N-domain (Corradi et al., 2006) and tACE (Natesh et al., 2003) have shown them to be homologous helical ellipsoids harbouring two large cavities connected by a narrow catalytic channel containing the catalytic zinc and the HexxH motif. Somatic ACE N- and C-domain active sites have distinct enzymatic specificities with respect to in vivo and/or synthetic substrates. Indeed, the C-domain is highly selective for Hippuryl-Histidyl-Leucine (HHL) (Wei et al., 1991a) whereas the N-domain efficiently cleaves the hemoregulatory peptide Ac-Ser-Asp-Lys-Pro (AcSDKP) (Rousseau et al., 1995) and Ang 1-7 (Deddish et al., 1998). In addition, ACE inhibitors (Dive et al., 1999) and chloride ion dependence (Jaspard et al., 1993) can discriminate between the two domains. Both sACE and tACE possess a C-terminal transmembrane domain and are membrane-anchored isoforms, though a post-translational shedding releases active solubilized ACE into the extracellular milieu (Eyries et al., 2001, Oppong and Hooper, 1993). A single domain ACE homologue, ACE2, has also been characterized in the human (Donoghue et al., 2000, Tipnis et al., 2000) and its structure determined (Towler et al., 2004). ACE2 is an essential regulator of heart function (Crackower et al., 2002) and has also been identified as a functional receptor for the SARS coronavirus (Li et al., 2003).
In invertebrates, however, functional ACE-related enzymes exist in insects and in annelids (which belong to the Ecdysozoa and Lophotrochozoa groups, respectively). Interestingly, and though in silico evidence suggest the presence of a two-domain ACE-related protein in mosquitoes (Burnham et al., 2005), all the cloned genes encode soluble, single active site enzymes (Tatei et al., 1995, Taylor et al., 1996, Wijffels et al., 1996). In the fruit fly Drosophila melanogaster, two homologues have been cloned and named AnCE (Cornell et al., 1995) and ACER (Tatei et al. 1995) (or Race), of which AnCE has had its structure determined (Kim et al., 2003). AnCE and ACER show about 40% amino-acid sequence identities with vertebrate ACEs, share similar enzymatic properties (Coates et al., 2000), acting on a broad range of substrates and play a key role in development (Crackower et al., 2002) and reproduction (Ekbote et al., 1999, Hurst et al., 2003). Molecular and biochemical data from the leech ACE homologue have related it to the N-domain of mammalian ACE (Riviere et al., 2004). However, no functional ACE is present in the genome of the nematode Caenorhabditis elegans (C. elegans), despite ACN-1, a homologue that lacks ACE-like proteolytic activity, is crucial in development (Brooks et al., 2003). Recently, a dicarboxypeptidase, LdDCP, was cloned in a pathogenic unicellular eukaryote, Leishmania donovani (Goyal et al., 2005). LdDCP, like ACE, is able to cleave a carboxyterminal dipeptide from HHL and is inhibited by captopril, suggesting that ACE may have already been present before the appearance of multicellular organisms. However, LdDCP was not reported to hydrolyse AngI and does not display sequence homology with ACE enzymes. These observations raise questions about the presence of a true ACE in more distant species.
Very surprisingly, in silico databank analysis revealed that DNA sequences in bacterial genomes have been annotated as encoding potentially functional ACE-like enzymes. All these sequences display a great similarity despite phylogenetic distance (for references see http://tolweb.org and (DeLong and Pace, 2001)). The striking sequence conservation in the active region of these putative ACE-like peptidases suggests that the protease activity would be conserved in these ancestral, prokaryotic species. Among them, Xanthomonas axonopodis pv. citri (X. citri) is a phytopathogen gamma-proteobacteria in whose genome an ACE-like encoding sequence has been annotated. In order to gain more insight into stages in ACE evolution and to investigate its putative activity in unicellular organisms, we report the cloning and biochemical characterization of the expressed recombinant enzyme. We have also generated a homology-based model of XcACE using the structure of tACE to understand the interactions of residues in the active site. This ACE-like enzyme in X. citri is, to our knowledge, the first bacterial ACE-like enzyme ever characterized.
2. Materials and methods
2.1. In silico analysis
The protein sequence of the leech ACE (TtACE, Genbank accession no. 45272589), the most distant known ACE characterized at the time of the present study, was submitted to similarity searches using the BLAST program suite (tblastn; default parameters) (http://www.nih.nlm.ncbi.gov/blast) against all bacterial genomic sequences available at the NCBI server at the time of the study. Alignments were performed using the ClustalW (default parameters) software (http://www.ebi.ac.uk/clustalw/). The distance matrix generated into the tree file of ClustalW was used in the drawtree program of the PHYLIP suite and an unrooted tree diagram was generated (http://bioweb.pasteur.fr/seqanal/interfaces/drawtree-simple).
2.2. Bacterial strain
X. citri strain N1 was from the culture collection of the Laboratory of Plant Pathology, Faculty of Agriculture, Shizuoka University, Shizuoka, Japan (Talaga et al., 1996).
2.3. DNA extraction
Bacterial DNA was obtained as previously described (Davis et al., 1980).
2.4. RNA extraction
Total RNA was extracted using the SV total RNA isolation system (Promega).
2.5. Protein extraction
X. citri was cultured in YP medium under agitation at 30 °C until an optical density of 0.3 at 620 nm and centrifuged (5000 ×g, 30 min). Cells were washed in water, centrifuged, and the pellet resuspended in water in one hundredth of the culture volume. The resulting suspension was passed twice through a French pressure cell at 15,000 lb/in.2 (108 Pa) and unbroken cells and debris were removed by 5 min centrifugation at 5000 ×g.
2.6. RT-PCR
To assess the presence of a messenger RNA correponding to the XcACE gene, total RNAs (30–40 ng) were treated with RNAse-free DNAse I and reverse transcribed with Superscript III Reverse Transcriptase (Invitrogen). The same amount of nucleic acids (170 ng) was further used in PCR reactions containing 2% DMSO using XcInt-S1 (5′-GCTGCAGCAGATCCCACAGA-3′) and XcInt-AS1 (5′-CTATTGCGTGGGCGTAGGC-3′) as primers. Water was used instead of cDNA as a negative control. PCR products were resolved on an agarose gel stained with ethidium bromide.
2.7. Cloning of XcACE, construction of the pXcACE vector and expression in mammalian cell cultures
2.7.1. Cloning
Full-length XcACE cDNA sequence (NP 641553; GenBank accession no. gi: 21241971) was obtained by PCR on X. citri DNA (see Section 2.4). Sense and antisense primers (XcACEFL-S1 5′-CTGACCTATTCGGATGCGCTCAAGGAT-3′ and XcACEFL-AS2 5′-CTCTAGCCTTCGGCGGACTTCACTGCAAAGGAC-3′) were used (2 pmol each) in a PCR reaction containing 400 ng DNA, 2% DMSO, 20 μM dNTPs, 5 U Pfu DNA polymerase (Promega) in 50 μL total volume. Reaction cycling parameters were 94 °C, 3 min; 94 °C, 40 s, 61 °C, 1 min, 72 °C, 4 min for 35 cycles. The expected 2.2 kb product was blunt-end subcloned into the EcoRV site of pcDNA3.1 (Invitrogen) and sequenced with XcACE and pcDNA3 specific primers.
2.7.2. Construct
Heterologous expression of XcACE was carried out using mammalian CHO-K1 cells (Invitrogen). Because the start codon is GTG for XcACE, it was necessary to change it into the standard ATG, a sequence recognized as an initiation codon by CHO-K1 cells. This substitution mutation (T11A) was realised by PCR using the XcFLAS2.2 (5′-GCT CTA GCC TTC GGC GGA CTT CAA TGC AAG-3′) antisense oligonucleotide and resulted in an in-frame ATG initiation codon. The PCR product was subcloned in pcDNA3.1 and the obtained pXcACE plasmid sequenced as described above (see Section 2.7.1).
2.7.3. Expression
CHO-K1 cells were stably transfected with pXcACE using lipofectamine 2000 reagent (Invitrogen), and were selected for resistance to Geneticin. Individual resistant colonies producing large amounts of XcACE were cloned by limiting dilution techniques as described (Wei et al., 1991b).
2.8. Biochemical characterization of XcACE
2.8.1. Semi-purification
Both native and recombinant XcACE were partially purified by ion exchange chromatography. Briefly, native XcACE was obtained from bacterial protein extract, and secreted recombinant XcACE from concentrated culture medium of transfected CHO cells (fraction A). The fraction A samples were first desalted on a HiPrep Fast Desalting Column (Amersham Pharmacia Biotech), then fractionated onto a FPLC MonoQ anion-exchange column system (Pharmacia). The resulting fractions showing specific ACE activity were pooled and concentrated. The resulting fractions were named native XcACE (nXcACE) and recombinant XcACE (rXcACE), respectively. For nXcACE purification steps, each pooled fractions containing specific ACE activity were analysed by SDS-PAGE.
2.8.2. Western blot
Both nXcACE and rXcACE (see Section 2.8.1) were submitted to western blot experiment using a polyclonal antibody (HKCE) raised against the human full-length sACE.
2.8.3. Enzymatic assays
ACE activity was assayed using various substrates: HHL (5 mM), as a human ACE C-domain specific substrate, AcSDAcKP (Acetyl-Serine-Aspartyl-Acetyl-Lysine-Proline) (2.5 mM), as an N-domain specific substrate, Ang I (100 μM), as both an N- and a C-domain ACE substrate. Assays were performed at 37 °C for 30 or 60 min. Hydrolysis of [Leu5]-enkephalinamide ([Leu5]-enk-NH2) (0.8 mM, incubated for 330 min), as an amidated substrate and [Leu5]-enkephalin ([Leu5]-enk) (0.8 mM, incubated for 30 min), were compared to determine whether XcACE also exhibits endopeptidase activity. The rate of hydrolysis of all substrates used was determined and quantified using reverse phase HPLC (Waters Co, Milford, MA, USA) as previously described (Azizi et al., 2000, Houard et al., 1998, Wei et al., 1991b). The specificity of the reaction was assessed in the presence of 10 μM captopril. The effect of chloride ions (NaCl concentrations from 0 to 1.0 M) on HHL hydrolysis was determined under standard conditions.
2.8.4. Inhibition of XcACE activity by various ACE inhibitors
The inhibition potentials of various tight binding ACE inhibitors (captopril, delaprilat, fosinoprilat, lisinopril, perindoprilat, RXP 407 and enalaprilat) were assessed by determining the IC50 values for the fluorogenic substrate Mca-Ala-Ser-Asp-Lys-DpaOH hydrolysis in standard conditions as described previously (Dive et al., 1999). Compounds were pre-incubated with nXcACE (see Section 2.8.1) for 1 h and reactions initiated by the addition of 12 μM substrate.
2.9. Modelling of XcACE
A model of XcACE was generated by the ESyPred3D server (http://www.fundp.ac.be/sciences/biologie/urbm/bioinfo/esypred/) (Lambert et al., 2002) using testis ACE (ProteinDataBank pdb code 1O8A) as a model. This model was compared with the testis ACE structures (Natesh et al., 2003, Natesh et al., 2004) and the N-domain structure (Corradi et al., 2006) and AnCE (Houard et al., 1998) using the program Coot (Emsley and Cowtan, 2004). Figures were generated with Pymol (DeLano, 2002).
2.10. XcACE activity during bacterial growth
To examine XcACE localization, ACE activity in medium and in bacterial extracts was monitored along during bacterial growth using HHL as substrate as described above.
3. Results
3.1. Presence of ACE orthologues in bacterial genomes
Among 465 bacterial strains genomes screened, 26 sequences produced significant similarity (E value < 0.5) with leech TtACE. Further analysis revealed that at least 22 sequences within 19 different bacterial strains within 8 generas potentially encode putative ACE-like enzymes. These species are ecologically as well as phylogenetically distant, since they are found amongst Cyanobacteria (Gloeobacter violaceus, gi: 37522712, gi: 35213714), Acidobacteria (Solibacter usitatus, gi: 67858770, gi: 66769433) and Proteobacteria of the subdivision alpha (Erythrobacter litoralis, gi: 61100845, gi: 60736774), delta (Anaeromyxobacter dehalogenans, gi:66855040) and gamma (X. citri, gi: 21241971, gi: 21107345; Xanthomonas oryzae, gi: 37931607, gi: 58583162; Xanthomonas campestris pv campestris, gi:21230574, gi: 21112152; Shewanella oneidensis, gi: 24374038, gi: 24348510) proteobacteria. All these predicted proteins share between 38 and 98% primary sequence identity between them, indicating a wide variety among the potential bacterial ACEs (Fig. 1 and Supplementary data). Nevertheless, all these sequences exhibit key ACE features such as the presence of two highly conserved consensus VCHASAWDFY and GANPGFHEA motifs surrounding a putative M2-catalytic site HexxH (data not shown).
Fig. 1.
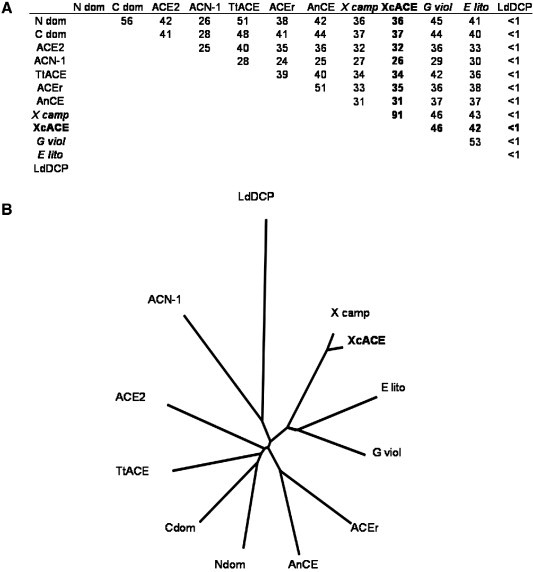
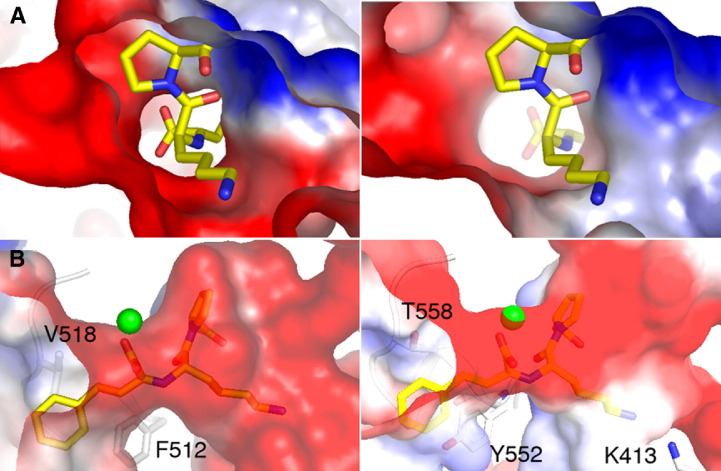
Homology of the bacterial ACE related to various ACEs in the phylogenetic tree. (A): Percentage of identity in amino acids between XcACE and various ACE-like enzymes primary sequences described or deduced from genomic sequences: Ndom, human N-domain ACE (gi: 4732026, residues 1–583); Cdom, human C-domain ACE (gi: 4732026, residues 641–1306); ACE2, human ACE2 (gi: 71658783); ACEr, Drosophila melanogaster ACEr (gi: 73921650); X camp, Xanthomonas campestris pv. campestris predicted dipeptidyl-carboxypeptidase (gi: 66769433); E lito, Erythrobacter litoralis predicted dipeptidyl-carboxypeptidase (gi: 61100845); G viol, Gloeobacter violaceus predicted dipeptidyl-carboxypeptidase (gi: 37522712). (B): unrooted cladogram indicating evolutionary relationships between the different ACE homologues among the living kingdom.
3.2. Molecular characterization of XcACE
We cloned an ACE homologue in the phytopathogenic bacterium X. citri corresponding to the sequence XAC1217 (gi: 21107356, gene ID 1155288). The transcription of the XcACE gene was suggested by the sequence conservation upstream the start codon and is assessed by the presence of a specific 437-bp RT-PCR product in DNAse-treated, reverse-transcribed total RNA (Fig. 2 ). The protein encoded by this gene, referred to as XcACE (Xanthomonas citri angiotensin-converting enzyme), is a 672 amino-acid protein. Hydrophobicity profile (SignalP V1.1 at www.cbs.dtu.dk/services/SignalP) indicates cleavage of a putative signal peptide between residues 26 and 27 of the native protein (…RDA↕AP…). There does not appear to be a motif encoding a transmembrane domain at the C-terminus or elsewhere in the mature molecule, suggesting that XcACE should exist only in a soluble form. Because of the mutation inserted, the recombinant proenzyme is slightly different from the native one in the N-terminal end (rXcACE: MQGPPVNPRLL vs nXcACE: MNPRLL). However, this difference does not change predictions about the signal peptide (hydrophobicity profile and cleavage site) (data not shown), indicating that the mature recombinant and native proteins have identical primary sequences. The mature enzyme has a theoretical molecular weight of 72 kDa and an isoelectric point of 5.72. Its primary structure includes all the residues implied in zinc (H429, H433 and E457), lisinopril (H391, A392, E430, K556, H558, Y565 and Y568) (Natesh et al., 2003) and captopril (Q290, K556, Y565, H391, H558 and Y568) (Natesh et al., 2004) binding. Thus the soluble, secreted enzyme should possess a single active site like the other ACEs found in invertebrates. Whereas two chloride atoms were observed bound to the human tACE, XcACE appears to be more similar to the N-domain in this respect, as it only has the chloride II binding site conserved (Y230 and R567), and only one amino acid among the three implicated in the binding of the first chloride atom (W530). The XcACE primary sequence has between 31 and 37% identity with the other known ACEs (human, Drosophila and leech) and 26% with C. elegans ACN-1 (Fig. 1 and Supplementary data).
Fig. 2.
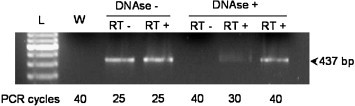
RT-PCR assay of the XcACE transcript. cDNA was generated and submitted to PCR (cycling parameters: 94 °C, 2 min; 94 °C, 45 s, 61 °C, 30 s, 72 °C, 1 min, for 25, 30 or 40 cycles, 72 °C, 4 min).Treatments of the different samples are indicated above the corresponding lane. The size of the expected band is indicated. L: DNA molecular weight marker, W: water control. The number of PCR cycles is indicated under each sample.
3.3. Biochemical characterization of XcACE
3.3.1. Semi-purification
The semi-purification of XcACE procedure using FPLC (Fig. 3A) resulted in a fraction containing a protein band at the expected size for XcACE (∼ 75 kDa), as revealed by SDS-PAGE analysis of the different fractions obtained after each purification step (data not shown).
Fig. 3.
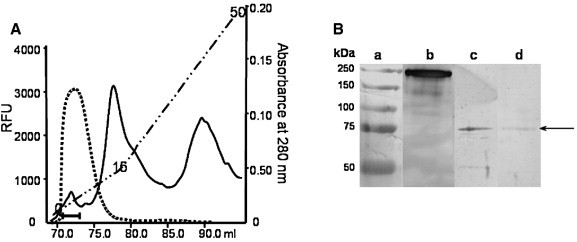
Semi-purification and Western blot of XcACE. (A): FPLC profile of the last purification step (see Section 2.8.1), absorbance at 280 nm (full line), the gradient (dashed line) was from 0 to 50 mS/cm, arbitrary units of ACE activity (dotted line), the horizontal bar indicates the fractions pooled for further analysis. (B): Native and recombinant XcACE were analysed by SDS-PAGE followed by Western blot with the antiserum HKCE. Lane a, molecular-mass standards; lane b, human somatic ACE; lane c, native XcACE; lane d, recombinant XcACE. The band corresponding to XcACE is indicated (arrow).
3.3.2. Immunological characterization
Both native and recombinant XcACE were analysed by Western blotting and migrated with an apparent molecular weight of 72 kDa, matching theoretical predictions (Fig. 3B), and have a similar mobility on SDS-PAGE to deglycosylated human tACE (Yu et al., 1997).
3.3.3. Enzymatic activity
XcACE shows dipeptidyl-carboxypeptidase activity, cleaving a carboxyterminal dipeptide from peptidic substrates. It generates AngII from AngI (Fig. 4A), hippuric acid from HHL (Fig. 4B), AcKP from AcSDAcKP (Fig. 4C), and Tyrosyl-glycyl-glycine (YGG) from Leu-enkephalin (data not shown). XcACE also displays an endopeptidase activity, as shown by its ability to hydrolyse the amidated substrate, Leu-enk-NH2 (Fig. 4D). As for AngI hydrolysis (Fig. 4E), the same results were obtained with recombinant XcACE (data not shown). XcACE activity depends on zinc binding. Indeed, hydrolysis of Mca-SDKP-DpaOH was abolished after zinc chelation with EDTA and dialysis but was restored by zinc addition (data not shown).
Fig. 4.
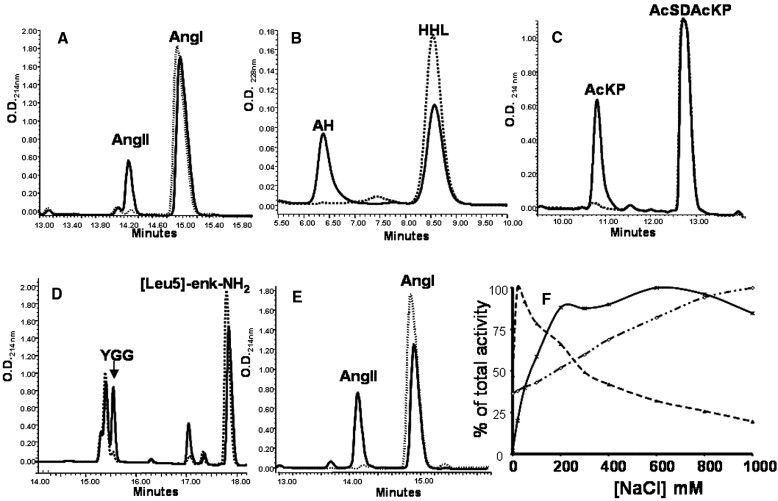
Biochemical characterization of XcACE. Hydrolysis of various ACE substrates by native (nXcACE) (A–D) and recombinant (rXcACE) (E). Hydrolysis of angiotensin I (A), hydrolysis of HHL (B), AcSDAcKP (C), [Leu5]-enkephalin (D) and [Leu5]-enkephalinamide (E) by native XcACE. Hydrolysis of angiotensin I by recombinant XcACE (F). The specificity of the reactions was assessed in the presence of 10 μM captopril (dashed line). AngI, angiotensin I; AngII, angiotensin II; AH, hippuric acid; YGG, tyrosyl-glycyl-glycine. Chloride sensitivity of XcACE (F). Effect of chloride ions concentration on HHL hydrolysis by native (◇) XcACE, C-(ж) and N-domain (▲) of human somatic ACE.
3.3.4. Effect of chloride concentration
Native XcACE is already active in the absence of chloride ions. However, its activity in the presence of increasing chloride ion concentrations increases 3-fold. In this respect, XcACE Cl− activation presents intermediate characteristics when compared to both N- and C-domain human ACE (Riviere et al., 2004). (Fig. 4F).
3.3.5. Effect of ACE inhibitors
Fosinoprilat is the most potent inhibitor of XcACE activity (IC50: 3.0 · 10− 8 ± 1.0 · 10− 8 M), whereas captopril is less efficient (1.6 · 10− 7 ± 0.1 · 10− 7 M). RXP 407 (1.9 · 10− 6 ± 1.5 · 10− 6 M) and Enalaprilat (3.8 · 10− 6 ± 2.8 · 10− 6 M) displayed IC50 values in the micromolar range. Perindoprilat (1.3 · 10− 4 ± 1.3 · 10− 4 M), delaprilat (3.7 · 10− 5 ± 1.7 · 10− 5 M) and lisinopril (4.7 · 10− 5 ± 4.3 · 10− 5 M), all containing carboxylate zinc-binding groups, were the least effective inhibitors of XcACE. Values are means ± standard error from three independent experiments.
3.4. Predicted structure of XcACE
As the crystal structures of human ACE (Corradi et al., 2006, Natesh et al., 2003), ACE2 (Towler et al., 2004) and the Drosophila AnCE (Kim et al., 2003) all have the same fold, the 34% sequence identity of human tACE and bacterial XcACE allowed us to model with confidence the overall fold of XcACE based on the tACE structure (pdb code 1O8A). We identified active site residues that could account for the differences in inhibitor affinities (see Section 4.2) (Fig. 5 ).
Fig. 5.
Comparison of the charge of the active site between tACE and XcACE. ACE is represented as a surface with the positive charge indicated in blue and the negative in red. The zinc atom is represented (green), and some of the residues are indicated. Lisinopril (in yellow) is shown to orientate the active site. A, The active site viewed as a surface from the lysyl pocket for tACE (left) and XcACE (right). B, The active site cavity viewed from the outside for tACE (left) and XcACE (right). tACE is shown to have a higher negative charge in the lysyl binding pocket.
3.5. XcACE expression during bacterial growth and localization
Only background ACE activity was detected in the culture medium, which remains constant over time. In contrast, ACE activity in bacterial extract correlates bacterial growth indicating that XcACE is constituvely expressed and is likely to be secreted and further localised in the periplasmic space (Fig. 6 ).
Fig. 6.
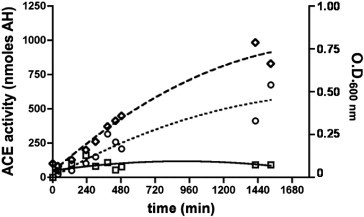
XcACE activity during bacterial growth. Follow-up of bacterial growth by O.D. at 600 nm (◇) and ACE activity (nmoles hippuric acid generated) in culture medium (□) and within bacteria (○). Error bars were omitted for clarity.
4. Discussion
4.1. ACE is an ancestral enzyme
Databank analysis revealed the presence of putative ACE-encoding genomic DNA sequences in a very wide range of bacterial species, corresponding to a broad range of ecological lifestyles and terminal electron acceptor for growth (DeLong and Pace, 2001), suggesting that ACE is an ancestral enzyme. Characterization of XcACE constitutes the first evidence of the presence of an ACE-like enzyme in more distant organisms than cellular metazoans. Indeed, in contrast to XcACE, the amino-acid sequence of L. donovani dicarboxypeptidase (LdDCP) (Goyal et al., 2005) does not show any significant alignment with all other known ACEs, was not reported to hydrolyse Ang I to Ang II, and lacks conserved consensus motifs found in almost all ACE primary sequences (for references see the MEROPS database available at http://merops.sanger.ac.uk/). However, in contrast with Leishmania, none of the bacterial species putatively possessing an ACE mentioned in this study is known to be a human pathogen, whereas xanthomonadacaea are phytopathogens among which X. citri was chosen for technical reasons.
4.2. Structure–activity relationship of XcACE, a functional angiotensin-converting enzyme in vivo
The XcACE possesses a single M2 catalytic site and a signal peptide but no membrane anchor, like the ACE characterized in invertebrates. Despite the fact that the recombinant enzyme was expressed in a heterologous system, both native and recombinant forms display almost identical features, indicating a satisfactory purity of XcACE and strongly suggesting that the ACE in X. citri is a functional ACE enzyme in vivo. XcACE, like Drosophila AnCE (data not shown), is active in the absence of chloride ions and less sensitive to variations in chloride ion concentration than human C-domain ACE (Houard et al., 1998). This characteristic is likely to be due to the absence in the primary sequence of XcACE, as in AnCE, of two key amino acids observed to bind ClI in human tACE (XcACE : R186H and R489K; AnCE : R1867Y and W485F, when compared to tACE). XcACE hydrolyses a broad range of substrates, including human N- and C-domain ACE specific substrates (AcSDAcKP and HHL, respectively). In addition to this dicarboxypeptidase activity, XcACE also acts as an endopeptidase, hydrolysing the C-terminal blocked amidated substrate [Leu5]-enkephalinamide. Western blot results using polyclonal antibody raised against the full-length human sACE protein show a unique band corresponding to XcACE, indicating that XcACE shares common epitopic features with the human enzyme. Molecular modelling allowed us to visualise the structural differences in the catalytic site. Key active site residues such as the zinc-binding motif, His513 and Tyr526 (tACE numbering) are conserved in XcACE and many other ACE-like enzymes. However, in line with the assay results, the non-conserved active site residues are neither identical to the C- or N-domain of human ACE, and are the presumed cause of the different substrate selectivity and inhibition profile of XcACE. Comparison of the XcACE model with either the human tACE or N-domain shows that lisinopril fits in the XcACE active site and retains most of the same binding interactions. The exception is in the lysyl pocket, which has features in common with AnCE or the human N-domain including a positively charged residue, lysine (equivalent to Arg356 in AnCE and Arg350 in the human N-domain) that appears to affect the overall charge of the pocket (Fig. 5). Indeed, similar to XcACE, neither AnCE nor the N-domain have as high affinity for lisinopril as tACE (Michaud et al., 1997, Williams et al., 1996). Furthermore, the difference in affinity of XcACE and tACE for inhibitors with hydrophobic P1 groups (i.e. lisinopril, delaprilat and perindoprilat) could be attributed to the difference in hydrophobicity of the S1 subsite. XcACE has a threonine and tyrosine in contrast to Phe512 and Val518 in tACE (Fig. 5). The N-terminal aspartate, N-acetyl groups and the C-terminal amide of RXP407 are the reported determinants required for its remarkable N-domain specificity (Dive et al., 1999). Of the four possible residues suggested for N-domain selective recognition of RXP 407 (Corradi et al., 2006, Tzakos and Gerothanassis, 2005), XcACE only retains the threonine (T496 in the N-domain) and has a lysine which replaces Arg500 of the N-domain. This, combined with the hydrophobic P1 group, may be the reason that RXP407 is not a strong inhibitor for XcACE. The affinity of XcACE for fosinoprilat is harder to rationalise as it has been modelled in different conformations for the N- and C-domains of human ACE (Tzakos and Gerothanassis, 2005). However, XcACE retains all the residues suggested to be involved in inhibitor binding with both domains of human ACE.
4.3. Putative physiological roles
The biological functions of XcACE remain unclear but could constitute the original, ancestral function of ACE. XcACE is soluble and its sequence exhibits a signal peptide indicating, along with the absence of ACE activity in the culture medium, that XcACE is a secreted enzyme which is trapped into the periplasmic space. The constitutive expression could indicate an important function for ACE in X. citri, as a possible role for the enzyme in peptidoglycan remodelling, a critical parameter in environmental adaptation. This is in line with the characterization in the periplasm of other gram negative bacteria of VanX d-alanyl-d-alanine dipeptidase, a regulated carboxypeptidase that is responsible for Vancomycin resistance (Lessard and Walsh, 1999). XcACE could also participate in X.citri pathogenicity within the cocktail of degradative enzymes required during plant infection. Interestingly, in this regard, ACE inhibitors are found in hydrolysates of plants sensitive to the “black rot” caused by Xanthomonas, including rice and soybean (He et al., 2005, Wu and Ding, 2001). The large number of substrates of XcACE does not exclude a possible function of protein degradation that might generate dipeptides for protein biosynthesis. To this extent, it is interesting to note that ACE is highly expressed in the digestive tract of all metazoan species presenting an active ACE.
4.4. Evolutionary and phylogenetic aspects
One of the most relevant questions raised by this study deals with phylogenetic considerations and evolution of ACE. Its presence in eukarya as well as in bacteria indicates that the enzyme could have appeared before the divergence between these two groups. This strongly suggests that XcACE is the most ancestral present-day representative of ACE characterized to date. However, as all completely sequenced genomes of plants, algaes and mycetes lack ACE, caution should be exercised with this hypothesis. The presence of an ACE-like enzyme in bacteria could also be the result of a horizontal gene transfer from eucaryotic to bacterial species. However, conservation in sequences surrounding the ACE gene, suggestive of mobile genetic elements, is not observed within 30 kb of the gene in seven distinct bacterial strains (data not shown). Furthermore, the parasitic bacterial species with an ACE-like gene are all hosted by organisms lacking the enzyme. In addition, not all bacterial species with a putative ACE gene are parasitic. Thus, the presence of ACE in bacteria, like the situation encountered in annelids, is unlikely to be related to parasitism, suggesting that bacterial ACE, represented herein by XcACE, corresponds to the ancestral, original enzyme.
4.5. Ancestral ACE is neither related to the N- nor the C-domain of mammalian ACE
XcACE exhibits a similar primary sequence identity with both N- and C-domains and, although XcACE cleaves the N-domain substrate AcSDAcKP, the N-domain specific inhibitor RXP407 is not a potent inhibitor of XcACE activity. In contrast, lisinopril, a C-domain-related inhibitor, is less efficient than captopril, related to the N-domain, though most of the residues for binding of both are conserved. Thus, from the biochemical and the structural properties of XcACE it is not possible to ascribe this bacterial enzyme to either the mammalian N-or C-domain of human ACE. The primitive ACE would then not be closer to either domain of the mammalian enzyme, in contrast to what was previously suggested (Coates et al., 2000, Hubert et al., 1991, Lattion et al., 1989).
5. Conclusion
This work describes the cloning, expression, purification, biochemical characterization, molecular modelling and localization of XcACE, the first ACE-like enzyme in a bacterium, X. citri. Taken together, our results bring the evidence of the presence of a functional ACE in prokaryotes, in contrast to the previous hypothesis about the latter appearance of ACE during phylogeny. It also appears that sequence/structure conservation of ACE enabled preservation of dipeptidase activity during evolutionary specialisation. XcACE is the most ancestral ACE present-day representative and has features in common to both the N- and the C-domain of mammalian somatic ACE. It is possible that this ancestral ACE was a poorly specialised protease which has acquired its present specialisation over a long evolutionary period. However, the biological function(s) as well as the endogenous substrates and/or inhibitors of bacterial ACE remain to be characterized. To clarify these issues, the use of ACE inhibitors in vivo and the generation of a mutant bacterial strain lacking ACE may be helpful. Some of these functions of ACE might also be conserved in humans, and their characterization could lead to a better understanding of the evolution of this crucial enzyme.
Acknowledgements
This work was supported by grants from INSERM (Institut National pour la Santé et la Recherche Médicale), Region Nord Pas de Calais and CNRS (Centre National pour la Recherche Scientifique) and Wellcome Trust (U.K.) grants 070060 (K.R.A.) and 071047 (E.D.S.).
Received by J.A. Engler
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.gene.2007.05.010.
Appendix A. Supplementary data
References
- Azizi M., Massien C., Michaud A., Corvol P. In vitro and in vivo inhibition of the 2 active sites of ACE by omapatrilat, a vasopeptidase inhibitor. Hypertension. 2000;35:1226–1231. doi: 10.1161/01.hyp.35.6.1226. [DOI] [PubMed] [Google Scholar]
- Brooks D.R., Appleford P.J., Murray L., Isaac R.E. An essential role in moulting and morphogenesis of Caenorhabditis elegans for ACN-1: a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J Biol. Chem. 2003;278:52340–52346. doi: 10.1074/jbc.M308858200. [DOI] [PubMed] [Google Scholar]
- Burnham S., Smith J.A., Lee A.J., Isaac R.E., Shirras A.D. The angiotensin-converting enzyme (ACE) gene family of Anopheles gambiae. BMC. Genomics. 2005;6:172. doi: 10.1186/1471-2164-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates D. Functional conservation of the active sites of human and Drosophila angiotensin I-converting enzyme. Biochemistry. 2000;39:8963–8969. doi: 10.1021/bi000593q. [DOI] [PubMed] [Google Scholar]
- Cornell M.J. Cloning and expression of an evolutionary conserved single-domain angiotensin converting enzyme from Drosophila melanogaster. J. Biol. Chem. 1995;270:13613–13619. doi: 10.1074/jbc.270.23.13613. [DOI] [PubMed] [Google Scholar]
- Corradi H.R., Schwager S.L., Nchinda A.T., Sturrock E.D., Acharya K.R. Crystal structure of the N domain of human somatic angiotensin I-converting enzyme provides a structural basis for domain-specific inhibitor design. J Mol. Biol. 2006;357:964–974. doi: 10.1016/j.jmb.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Corvol P., Eyries M. Peptidyl dipeptidase A: angiotensin I converting enzyme. In: Rawlings M.D., Woessner F.J., editors. , Handbook of Proteolytic Enzymes. Academic Press; London: 2004. pp. 332–346. [Google Scholar]
- Crackower M.A. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- Davis R., Botstein D., Roth J. Manual for Genetic Engineering. Cold Spring Harbor Laboratory Press; New York: 1980. Advanced bacterial genetics. [Google Scholar]
- Deddish P.A., Marcic B., Jackman H.L., Wang H.Z., Skidgel R.A., Erdos E.G. N-domain-specific substrate and C-domain inhibitors of angiotensin-converting enzyme : angiotensin-(1-7) and keto-ACE. Hypertension. 1998;31:912–917. doi: 10.1161/01.hyp.31.4.912. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos CA USA. http://www.pymol.org. [Google Scholar]
- DeLong E.F., Pace N.R. Environmental diversity of bacteria and archaea. Syst. Biol. 2001;50:470–478. [PubMed] [Google Scholar]
- Dive V. RXP 407, a phosphinic peptide, is a potent inhibitor of angiotensin I converting enzyme able to differentiate between its two active sites. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4330–4335. doi: 10.1073/pnas.96.8.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Ekbote U., Coates D., Isaac R.E. A mosquito (Anopheles stephensi) angiotensin I-converting enzyme (ACE) is induced by a blood meal and accumulates in the developing ovary. FEBS Lett. 1999;455:219–222. doi: 10.1016/s0014-5793(99)00870-4. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eyries M. Increased shedding of angiotensin-converting enzyme by a mutation identified in the stalk region. J Biol. Chem. 2001;276:5525–5532. doi: 10.1074/jbc.M007706200. [DOI] [PubMed] [Google Scholar]
- Goyal N., Duncan R., Selvapandiyan A., Debrabant A., Baig M.S., Nakhasi H.L. Cloning and characterization of angiotensin converting enzyme related dipeptidylcarboxypeptidase from Leishmania donovani. Mol. Biochem. Parasitol. 2005 doi: 10.1016/j.molbiopara.2005.09.014. [DOI] [PubMed] [Google Scholar]
- He G.Q., Xuan G.D., Ruan H., Chen Q.H., Xu Y. Optimization of angiotensin I-converting enzyme (ACE) inhibition by rice dregs hydrolysates using response surface methodology. J Zhejiang. Univ Sci. B. 2005;6:508–513. doi: 10.1631/jzus.2005.B0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houard X. The Drosophila melanogaster-related angiotensin-I-converting enzymes Acer and Ance—distinct enzymic characteristics and alternative expression during pupal development. Eur. J. Biochem. 1998;257:599–606. doi: 10.1046/j.1432-1327.1998.2570599.x. [DOI] [PubMed] [Google Scholar]
- Howard T.E., Shai S.Y., Langford K.G., Martin B.M., Bernstein K.E. Transcription of testicular angiotensin-converting enzyme (ACE) is initiated within the 12th intron of the somatic ACE gene. Mol. Cell Biol. 1990;10:4294–4302. doi: 10.1128/mcb.10.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert C., Houot A.M., Corvol P., Soubrier F. Structure of the angiotensin I-converting enzyme gene. Two alternate promoters correspond to evolutionary steps of a duplicated gene. J Biol. Chem. 1991;266:15377–15383. [PubMed] [Google Scholar]
- Hurst D., Rylett C.M., Isaac R.E., Shirras A.D. The drosophila angiotensin-converting enzyme homologue Ance is required for spermiogenesis. Dev. Biol. 2003;254:238–247. doi: 10.1016/s0012-1606(02)00082-9. [DOI] [PubMed] [Google Scholar]
- Jaspard E., Wei L., Alhenc-Gelas F. Differences in the properties and enzymatic specificities of the two active sites of angiotensin I-converting enzyme (kininase II). Studies with bradykinin and other natural peptides. J. Biol. Chem. 1993;268:9496–9503. [PubMed] [Google Scholar]
- Kim H.M., Shin D.R., Yoo O.J., Lee H., Lee J.O. Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril. FEBS Lett. 2003;538:65–70. doi: 10.1016/s0014-5793(03)00128-5. [DOI] [PubMed] [Google Scholar]
- Lambert C., Leonard N., De B.X., Depiereux E. ESyPred3D: prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- Lattion A.L., Soubrier F., Allegrini J., Hubert C., Corvol P., Alhenc-Gelas F. The testicular transcript of the angiotensin I-converting enzyme encodes for the ancestral, non-duplicated form of the enzyme. FEBS Lett. 1989;252:99–104. doi: 10.1016/0014-5793(89)80897-x. [DOI] [PubMed] [Google Scholar]
- Lessard I.A., Walsh C.T. VanX, a bacterial d-alanyl-d-alanine dipeptidase: resistance, immunity, or survival function? Proc. Natl Acad. Sci. U.S.A. 1999;96:11028–11032. doi: 10.1073/pnas.96.20.11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud A., Williams T.A., Chauvet M.T., Corvol P. Substrate dependence of angiotensin I-converting enzyme inhibition: captopril displays a partial selectivity for inhibition of N-acetyl-seryl-aspartyl-lysyl-proline hydrolysis compared with that of angiotensin I. Mol. Pharmacol. 1997;51:1070–1076. doi: 10.1124/mol.51.6.1070. [DOI] [PubMed] [Google Scholar]
- Natesh R., Schwager S.L., Evans H.R., Sturrock E.D., Acharya K.R. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry. 2004;43:8718–8724. doi: 10.1021/bi049480n. [DOI] [PubMed] [Google Scholar]
- Natesh R., Schwager S.L., Sturrock E.D., Acharya K.R. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature. 2003;421:551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- Oppong S.Y., Hooper N.M. Characterization of a secretase activity which releases angiotensin-converting enzyme from the membrane. Biochem. J. 1993;292(Pt 2):597–603. doi: 10.1042/bj2920597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere G. Characterization of the first non-insect invertebrate functional angiotensin-converting enzyme (ACE): leech TtACE resembles the N-domain of mammalian ACE. Biochem. J. 2004;382:565–573. doi: 10.1042/BJ20040522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau A., Michaud A., Chauvet M.T., Lenfant M., Corvol P. The hemoregulatory peptide N-acetyl-Ser-Asp-Lys-Pro is a natural and specific substrate of the N-terminal active site of human angiotensin-converting enzyme. J. Biol. Chem. 1995;270:3656–3661. doi: 10.1074/jbc.270.8.3656. [DOI] [PubMed] [Google Scholar]
- Talaga P. Cell-associated glucans of Burkholderia solanacearum and Xanthomonas campestris pv. citri: a new family of periplasmic glucans. J. Bacteriol. 1996;178:2263–2271. doi: 10.1128/jb.178.8.2263-2271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatei K., Cai H., Ip Y.T., Levine M. Race: a Drosophila homologue of the angiotensin converting enzyme. Mech. Dev. 1995;51:157–168. doi: 10.1016/0925-4773(95)00349-5. [DOI] [PubMed] [Google Scholar]
- Taylor C.A., Coates D., Shirras A.D. The Acer gene of Drosophila codes for an angiotensin-converting enzyme homologue. Gene. 1996;181:191–197. doi: 10.1016/s0378-1119(96)00503-3. [DOI] [PubMed] [Google Scholar]
- Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- Towler P. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzakos A.G., Gerothanassis I.P. Domain-selective ligand-binding modes and atomic level pharmacophore refinement in angiotensin I converting enzyme (ACE) inhibitors. Chembiochem. 2005;6:1089–1103. doi: 10.1002/cbic.200400386. [DOI] [PubMed] [Google Scholar]
- Wei L., Alhenc-Gelas F., Corvol P., Clauser E. The two homologous domains of human angiotensin I-converting enzyme are both catalytically active. J. Biol. Chem. 1991;266:9002–9008. [PubMed] [Google Scholar]
- Wei L., Alhenc-Gelas F., Soubrier F., Michaud A., Corvol P., Clauser E. Expression and characterization of recombinant human angiotensin I-converting enzyme. Evidence for a C-terminal transmembrane anchor and for a proteolytic processing of the secreted recombinant and plasma enzymes. J. Biol. Chem. 1991;266:5540–5546. [PubMed] [Google Scholar]
- Wijffels G. Cloning and characterisation of angiotensin-converting enzyme from the dipteran species, Haematobia irritans exigua, and its expression in the maturing male reproductive system. Eur. J. Biochem. 1996;237:414–423. doi: 10.1111/j.1432-1033.1996.0414k.x. [DOI] [PubMed] [Google Scholar]
- Williams T.A., Michaud A., Houard X., Chauvet M.T., Soubrier F., Corvol P. Drosophila melanogaster angiotensin I-converting enzyme expressed in Pichia pastoris resembles the C domain of the mammalian homologue and does not require glycosylation for secretion and enzymic activity. Biochem. J. 1996;318(Pt 1):125–131. doi: 10.1042/bj3180125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Ding X. Hypotensive and physiological effect of angiotensin converting enzyme inhibitory peptides derived from soy protein on spontaneously hypertensive rats. J. Agric. Food Chem. 2001;49:501–506. doi: 10.1021/jf000695n. [DOI] [PubMed] [Google Scholar]
- Yu X.C., Sturrock E., Wu Z., Biemann K., Ehlers M., Riordan J. Identification of N-linked glycosylation sites in human testis angiotensin-converting enzyme and expression of an active deglycosylated form. J. Biol. Chem. 1997;272:3511–3519. doi: 10.1074/jbc.272.6.3511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.