Abstract
Hibiscus latent Singapore virus (HLSV) is a member of Tobamovirus and its full-length cDNA clones were constructed. The in vitro transcripts from two HLSV full-length cDNA clones, which contain a hepta-adenosine stretch (pHLSV-7A) and an octo-adenosine stretch (pHLSV-8A), are both infectious. The replication level of HLSV-7A in Nicotiana benthamiana protoplasts was 5-fold lower, as compared to that of HLSV-8A. The replicase proteins of HLSV-7A were produced through programmed −1 ribosomal frameshift (−1 PRF) and the 7A stretch was a slippery sequence for −1 PRF. Mutations to the downstream pseudoknot of 7A stretch showed that the pseudoknot was not required for the frameshift in vitro. The stretch was found to be extended to 8A after subsequent replication cycles in vivo. It is envisaged that HLSV employs the monotonous runs of A and −1 PRF to convert its 7A to 8A to reach higher replication for its survival in plants.
Keywords: Tobamovirus, Biologically active cDNA clone, Hepta-adenosine stretch, −1 PRF, Downstream pseudoknot
Highlights
-
•
Successful construction of HLSV full-length cDNA clone.
-
•
First report of a programmed −1 frameshift function in Tobamovirus.
-
•
7A stretch is the sole signal for programmed −1 frameshift in HLSV.
-
•
7A stretch can be extended to 8A stretch during HLSV replication.
Introduction
The genomic RNAs of viruses are often polycistronic and need to use strategies to express their downstream open reading frames (ORFs). Production of subgenomic RNAs, readthrough of the stop codon, ribosome leaky scanning and programmed −1, −2 or +1 ribosomal frameshift (PRF) are strategies used by viruses (Plant, 2012). PRF has been reported in some plant virus groups, including Luteovirus, Dianthovirus, Umbravirus, Polerovirus and Enamovirus, and most proteins translated via ribosomal frameshifting are the RNA-dependent RNA polymerase (RdRp) (Miller and Giedroc, 2010). Typically, −1 PRF requires two cis-acting RNA elements. The first element is a hepta-nucleotide sequence where the reading frame shifts. This sequence usually fits the consensus XXXYYYN (X is any identical base, Y is A or U, and N is not G; Dreher and Miller, 2006, Jacks et al., 1988). A second element is an RNA secondary structure, a pseudoknot or a very stable RNA structure (Giedroc and Cornish, 2009), immediately downstream from the shift site (Brierley et al., 1989, Brierley and Pennell, 2001). This RNA structure is regarded as a physical barrier to stop translating ribosomes and to shift the reading frame (Namy et al., 2006). There are examples in which slippery sequence could trigger frameshift, independent of the adjacent downstream RNA secondary structure (Wang et al., 2006, Wilson et al., 1988). In addition, far downstream RNA elements of the slippery sequences in Barley yellow dwarf virus (BYDV) and Red clover necrotic mosaic virus (RCNMV) required for −1 PRF through base pairing with the immediate downstream RNA sequence were also reported (Barry and Miller, 2002, Paul et al., 2001, Tajima et al., 2011).
The genus Tobamovirus consists of 33 species reported by the International Committee on Taxonomy of Viruses in 2012 (http://ictvonline.org/virusTaxonomy.asp?version=2012). Hibiscus latent Singapore virus (HLSV), discovered in Singapore (Srinivasan et al., 2002, Srinivasan et al., 2005), is one of two members of Tobamovirus infecting Hibiscus (Kamenova and Adkins, 2004). It has a similar genome structure to that of other tobamoviruses with a 5′ untranslated region (UTR), four open reading frames (ORFs) encoding viral replicase components (128 kDa and 184 kDa), movement protein (MP, 31 kDa), coat protein (CP, 18 kDa) and a 3′ UTR (Ishikawa and Okada, 2004, Zaitlin, 1999). Different from other tobamoviruses, only Hibiscus latent Fort Pierce virus (HLFPV) and HLSV possess a unique internal poly(A) tract (personal communication, Wong and Adkins). HLSV contains a variable length ranging from 77 to 96 nucleotides (nt) upstream of the tRNA-like structure (TLS) in the 3′ UTR (Srinivasan et al., 2005). There is no report about viral protein expression by −1 PRF in tobamoviruses.
In this study, HLSV full-length cDNA clones were constructed and were found to be infectious. A full-length cDNA clone of HLSV with a 7A stretch was located in its replicase gene replicated with a lower level, compared to another HLSV full-length cDNA clone with an 8A stretch. Mutational analyses showed that the 7A stretch is the slippery sequence required for −1 PRF and the downstream pseudoknot is dispensable. An extension from 7A to 8A stretch was observed during subsequent HLSV replication.
Results
In vitro transcripts of HLSV full-length cDNA clones are infectious in Nicotiana benthamiana and the replication level of HLSV-7A is lower than that of HLSV-8A
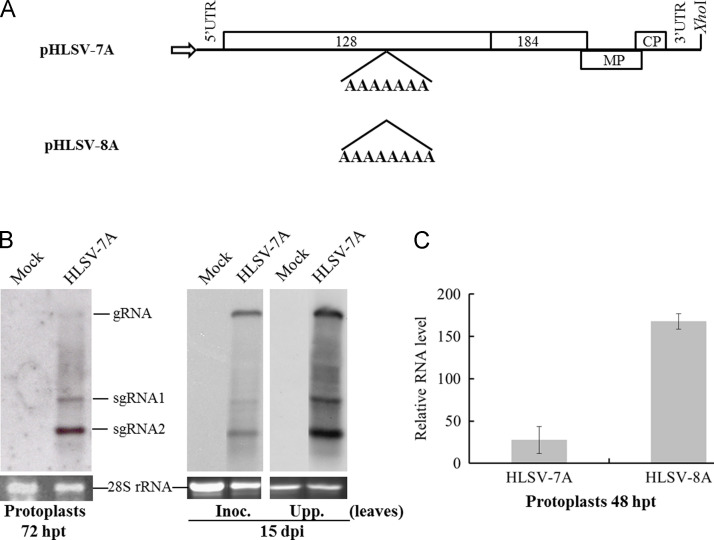
To study the replication of HLSV, a biologically active full-length cDNA clone has to be constructed. According to the HLSV complete genome sequence (GenBank Accession No. GI 113460144), primers were designated for construction of HLSV cDNA full-length clone. During cloning, we found that one of the clones, designated as pHLSV-7A, possessed a hepta-adenosine stretch, in contrast to an octo-adenosine stretch of another clone at the position nt 2078, designated as pHLSV-8A ( Fig. 1A). The in vitro transcripts of HLSV-7A were able to replicate in Nicotiana benthamiana protoplasts and infect N. benthamiana systemically by Northern blot analysis (Fig. 1B), although its RNA accumulation level was ~5-fold lower than that of HLSV-8A, as determined by quantitative real-time RT-PCR (qRT-PCR) analysis in transfected protoplasts (Fig. 1C).
Fig. 1.
In vitro transcripts of HLSV full-length cDNA clones are infectious and viral RNA accumulation in HLSV-7A transfected Nicotiana benthamiana protoplasts was lower than that in HLSV-8A at 48 h post-transfection (hpt). (A) Schematic representation of HLSV full-length cDNA clone pHLSV-7A and pHLSV-8A. (B) Northern blot of HLSV RNA from HLSV-7A infected N. benthamiana protoplasts at 72 hpt and plants at 15 days post-inoculation (dpi). Probe used was DIG-labeled HLSV CP gene PCR products. Inoculated and upper leaves are abbreviated as Inoc. and Upp. leaves, respectively. (C) Comparison of viral RNA accumulation in HLSV-7A and HLSV-8A transfected N. benthamiana protoplasts at 48 hpt by quantitative real-time RT-PCR in triplicates. Actin was used as an internal control. The relative HLSV RNA level was calculated as 2^ΔCt (actin-virus).
Hepta-adenosine stretch in HLSV is the slippery sequence for −1 PRF
According to the deduced protein sequence of HLSV-7A, only a premature 78 kDa protein was translated from the 5′ replicase gene sequence. Since both replicase proteins of tobamoviruses are essential for efficient virus replication (Bao et al., 1996, Mizumoto et al., 2010, Ogawa et al., 1991), we wondered how HLSV-7A was able to replicate without both functional replicases. Surprisingly, two additional bands, a clear 128 and a faint 184 kDa replicase proteins, were detected, in addition to the prominent 78 kDa protein in in vitro translation products using wheat germ extract. The level of the two additional proteins was much lower as compared to those generated from HLSV wild-type virions RNA ( Fig. 2B) and in vitro transcripts of HLSV-8A (Fig. 2C, first lane from the right). These two proteins must have been produced by −1 PRF due to the absence of an adenosine residue at nt 2078 in HLSV-7A.
Fig. 2.
Hepta-adenosine stretch in HLSV is the slippery sequence for programmed −1 ribosome frameshift (−1 PRF). (A) Schematic representation of the slippery sequence mutants pHLSV-AAGAAGAA and pHLSV-AAGAAGA. (B) In vitro translation from in vitro transcripts of HLSV-7A and RNA extracted from wild type HLSV virions in wheat germ extract. (C) In vitro translation from in vitro transcripts of HLSV-7A mutants in wheat germ extract. This experiment was repeated three times with similar results. The in vitro translation products were labeled with l-[35S]-methionine.
To determine if the 7A stretch is the slippery sequence for −1 PRF, two other HLSV mutated subclones pHLSV-AAGAAGA and pHLSV-AAGAAGAA, in which 7A and 8A stretches were mutated to non-slippery sequences AAGAAGA and AAGAAGAA, respectively, were constructed (Fig. 2A) and confirmed by DNA sequencing. The deduced −1 PRF product 108 kDa protein was expected when pHLSV-7A was digested with BspHI (at nt 2940 of HLSV genome) (Fig. 2C, fourth lane from the right), as compared to the full-length 128 kDa protein produced in pHLSV-7A when digested with XhoI (Fig. 2C, second lane from the right). If the 7A stretch is a slippery sequence, only a 78 kDa or a 108 kDa target protein will be produced from in vitro transcripts of HLSV-AAGAAGA or HLSV-AAGAAGAA. In vitro translation experiment showed that 78 kDa (Fig. 2C, second lane from the left) and 108 kDa proteins (Fig. 2C, third lane from the left) were detected from HLSV-AAGAAGAA and HLSV-AAGAAGA, respectively (Fig. 2C). Therefore, we conclude that the 7A stretch is the slippery sequence for −1 PRF in HLSV-7A.
Immediate downstream pseudoknot of the 7A stretch in HLSV-7A is dispensable for −1 PRF
In most cases, slippery sequence and its immediate downstream RNA secondary structure are needed for efficient −1 PRF (Brierley et al., 1989, Brierley and Pennell, 2001, Tsai et al., 1999). The immediate downstream 80 nt RNA sequence of the hepta-adenosine stretch of HLSV was analyzed with the online software KnotInFrame (http://bibiserv.techfak.uni-bielefeld.de/knotinframe) and a predicted 60 nt pseudoknot composed of 3 stems was obtained ( Fig. 3A, the first panel from left). To investigate the function of the pseudoknot on −1 PRF, the 3 stems were mutated partially or completely in pHLSV-7A (Fig. 3A) and the pseudoknot structure was not detected using the software, when all three stems were mutated (data not shown). Similar level of −1 PRF product, 128 kDa protein, was detected in each mutant, as compared to that in HLSV-7A, which indicates that the pseudoknot is not required for the −1 PRF in HLSV-7A (Fig. 3B, left panel). As expected, the 128 kDa protein was not detected in HLSV-7A-mS1+mS2+mS3stop in which a stop codon was introduced into the third stem of the pseudoknot (Fig. 3B, right panel).
Fig. 3.
A downstream pseudoknot structure of HLSV hepta-adenosine stretch is dispensable for −1 PRF. (A) Schematic representation of the pseudoknot mutants. S1, S2 and S3 (boxed) are the first, second and third stems within the pseudoknot, respectively. The mutated nucleotides are circled and shadowed. (B) In vitro translation from transcripts of the pseudoknot mutants in wheat germ extract. The frame shift (fs in %) efficiency was calculated as the mean value of p128 to p78 ratio from three independent experiments, analyzed by the ImageJ software (National Institutes of Health). The standard deviation (SD) and T-test P (comparison of fs% between each pseudoknot mutant and HLSV-7A) are indicated. The in vitro translation products were labeled with biotinylated lysine.
Far downstream HLSV sequence is not required for −1 PRF in vitro
In BYDV and RCNMV, the far downstream RNA elements of the slippery sequences are required for −1 PRF through base pairing with the immediate downstream RNA sequence (Barry and Miller, 2002, Paul et al., 2001, Tajima et al., 2011). In HLSV, a −1 PRF product, 108 kDa protein, was still detected using transcripts derived from BspHI digested pHLSV-7A, in which the far downstream HLSV sequence was cut off (Fig. 2C, fourth lane from the right). Therefore, the far downstream RNA sequence is not required for the −1 PRF in HLSV-7A in vitro.
7A stretch could extend to 8A stretch in HLSV during its replication
To determine whether the 7A stretch in HLSV could extend to 8A stretch during its replication, N. benthamiana leaves inoculated with HLSV-7A were collected at 2 and 6 days post-inoculation (dpi) and total RNAs were extracted from these inoculated leaves. RT-PCR to amplify the region containing the stretch was carried out, followed by insertion of these PCR products into pGEM®-T Easy Vector. Sequence analyses of the transformants showed that only 3 out of 30 clones were added with 1A at 2 dpi. However, a large number of clones were found to have 1A added (24 out of 30) at 6 dpi ( Table 1). This result indicates that the 7A stretch can extend to 8A stretch by 1A insertion during its replication.
Table 1.
Distribution of 7A versus 8A recovered from HLSV-7A inoculated Nicotiana enthamiana.
| dpia |
||||
|---|---|---|---|---|
| 2 | 6 | |||
| Length of adenosine stretch | 7A | 8A | 7A | 8A |
| Number of clones randomly selected and sequenced (total=30) | 27 | 3 | 6 | 24 |
In vitro transcripts derived from HLSV-7A were mechanically inoculated onto N. benthamiana leaves and total RNA was extracted from inoculated leaves at 2 and 6 days post-inoculation (dpi), respectively, followed by RT-PCR, cDNA cloning and sequencing.
Discussion
We report here the construction of HLSV biologically active full-length cDNA clones and the identification of a hepta-adenosine stretch in HLSV genome as a slippery sequence responsible for −1 PRF, independent of its downstream pseudoknot structure and the far downstream RNA sequence in vitro. The virus is able to generate functional viral replicases through the −1 PRF and extension from 7A to 8A stretch during replication in plants.
According to earlier studies on −1 PRF, most of the frameshifting signals consist of a slippery sequence and a downstream RNA secondary structure (pseudoknot or stem-loop) (Biswas et al., 2004, Brierley et al., 1992, Jacks et al., 1988, Liao et al., 2011, ten Dam et al., 1990). In this study, a downstream 60 nt RNA pseudoknot sequence of the 7A stretch in HLSV was predicted by using the online software KnotInFrame (Theis et al., 2008). Mutation analyses of the three stems within the pseudoknot showed that they were not required for −1 PRF (Fig. 3B) in vitro. Translation analysis of BspHI digested pHLSV-7A showed that the far downstream RNA sequences are also not required for the −1 PRF (Fig. 2C, fourth lane from the right). These results indicate that the 7A stretch can function solely for the −1 PRF in vitro. Similarly, a 7U stretch has been reported to be a sole signal for efficient −1 PRF in coronavirus (Wang et al., 2006). Earlier study on HIV also showed that frameshift was independent of the downstream pseudoknot structure (Wilson et al., 1988). The slippery sequences in the two cases mentioned above are all monotonous runs of U. Taken together, we believe that, compared to other slippery sequences, the monotonous runs of A or U are the sole signals for −1 PRF.
Since monotonous runs of nucleotides can also cause −2 or +1 PRF (Brierley et al., 1992), we noticed that in the in vitro translation products using HLSV-8A transcripts, a 78 kDa protein was detected (Fig. 2C, first lane from the right). One possible explanation is that +1 PRF has occurred in the 8A stretch during translation. However, in the translation products from purified viral RNA from infected N. benthamiana leaves, there was no detectable 78 kDa protein band (Fig. 2B, first lane from the right). It has been reported that capping in the 5′ end of mRNA reduces the efficiency of +1 PRF (Charbonneau et al., 2012, Gendron et al., 2008). All the viral RNA molecules extracted from purified HLSV virions should be capped. However, only ~70% of the in vitro transcripts derived from pHLSV-8A/XhoI were capped (according to the transcription kit manual). We also noticed larger size of translation products in HLSV-AAGAAGAA and HLSV digested with BspHI (Fig. 2C, third and fifth lanes from the left, respectively). It may have resulted from incomplete digestion of these two plasmids, although DNA bands may appear completely digested after gel electrophoresis (data not shown).
Among plant viruses, reversions of mutated sites to wild-type sequences are not uncommon (Arguello-Astorga et al., 2007, Shepherd et al., 2005, Wang et al., 2004). In HLSV, the 7A stretch was extended to 8A during its replication. We believe that HLSV replicases can slide backward or forward on the slippery sequence of the positive or negative strand RNA such that one additional A or U is added on the respective strands. Once an A is added, the virus will replicate faster through more replicase production. Therefore, HLSV with 8A becomes dominant in HLSV-7A inoculated leaves at 6 dpi. For tobamoviruses such as Tobacco mosaic virus and Odontoglossum ringspot virus, their replicase proteins produced by a replication-competent mutant are able to complement replication-defective mutants in trans but the efficiency is low (Ogawa et al., 1991, Wang et al., 2004). Thus, the dominance of HLSV with 8A is due to its faster replication through production of higher amount of replicase proteins.
To detect the distribution of 7A and 8A stretch in wild-type HLSV infected plants, in vitro transcripts derived from XhoI digested pHLSV-8A were inoculated onto N. benthamiana leaves. The upper leaves were collected at 21 dpi for virions purification, followed by viral RNA extraction, RT-PCR, cDNA cloning and sequencing. In randomly selected 61 clones, 60 clones contained 8A stretch and one clone contained 11A stretch, but no 7A stretch was detected (data not shown). This may be due to limited number of colonies sequenced. According to this result, the possibility of the 7A stretch in pHLSV-7A resulting from PCR error is very low. While we cannot rule out the possibility of slippery sequence occurring during in vitro transcription, it will be logistically difficult to check the rare occurrence. Since we have observed an 11A sequence in the 8A inoculated plants, it is evident that the slippery event does occur in vivo. The error rate (mismatch) for T7 RNA polymerase during transcription is 6×10−6 and no insertion or deletion event was detected (Brakmann and Grzeszik, 2001, Rong et al., 1998). The existence of 11A stretch in HLSV infected plant reveals that virus replicase may have been slided backward 3A in the 8A stretch during replication.
Earlier study on the PRF and programmed transcriptional realignment events in bacterial genomes showed that the most frequent hepta nucleotide stretch for −1 PRF is AAAAAAC, followed by AAAAAAG and AAAAAAA. These results indicate that −1 PRF tends to be evolutionarily conserved, and also some genes tend to select monotonous runs of nucleotides to ensure the right protein production once deletion or insertion happens (Sharma et al., 2011). We believe that HLSV also uses the A stretch to ensure its replicase proteins production once deletion happens. In conclusion, the HLSV-7A uses monotonous runs of 7A, −1 PRF and 1A insertion to reach its optimal replication through the functional replicase production in plants.
Materials and methods
Plasmid constructions
To construct HLSV full length cDNA clone, reverse transcription was carried out using purified HLSV RNA as template by Superscript III Reverse Transcriptase (Life Technologies, Invitrogen), by using primer HL-R1. All the primers used in this study are listed in Supplementary Table S1. The synthesized cDNA was used to amplify the HLSV 5′-end (nt 1-3333) and 3′-end (nt 3334-6474) fragments using primer pairs HL-F1/HL-R2 and HL-F2/HL-R1, respectively. The 3′-end fragment was first digested with XbaI and MluI and inserted into modified pBlueScript II KS(+), resulting in p3′HLSV. The 5′-end fragment was inserted into pGEM®-T vector (Promega) to generate p5′HLSV which was cut with KpnI and XbaI and inserted into the p3′HLSV, resulting in HLSV full-length clones pHLSV containing 87 nt internal poly(A) tract. To construct the slippery sequence mutants, a subclone pHLSV2960 was constructed by inserting the PCR product amplified by primers HL-F22 and HL-R18 using pHLSV as template into pGEM®-T Easy Vector (Promega). The following two plasmids were constructed using pHLSV2960 as a template by Quick Change mutagenesis (Stratagene) and verified by sequencing. Primers HL-F25 and HL-R20 were used for 7A stretch mutants, designated as pHLSV-AAGAAGA. Primers HL-F19 and HL-R16 were used for 8A stretch mutant, designated as pHLSV-AAGAAGAA. To construct the downstream pseudoknot mutants, in which partial or all three stems within the pseudoknot were disrupted by nucleotide substitution, primer pairs were designed to amplify pHLSV-7A. The designated plasmid names and their corresponding primers were as follows: HL-F39 and HL-R33, HL-F40 and HL-R34, HL-F41 and HL-R35, HL-F44 and HL-R38, HL-F45 and HL-R39 and HL-F43 and HL-R37 were for pHLSV-7AmS1+S2+S3, pHLSV-7A-mS1+mS2+S3, pHLSV-7A-mS1+mS2+mS3, pHLSV-7A-mS1+S2+mS3, pHLSV-7A-S1+mS2+mS3 and pHLSV-7A-S1+S2+mS3, respectively. To introduce a stop codon in the third stem in pHLSV-7A, primers HL-F46 and HL-R40 were used to amplify pHLSV-7A and this mutant was designated as pHLSV-7A-mS1+mS2+mS3stop.
In vitro transcription and in vitro translation assay
HLSV full-length cDNA clones pHLSV-7A and pHLSV-8A were linearized with either XhoI or BspHI, followed by phenol/chloroform extraction and ethanol precipitation. pHLSV-AAGAAGA and pHLSV-AAGAAGAA were digested with BspHI, and the downstream pseudoknot mutants were digested with XhoI, followed by phenol/chloroform extraction and ethanol precipitation. The purified DNAs were used for in vitro transcription by using mMessage mMachine in vitro transcription kit (Life Technologies, Ambion). Transcripts purified by LiCl precipitation were used for in vitro translation. In vitro translation was carried out by using a wheat germ extract system (Promega) and products were labeled with EasyTag™ l-[35S]-methionine (NEG709A001MC, Perkin-Elmer) or biotinylated lysine (Transcend™ tRNA, Promega).
Protoplast transfection, total RNA extraction and qRT-PCR
N. benthamiana protoplasts were isolated and transfected with in vitro transcripts derived from pHLSV-7A and pHLSV-8A, respectively (Qiao et al., 2009). Total RNAs were extracted by the SDS–phenol method (Hans et al., 1992) from harvested protoplasts at 48 hpt, followed by cDNA synthesis using primers HL-R8 and Actin-R. Synthesized cDNA was used for qRT-PCR in triplicates with KAPA SYBR® on a CFX384 Real-Time PCR system (Bio-Rad) by using actin as an internal control. Gene specific primers HL-F9 and HL-R9 were used for qRT-PCR and primers Actin-F and Actin-R were used for actin gene amplification.
Plant inoculation and Northern blot
In vitro transcripts (0.5 µg for each leaf) derived from XhoI digested pHLSV-7A were inoculated onto N. benthamiana leaves and total RNAs were extracted from inoculated and upper leaves at 15 dpi. RNA (5 µg each) was separated in 1.2% agarose gel and transferred onto positively charged nylon membranes, followed by hybridization with probe (covering HLSV CP region) labeled with a PCR DIG Probe synthesis kit (Roche) with primers HL-CP-F and HL-CP-R, using pHLSV-7A as template.
Detection of population of adenosine stretches by RT-PCR
Total RNAs were extracted from N. benthamiana leaves inoculated with HLSV-7A at 2 and 6 dpi, respectively; and viral RNA was extracted from wild-type virions purified from HLSV-8A infected plants at 21 dpi. The RNAs were used for cDNA synthesis with primer HL-R6, followed by PCR with primers HL-F12 and HL-R18. The RT-PCR products were cloned into pGEM®-T Easy Vector. Selected colonies were cultured and purified plasmids were sequenced with primer HL-F12.
Acknowledgment
This work was supported by National University of Singapore Research Grant R-154-000-552-112.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2013.11.021.
Appendix A. Supplementary materials
Supplementary data
References
- Arguello-Astorga G., Ascencio-Ibanez J.T., Dallas M.B., Orozco B.M., Hanley-Bowdoin L. High-frequency reversion of geminivirus replication protein mutants during infection. J. Virol. 2007;81:11005–11015. doi: 10.1128/JVI.00925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Carter S.A., Nelson R.S. The 126- and 183-kilodalton proteins of tobacco mosaic virus, and not their common nucleotide sequence, control mosaic symptom formation in tobacco. J. Virol. 1996;70:6378–6383. doi: 10.1128/jvi.70.9.6378-6383.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J.K., Miller W.A. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. USA. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas P., Jiang X., Pacchia A.L., Dougherty J.P., Peltz S.W. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 2004;78:2082–2087. doi: 10.1128/JVI.78.4.2082-2087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakmann S., Grzeszik S. An error-prone T7 RNA polymerase mutant generated by directed evolution. ChemBioChem. 2001;2:212–219. doi: 10.1002/1439-7633(20010302)2:3<212::AID-CBIC212>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- Charbonneau J., Gendron K., Ferbeyre G., Brakier-Gingras L. The 5′ UTR of HIV-1 full-length mRNA and the Tat viral protein modulate the programmed −1 ribosomal frameshift that generates HIV-1 enzymes. RNA. 2012;18:519–529. doi: 10.1261/rna.030346.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T.W., Miller W.A. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron K., Charbonneau J., Dulude D., Heveker N., Ferbeyre G., Brakier-Gingras L. The presence of the TAR RNA structure alters the programmed −1 ribosomal frameshift efficiency of the human immunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res. 2008;36:30–40. doi: 10.1093/nar/gkm906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc D.P., Cornish P.V. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F., Fuchs M., Pinck L. Replication of grapevine fanleaf virus satellite RNA transcripts in Chenopodium quinoa protoplasts. J. Gen. Virol. 1992;73:2517–2523. doi: 10.1099/0022-1317-73-10-2517. [DOI] [PubMed] [Google Scholar]
- Ishikawa M., Okada Y. Replication of tobamovirus RNA. Proc. Jpn. Acad. Ser. B. 2004;80:215–224. [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenova I., Adkins S. Transmission, in planta distribution, and management of Hibiscus latent Fort Pierce virus, a novel tobamovirus isolated from Florida hibiscus. Plant Dis. 2004;88:674–679. doi: 10.1094/PDIS.2004.88.6.674. [DOI] [PubMed] [Google Scholar]
- Liao P.Y., Choi Y.S., Dinman J.D., Lee K.H. The many paths to frameshifting: kinetic modelling and analysis of the effects of different elongation steps on programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2011;39:300–312. doi: 10.1093/nar/gkq761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W.A., Giedroc D.P. Ribosomal frameshifting in decoding plant viral RNAs. In: Gesteland J.F., Raymond F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; Heidelberg: 2010. pp. 193–220. [Google Scholar]
- Mizumoto H., Kimura K., Kiba A., Hikichi Y. The 126- and/or 183-kDa replicases or their coding regions are responsible both for inefficient local and for systemic movements of Paprika mild mottle virus Japanese strain in tomato plants. Virus Res. 2010;153:205–211. doi: 10.1016/j.virusres.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Namy O., Moran S.J., Stuart D.I., Gilbert R.J., Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Watanabe Y., Meshi T., Okada Y. Trans complementation of virus-encoded replicase components of tobacco mosaic virus. Virology. 1991;185:580–584. doi: 10.1016/0042-6822(91)90528-j. [DOI] [PubMed] [Google Scholar]
- Paul C.P., Barry J.K., Dinesh-Kumar S.P., Brault V., Miller W.A. A sequence required for −1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol. 2001;310:987–999. doi: 10.1006/jmbi.2001.4801. [DOI] [PubMed] [Google Scholar]
- Plant E.P. Ribosomal frameshift signals in viral genomes. In: Garcia M.L., Romanowski V., editors. Viral Genomes—Molecular Structure, Diversity, Gene Expression Mechanisms and Host–Virus Interactions. InTech; Rijeka, Croatia: 2012. pp. 91–122. [Google Scholar]
- Qiao Y., Li H.F., Wong S.M., Fan Z.F. Plastocyanin transit peptide interacts with Potato virus X coat protein, while silencing of plastocyanin reduces coat protein accumulation in chloroplasts and symptom severity in host plants. Mol. Plant Microbe Interact. 2009;22:1523–1534. doi: 10.1094/MPMI-22-12-1523. [DOI] [PubMed] [Google Scholar]
- Rong M., Durbin R.K., McAllister W.T. Template strand switching by T7 RNA polymerase. J. Biol. Chem. 1998;273:10253–10260. doi: 10.1074/jbc.273.17.10253. [DOI] [PubMed] [Google Scholar]
- Sharma V., Firth A.E., Antonov I., Fayet O., Atkins J.F., Borodovsky M., Baranov P.V. A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. Mol. Biol. Evol. 2011;28:3195–3211. doi: 10.1093/molbev/msr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd D.N., Martin D.P., McGivern D.R., Boulton M.I., Thomson J.A., Rybicki E.P. A three-nucleotide mutation altering the Maize streak virus Rep pRBR-interaction motif reduces symptom severity in maize and partially reverts at high frequency without restoring pRBR–Rep binding. J. Gen. Virol. 2005;86:803–813. doi: 10.1099/vir.0.80694-0. [DOI] [PubMed] [Google Scholar]
- Srinivasan K.G., Min B.E., Ryu K.H., Adkins S., Wong S.M. Determination of complete nucleotide sequence of Hibiscus latent Singapore virus: evidence for the presence of an internal poly(A) tract. Arch. Virol. 2005;150:153–166. doi: 10.1007/s00705-004-0404-x. [DOI] [PubMed] [Google Scholar]
- Srinivasan K.G., Narendrakumar R., Wong S.M. Hibiscus virus S is a new subgroup II tobamovirus: evidence from its unique coat protein and movement protein sequences. Arch. Virol. 2002;147:1585–1598. doi: 10.1007/s00705-002-0829-z. [DOI] [PubMed] [Google Scholar]
- Tajima Y., Iwakawa H., Kaido M., Mise K., Okuno T. A long-distance RNA–RNA interaction plays an important role in programmed −1 ribosomal frameshifting in the translation of p88 replicase protein of Red clover necrotic mosaic virus. Virology. 2011;417:169–178. doi: 10.1016/j.virol.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E.B., Pleij C.W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis C., Reeder J., Giegerich R. KnotInFrame: prediction of −1 ribosomal frameshift events. Nucleic Acids Res. 2008;36:6013–6020. doi: 10.1093/nar/gkn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.H., Cheng C.P., Peng C.W., Lin B.Y., Lin N.S., Hsu Y.H. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. J. Virol. 1999;73:2703–2709. doi: 10.1128/jvi.73.4.2703-2709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.H., Yu H.H., Wong S.M. Mutation of Phe50 to Ser50 in the 126/183-kDa proteins of Odontoglossum ringspot virus abolishes virus replication but can be complemented and restored by exact reversion. J. Gen. Virol. 2004;85:2447–2457. doi: 10.1099/vir.0.80070-0. [DOI] [PubMed] [Google Scholar]
- Wang X., Wong S.M., Liu D.X. Identification of Hepta- and Octo-Uridine stretches as sole signals for programmed +1 and −1 ribosomal frameshifting during translation of SARS-CoV ORF 3a variants. Nucleic Acids Res. 2006;34:1250–1260. doi: 10.1093/nar/gkl017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S.E., Rathjen P.D., Kingsman S.M., Kingsman A.J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Zaitlin M. Elucidation of the genome organization of tobacco mosaic virus. Philos. Trans. R. Soc. London Ser. B. 1999;354:587–591. doi: 10.1098/rstb.1999.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data