Abstract
Three reverse transcription recombinase polymerase amplification assays with lateral flow dipsticks (RT-RPA-LFD) were developed for identification of the matrix and hemagglutinin (HA) genes to detect influenza A virus and distinguish subtypes H1 and H3. Assessment of the assays’ specificity showed that there was no cross-reactivity with other targets. Their limits of detection were 123.6 copies per reaction for the matrix gene, 677.1 copies per reaction for the H1 HA gene, and 112.2 copies/reaction for the H3 HA gene. Of 111 samples tested by RT-RPA-LFD assays, 27 were positive for influenza A virus, 14 were positive for H1, and 10 were positive for H3. Compared to the results obtained from real-time RT-PCR assays, the sensitivity of RT-RPA-LFD assays was 75%, 93.33% and 71.43% for the matrix, H1, and H3, with 100% specificity. The sensitivity of RT-RPA-LFD assays is lower than that of real-time RT-PCR, comparable or better than that of conventional RT-PCR, and much better than that of RIDTs. In conclusion, these assays offer an efficient and reliable tool for identification and subtyping of influenza A virus (subtype H1 and H3) in the resource-limited setting.
Keywords: Influenza A virus, Recombinase polymerase amplification, Detection, Subtyping, Lateral flow dipstick
Highlights
-
•
Three RT-RPA-LFD assays are developed for the detection and subtyping of influenza A virus.
-
•
RT-RPA-LFD assays used for the detection of influenza A virus is rapid and sensitive.
-
•
The results from RT-RPA-LFD assays are in a good agreement with those from real-time RT-PCR assays.
1. Introduction
Influenza viruses are common and major pathogens that cause viral respiratory infections. These viruses are a serious threat to the health of people worldwide as the severe morbidity and mortality they cause. According to the World Health Organization (WHO), seasonal influenza viruses cause about 290,000 to 650,000 deaths each year. Influenza viruses belong to the Orthomyxoviridae family and are divided into four types: type A, B, C, and D [1]. Influenza A virus (IAV) is the most virulent of these pathogens. Based on two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), IAVs are further divided into many subtypes, also known as serotypes. So far, 18 serotypes of HA (H1–H18) and 11 serotypes of NA (N1–N11) have been described and verified [2]. Due to reassortment or mutation, IAV subtypes evolve constantly and can cause pandemics, with a recent example being the influenza A(H1N1)pdm09 virus [3]. Among the IAV subtypes, H1N1 and H3N2 are the most common subtypes causing disease in humans around the world.
Although vaccination is an effective countermeasure against infection by influenza virus, some limitations still exist [4,5]. Clinically, NA inhibitors and adamantanes are approved for treatment and prevention of influenza virus infection. However, some subtypes with high-level antiviral resistance currently circulate which are resistant to all adamantanes or NA inhibitors [6]. Moreover, effective antiviral treatment needs to be initiated within 48 h of illness onset. Hence, it is necessary to rapidly identify IAVs for accurate treatment and infection control.
Infections of IAVs can be clinically identified and diagnosed based on patient's signs and symptoms. However, approximately 33% of individuals infected are asymptomatic [7], and others infected by bacteria or other viruses, such as respiratory syncytial virus, parainfluenza virus, and rhinovirus, also show influenza-like signs and symptoms. Compared to laboratory diagnostic technology, clinical diagnosis is estimated to have an 18–87% positive predictive value [8]. In the laboratory, the long turnaround time of viral culture is unfavorable for accurate treatment and control of IAVs. Rapid influenza diagnostic tests (RIDTs) are widely used to identify IAVs in healthcare services because they are simple and swift [[8], [9], [10]]. Although the specificity of RIDTs is up to 90%, the defect is an inconsistent sensitivity (10–80%) [8,9,11]. More recently, nucleic acid amplification tests (NAATs), such as reverse transcription polymerase chain reaction (RT-PCR) [[12], [13], [14], [15]], real-time RT-PCR [16,17], and reverse transcription loop-mediated isothermal amplification (RT-LAMP) [18,19], have been used for rapid and sensitive diagnosis or subtyping of IAVs. Nevertheless, these methods require expensive equipment and/or skilled technicians, making them inappropriate for use in developing countries.
Recombinase polymerase amplification (RPA) is a low-resource diagnostic tool that depends on several enzymes running at a constant temperature [20]. Previously, RPA has been used for diagnosis of many pathogens, such as human immunodeficiency virus type 1 [21], dengue virus [22], and Brucella [23], with great success. Therefore, detection of IAVs and differentiation of their subtypes could be achieved with RT-RPA with lateral flow dipstick, providing an efficient and rapid assay for clinical diagnosis and epidemiological surveys.
In this study, we developed three reverse transcription-RPA assays with lateral flow dipsticks (RT-RPA-LFD) for detecting IAVs and distinguishing the H1 and H3 subtypes. One assay targeted to the matrix gene was used for diagnosis of IAVs, and IAV-positive samples were further subtyped using the other two assays.
2. Materials and methods
2.1. Clinical specimen collection and virus isolates
Eighty-seven throat swabs were collected from children with influenza-like illness at Jinling Hospital (Nanjing, Jiangsu, China) using Virocult swabs (Yocon Biotech. Co., Beijing, China) and stored at −80 °C within 2 h. These specimens were collected between February 2016 and March 2017.
Respiratory pathogens used in this study are listed in Table 1 . Clinical isolates including H1N1, H3N2, influenza B virus (Flu B), and respiratory syncytial virus (RSV) subgroup A and B were previously identified by RT-PCR and sequencing [24]. H1N1, H3N2, and influenza B virus were designated as A/Nanjing/37/2015(H1N1), A/Nanjing/46/2015(H3N2), and B/Victoria/117/2015, respectively. Two subtypes of IAV strains, A/Michigan/45/2015(H1N1) and A/Hong Kong/4801/2014(H3N2), were provided by Shanghai Institute of Biological Products Co., Ltd. Staphylococcus aureus (ATCC 25923), Haemophilus influenzae (ATCC 49247), Streptococcus pneumoniae (ATCC 49619), Streptococcus hemolyticus, Chlamydia pneumonia, Mycoplasma pneumoniae were stored in our laboratory.
Table 1.
Influenza viruses and other respiratory pathogens used in this study.
| Respiratory pathogensa | Subtype/subgroup | Strain name | Sample | Sourceb | RT-RPA-LFD |
||
|---|---|---|---|---|---|---|---|
| matrix | H1 | H3 | |||||
| influenza A virus | H1N1 | A/Nanjing/37/2015(H1N1) | throat swabs | Our laboratory | + | + | – |
| H1N1 | A/Michigen/45/2015(H1N1) | cell supernatant | SIBPC | + | + | – | |
| H3N2 | A/Nanjing/46/2015(H3N2) | throat swabs | Our laboratory | + | – | + | |
| H3N2 | A/HongKong/480/2014(H3N2) | cell supernatant | SIBPC | + | – | + | |
| influenza B virus | Victoria lineage | B/Victoria/117/2015 | throat swabs | Our laboratory | – | – | – |
| respiratory syncytial virus | A | / | throat swabs | Our laboratory | – | – | – |
| B | / | throat swabs | Our laboratory | – | – | – | |
| human metapneumovirus | unknown | / | nucleic acid | NHB | – | – | – |
| herpes simplex virus-1 | unknown | / | nucleic acid | NHB | – | – | – |
| human coronavirus 229E | unknown | / | nucleic acid | NHB | – | – | – |
| human adenovirus | unknown | / | nucleic acid | NHB | – | – | – |
| parainfluenza virus 1 | unknown | / | nucleic acid | NHB | – | – | – |
| parainfluenza virus 2 | unknown | / | nucleic acid | NHB | – | – | – |
| parainfluenza virus 3 | unknown | / | nucleic acid | NHB | – | – | – |
| human rhinovirus | unknown | / | nucleic acid | NHB | – | – | – |
| Staphylococcus aureus | / | ATCC 25923 | culture | ATCC | – | – | – |
| Haemophilus influenzae | / | ATCC 49247 | culture | ATCC | – | – | – |
| Streptococcus pneumoniae | / | ATCC 49619 | culture | ATCC | – | – | – |
| Streptococcus hemolyticus | / | clinical isolate | sputum | Our laboratory | – | – | – |
| Chlamydia pneumoniae | / | clinical isolate | sputum | Our laboratory | – | – | – |
| Mycoplasma pneumoniae | / | clinical isolate | sputum | Our laboratory | – | – | – |
Human metapneumovirus, herpes simplex virus-1, human coronavirus 229E, human adenovirus, parainfluenza virus 1–3, and human rhinovirus were collected by Ningbo Health BioMed Co., Ltd, and identified in Institut Pasteur of Shanghai. Streptococcus hemolyticus, Chlamydia pneumonia, and Mycoplasma pneumonia were isolated from patients with acute respiratory infections.
SIBPC, Shanghai Institute of Biological Products Co., Ltd.; NHB, Ningbo Health BioMed Co., Ltd.
Human metapneumovirus (hMPV), herpes simplex virus 1 (HSV-1), human coronavirus 229E (hCoV-229E), human adenovirus (hADV), parainfluenza virus 1–3 (PIV1-3), human rhinovirus (hRV), and twenty-four viral RNAs extracted from nasopharyngeal aspirates were supplied by Ningbo Health BioMed Co., Ltd (Ningbo, Zhejiang, China).
This study was approved by the Human Use Ethical Committee at Jinling Hospital. The informed consent was obtained from all patients or guardians.
2.2. RPA primer and probe design
Since the matrix gene is conserved and usually used for detecting IAVs in the previous reports [12,[15], [16], [17]], primers and nfo (Escherichia coli endonuclease IV) probe designed for the matrix gene were used to detect IAVs. Also, primers and probes were designed for subtyping H1 and H3. Due to antigen drift and genetic variability of IAVs, many isolates are collected and identified in the influenza surveillance annually. This makes it difficult to align all the published sequences of IAVs. Consequently, we firstly classified the sequences in the database based on both the geographically country/region and collection date/release date (Influenza Virus Resource, https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi#mainform). Secondly, a consensus sequence was obtained by alignment of the sequences isolated from the same country within a five-year span. Finally, all the consensus sequences were aligned using the clustalW (http://www.ebi.ac.uk/Tools/msa/clustalo/). The primers and nfo probes were designed according to the most conserved sequence, were synthesized by Sangon Biotech (Shanghai, China), and are shown in Table 2 and Table S1. Each nfo Probe was modified with fluorescein isothiocyanate (FITC) at the 5′ end, an internal abasic nucleotide analogue (tetrahydrofuran, THF) and a 3′-polymerase extension blocking group C3-spacer. Opposing to nfo probe, the primer was labeled with biotin at the 5′ end.
Table 2.
Primers and probes used in this study.
| Gene | Name | Sequence (5′-3′)a | Accession no. | Reference |
|---|---|---|---|---|
| matrix | AMPB | GATCACtaatacgactcactatagggCAGAGACTTGAAGATGTCTT | NC_026431 | [15] |
| AMPC | TGCTGGGAGTCAGCAATCTG | |||
| MF146 | GGCTCTCATGGAATGGCTAAAGACAAGAC | This study | ||
| MR425 | Biotin-TTGTATATGAGGCCCATGCAACTGGCAAGTG | |||
| A-p | FITC-TTCACGCTCACCGTGCCCAGTGAGCGAGGAC-THF-GCAGCGTAGACGCTTTG-spacer(C3) | |||
| H1 | H1F1073 | GATCACtaatacgactcactatagggGGTAGATGGATGGTACGGTT | CY225830 | This study |
| H1F1325 | Biotin-TGTTGGTTCTATTGGAAAATGAAAGAACTTT | |||
| H1-P | FITC-TCATAAGTCCCATTTTTGACACTTTCCATGC-THF-CGTGTTATCGCATTTG-spacer(C3) | |||
| H1R1540 | TGTTTAATTTTGCTTCCTCTGAGTATTTTGG | |||
| H3 | H3F376 | GATCACtaatacgactcactatagggTTATGCCTCCCTTAGGTCAC | CY225422 | This study |
| H3F763 | Biotin-GAATAAGCATCTATTGGACAATAGTAAAAC | |||
| H3-P | FITC-ATGGGTGCATCTGATCTCATTATTGAGCTTT-THF-CCCACTTCGTATTTTG-spacer(C3) | |||
| H3R1156 | CCCTCAGAATTTTGATGCCTGAAACCGTACCA |
Protecting bases are shown in boldface, and the T7 promoter at the 5′ end of forward primer is shown in lowercase.
2.3. RNA extraction and cDNA synthesis
Viral nucleic acid was extracted from 140 μl of viral transport medium and eluted into 50 μl of elution buffer using the TIANamp Virus DNA/RNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) as recommended by the manufacturer.
The complementary DNA (cDNA) was synthesized using the Reverse Transcription System (A3500, Promega, Madison, WI, USA). Briefly, in a 0.2 ml tube, 5 μl of RNA, 4 μl of MgCl2 (25 mM), 2 μl of 10× Reverse Transcription Buffer, 2 μl of dNTP Mixture (10 mM), 0.5 μl of Recombinant RNasin Ribonuclease Inhibitor (40 U/μl), 1 μl of Random Primers (0.5 μg/μl), and 0.5 μl of AMV Reverse Transcriptase (5 U/μl) were mixed. The total volume was increased to 20 μl by adding nuclease-free water. The mixture was incubated for 10 min at room temperature and subsequently at 42 °C for 30 min.
2.4. PCR and real-time PCR
PCR was performed using Ex Taq Version 2.0 plus dye kit (Takara, Dalian, China) in a total volume of 25 μl, which contained 2 μl of cDNA, 12.5 μl of Premix Taq, 0.5 μl of 10 μM forward primer (AMPB, H1F1073, or H3F376), and 0.5 μl of 10 μM reverse primer (AMPC, H1R1540, or H3R1156). Amplification cycles were performed as follows: 95 °C for 5 min; 10 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s; 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; and 72 °C for 5 min. PCR products were directly analyzed by gel electrophoresis on a 1.5% agarose gel.
Real-time PCR was performed on Applied Biosystems 7500 Real-Time PCR System (Thermo Scientific, Waltham, MA, USA) using Premix Ex Taq™ (Probe qPCR) kit (Takara, Dalian, China). The primers and TaqMan probes used for qPCR were reported in WHO information for the molecular detection of influenza viruses-update (Table S2) [25]. In a 0.2 ml tube, 2 μl of cDNA, 12.5 μl of Premix Ex Taq, 0.5 μl of each forward and reverse primers (10 μM), 0.3 μl of TaqMan probe (10 μM), 0.25 μl of 50× ROX Reference Dye II, and 8.75 μl of nuclease-free water was added and vortex mixed. The reaction was run as follows: 95 °C for 1 min; 40 cycles of 95 °C for 5 s, 60 °C for 30 s. Ct values, which are defined as quantification cycle representing the crossing point between fluorescent value and threshold, were calculated automatically by the software v2.0.5 (Applied Biosystems, Foster City, CA, USA). The positive sample was defined as its Ct value less than 35, or a sample with Ct value between 35 and 40 was confirmed again.
2.5. In vitro transcription
The forward primer with a T7 promoter at the 5’ terminus was used to amplify the target gene (matrix, H1 HA, or H3 HA) in PCR. After gel purification, the linear products bearing T7 promoters were blunted with T4 DNA Polymerase, transcribed using the In vitro Transcription T7 Kit (Takara, Dalian, China), and digested using DNase I at 37 °C for 30 min. Finally, the single-stranded RNAs were purified using phenol-chloroform extraction, and their concentration was measured by a NanoDrop Nano-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).
2.6. Preparation of lateral flow dipstick
An LFD was prepared according to the previous reports [26,27], which was composed of a sample pad, conjugate pad, nitrocellulose membrane, absorbent pad, and plastic adhesive backing. Streptavidin-coated gold colloid was dispensed onto the conjugate pad, and then dried at 37 °C overnight. Anti-FITC antibody (Test line, Abcam, Cambridge, MA, USA) and biotinylated bovine serum albumin (Control line, Nanjing Runyan Biotechnology Co., Ltd, China) were striped onto the nitrocellulose membrane, and then dried at 37 °C for 1 h. Finally, LFD was assembled and cut into 4-mm width strips using microcomputer automatic cutting machine (Shanghai Goldbio Co., Ltd., China).
2.7. RPA-LFD
RPA was performed in a total volume of 25 μl using TwistDx nfo kit (TwistDX, Cambridge, UK). For each reaction, 14.75 μl of rehydration buffer, 1 μl of each forward and reverse primers (10 μM), 0.3 μl of nfo probe (10 μM), 2 μl of cDNA, and 4.7 μl of nuclease-free water were mixed. Finally, 1.25 μl of 280 mM magnesium acetate was added. The mixture was incubated at 39 °C for 20 min. RPA products were analyzed using LFDs. Five microliters of RPA product was added into a tube containing 70 μl of phosphate-buffered saline with Tween-20 (PBST; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4; 0.05% Tween-20). LFDs were inserted into the tube and incubated for 5 min. Images were recorded using a smartphone and analyzed using ImageJ software (National Institutes of Health, MD, USA). The images were converted into 8-bit greyscale, and subsequently, the mean optical density of a fixed area was determined. Relative optical density was calculated by dividing the mean optical density by the maximum grey value (255). The sample was defined as positive when relative optical density was greater than the threshold (0.153), which was calculated by the average of twenty negative samples plus three times standard deviation.
2.8. Statistical analysis
Experimental data were analyzed by SPSS Statistics 19 (SPSSInc., Chicago, USA), and the significance level was set at 0.05. Diagnostic performance of RT-RPA-LFD assays was evaluated by comparison with real-time RT-PCR. Clinical sensitivity and specificity were calculated using standard formulas. Clinical test results obtained from RT-RPA-LFD and real-time RT-PCR were analyzed with McNemar's test.
3. Results
3.1. Specificity analysis
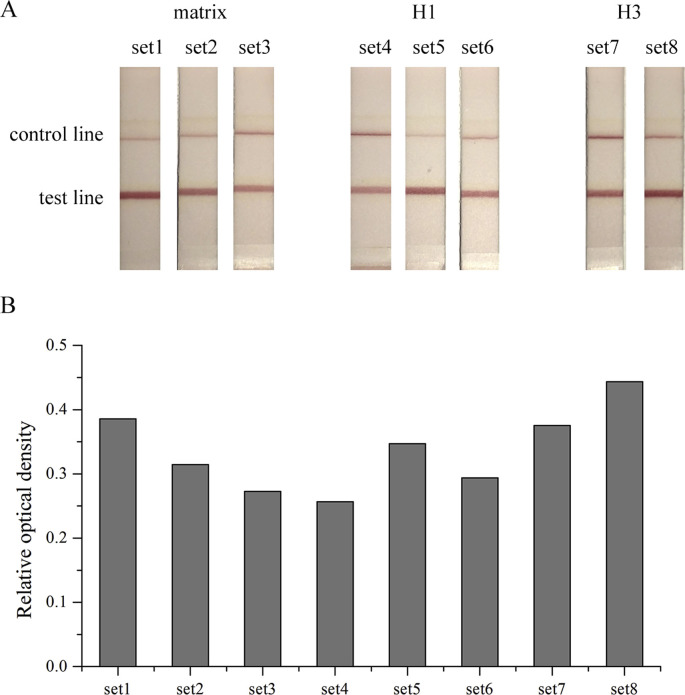
To optimize the performance of the detection and subtyping of IAVs, we designed and synthesized eight sets of primer and probe (Table S1). Viral RNAs extracted from two clinical isolates, H1N1 and H3N2, were detected by RT-RPA-LFD assays. The results indicated that set1 (matrix), set5 (H1), and set8 (H3) show the best performance (Fig. S1).
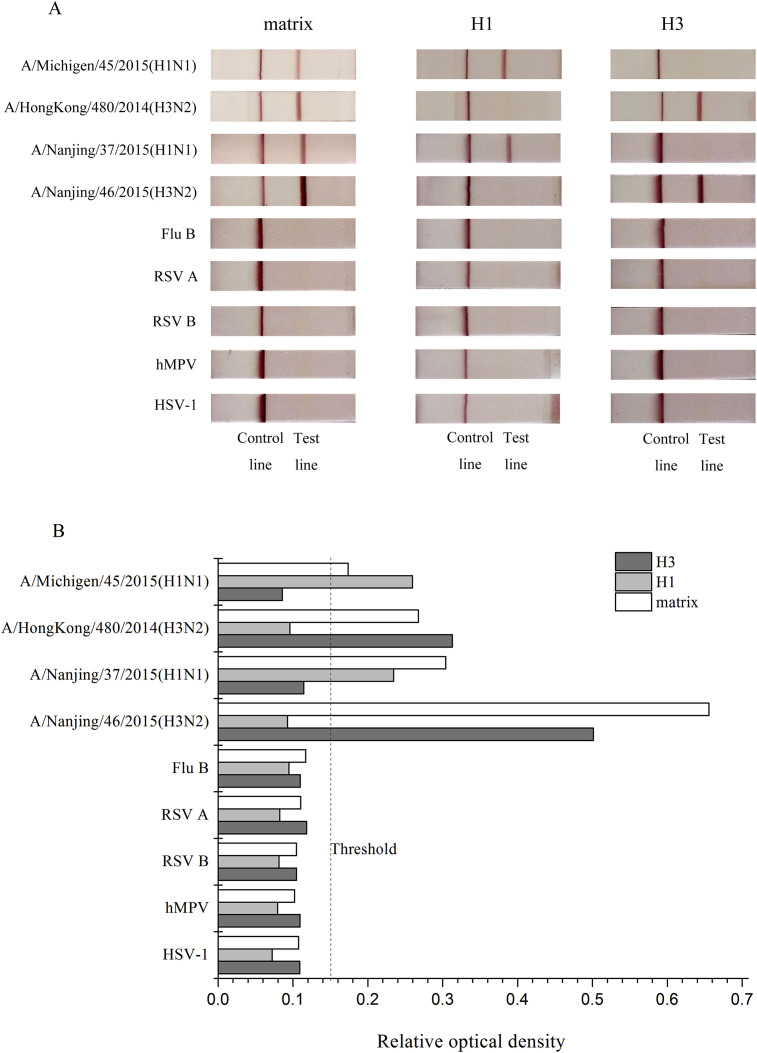
IAV subtype H1N1 and H3N2, influenza B virus (Flu B), and other respiratory pathogens were used to evaluate the specificity of three RT-RPA-LFD assays (Table 1; Fig. 1 ). As shown in Fig. 1, clear bands emerge on the test line for detecting both H1N1 and H3N2 by matrix RT-RPA-LFD assay, and no other visible bands are present on the test line for detecting other respiratory pathogens. The positive results obtained from detection of either H1N1 or H3N2 indicated that the other two assays were specific, and there was no cross-reactivity with other pathogens and between these two subtypes (Fig. 1). Although the positive and negative results are visible with the naked eye, the results of RT-RPA-LFD quantitatively analyzed using image software to diminish the influence of subjective factors (Fig. 1B).
Fig. 1.
Specificity of RT-RPA-LFD assays. (A) Nucleic acid extracted from H1N1, H3N2, influenza B virus (Flu B), respiratory syncytial virus (RSV) subgroup A and B, human metapneumovirus (hMPV) and herpes simplex virus 1 (HSV-1) was detected by three RT-RPA-LFD assays corresponding to the matrix, H1, and H3. (B) Quantitatively measurement of results of RT-RPA-LFD assays.
3.2. Sensitivity analysis
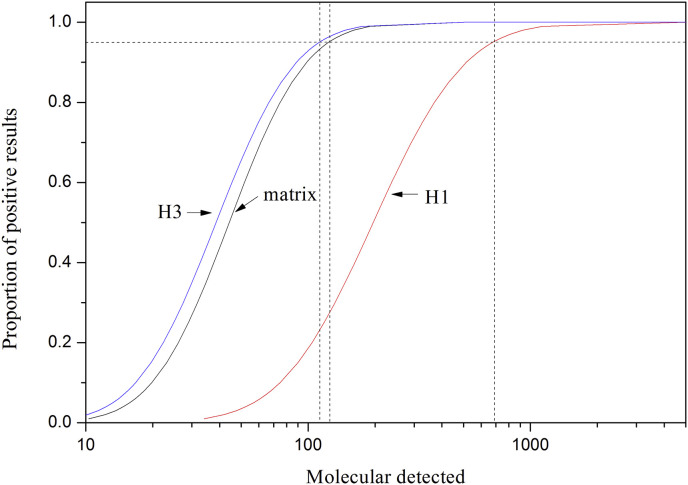
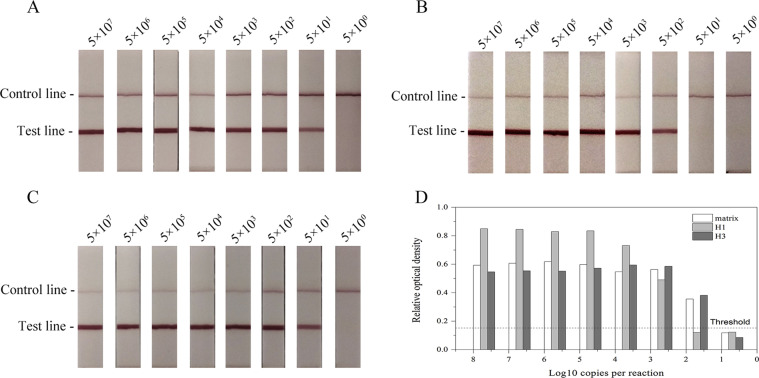
To determine the detection limits of RT-RPA-LFD assays, RNA transcripts (matrix, H1, and H3) were diluted by nuclease-free water and tested (Table 3 ; Fig. 2 ; Fig. S2). By calculating the copy number of each transcribed RNA in RPA reaction, probit analysis indicated that RT-RPA-LFD assays at 95% probability were able to detect 123.6 copies of the matrix gene, 677.1 copies of H1 HA gene, and 112.2 copies of H3 HA gene (Table 3; Fig. 2). The sensitivity of RT-RPA-LFD assays is lower than that of real-time RT-PCR [16,17], comparable or better than that of conventional RT-PCR [13,14], and much better than that of RIDTs [10].
Table 3.
Probit regression to calculate the detection limits of RT-RPA-LFD assays.
| Amount of RNA transcripts (Copies/reaction) | Replicates detected/Replicates tested by RT-RPA-LFD assays |
||
|---|---|---|---|
| matrix | H1 | H3 | |
| 5000 | 8/8 | 8/8 | 8/8 |
| 500 | 8/8 | 7/8 | 8/8 |
| 100 | 7/8 | 2/8 | 7/8 |
| 50 | 5/8 | 0/8 | 6/8 |
| 10 | 0/8 | 0/8 | 0/8 |
| LOD (95% probability) | 123.6 | 677.1 | 112.2 |
Fig. 2.
Probit regression of RT-RPA-LFD assays (matrix, H1, and H3) using the data of 8 independent assays.
3.3. Performance of LF-RT-RPA assays on clinical specimens
Both Eighty-seven throat swab specimens and twenty-four nucleic acids were tested using RT-RPA-LFD assays and real-time RT-PCR assays (Table 4 ; Table S3). Compared to real-time RT-PCR assays as the reference method, diagnostic parameters of RT-RPA-LFD assays, including sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV), were calculated as shown in Table 4. Overall, twenty-seven samples were positive for IAVs by matrix RT-RPA-LFD assay, while thirty-six samples were positive by matrix real-time RT-PCR. We further found that Ct values of nine discordant samples were greater than 37. Fourteen samples were H1-positive by RT-RPA-LFD, while fifteen samples were positive by real-time RT-PCR. Only one sample with a Ct value of 36.9 was negative by H1 RT-RPA-LFD assay. Additionally, four of fourteen H3-positive samples confirmed by real-time RT-PCR were negative by H3 RT-RPA-LFD assay. Ct values of four discordant samples were greater than 37. McNemar's test indicated that the difference between RT-RPA-LFD and real-time RT-PCR for distinguishing H1 and H3 subtypes is not significant (P > 0.05), but for detection of IAVs (P = 0.004). Agreement of RT-RPA-LFD and real-time RT-PCR was evaluated by kappa test. The correlation kappa values were 0.802 (matrix), 0.960 (H1) and 0.814 (H3), demonstrating that these assays had a good agreement for the detection of IAV and subtyping of H1 and H3.
Table 4.
Comparison of diagnostic performance between RT-RPA-LFD and real-time RT-PCR on clinical samples.
| Real-time RT-PCR |
Sensitivity | Specificity | PPVa | NPVb | P value | Kappa value | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | ||||||||
| matrix RT-RPA-LFD | Positive | 27 | 0 | 27 | 75.00% | 100% | 100% | 89.29% | 0.004 | 0.802 |
| Negative | 9 | 75 | 84 | |||||||
| Total (n = 111) | 36 | 75 | 111 | |||||||
| H1 RT-RPA-LFD | Positive | 14 | 0 | 14 | 93.33% | 100% | 100% | 98.97% | 1 | 0.96 |
| Negative | 1 | 96 | 97 | |||||||
| Total (n = 111) | 15 | 96 | 111 | |||||||
| H3 RT-RPA-LFD | Positive | 10 | 0 | 10 | 71% | 100% | 100% | 96.04% | 0.125 | 0.814 |
| Negative | 4 | 97 | 101 | |||||||
| Total (n = 111) | 14 | 97 | 111 | |||||||
PPV, positive predictive value.
NPV, negative predictive value.
In order to comprehensively evaluate the performance of RT-RPA-LFD, 28 positive throat swab specimens by matrix real-time RT-PCR were tested by RIDTs (Rapid influenza A virus antigen test kits, Guangzhou Wondfo Biotechnology Co., Ltd, China). Although commercial antigen test kit is rapid, only fourteen samples are positive for IAV (Table 5 ; Table S3), and its positive rate is much lower than that of matrix RT-RPA-LFD assay. McNemar's Test suggested that there was a significant difference between matrix RT-RPA-LFD and RIDTs (P = 0.008). Matrix RT-RPA-LFD assay could be sensitively used for diagnosis of IAV in comparison to RIDTs.
Table 5.
Comparison of the performance of matrix RT-RPA assay and rapid influenza A virus antigen test kits on 28 clinical samples.
| Rapid influenza A virus antigen test kits |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| matrix RT-RPA-LFD | Positive | 14 | 8 | 22 |
| Negative | 0 | 6 | 6 | |
| Total | 14 | 14 | 28 | |
4. Discussion
In this study, we developed three RT-RPA-LFD assays for detection and subtyping of IAVs. The targets of these three assays were the matrix, H1 HA, and H3 HA genes. The matrix gene is considered to be conserved among IAVs and was selected for identification of IAVs [12,[15], [16], [17]]. Primers and nfo probe were designed according to the alignment of matrix gene sequences. During the past several decades, subtypes H1N1 and H3N2 have been circulating worldwide [28]. According to the latest influenza surveillance [29], H3N2 and A(H1N1)pdm09 accounted for the majority of circulating viruses. Therefore, we designed another two assays for subtyping H1 and H3, thus providing a rapid and convenient method for subtyping of H1 and H3.
Specificity of the RT-RPA-LFD assays indicated that the expected signals were generated on the test line and no cross-reactivity was observed. The sensitivity of RT-RPA-LFD assays was evaluated using each single-stranded RNA transcripts of the matrix, H1, and H3 genes, and the limits of detection were determined to be 123.6, 677.1, and 112.2 copies per reaction, respectively. Clinical samples were used to evaluate the performance of RT-RPA-LFD assay and to compare its performance with real-time RT-PCR. A total of 111 samples were tested. Although the sensitivity of RT-RPA-LFD assays was slightly worse than real-time RT-PCR, the former is a better choice for the point-of-care test in resource-limited settings.
RIDTs are widely used for screening the influenza virus in official clinics or primary healthcare services. However, the detection limit of RIDTs is 10000 times higher than that of real-time RT-PCR [30]. That induces a false-negative result, which can result in inappropriate antibiotic use, treatment failure for patients, and further viral spread. The negative result of RIDTs does not rule out influenza infection, particularly in the season of high influenza activity. When a negative result of RIDT is inconsistent with clinical signs and symptoms, RT-RPA-LFD assays can be used as an effective supplement for IAV diagnosis. In addition, RT-RPA-LFD assays can be used to identify the subtype for IAV epidemiological investigations.
Although it is relatively rapid, specific and sensitive for diagnosis of IAVs and subtyping of H1/H3 by RT-RPA-LFD assays, there are two major drawbacks for these assays. One is that reverse transcription and RPA are two separate reactions. To further simplify the process, one-step RT-RPA was performed in a single tube containing TwistDx nfo kit (TwistDX, Cambridge, UK), reverse transcriptase XL (AMV, Takara, Dalian, China) and RNase Inhibitor (Takara, Dalian, China), and used to detect IAV and discriminate H1 and H3. Compared to two-step RT-RPA, the performance of one-step RT-RPA is unsatisfactory. This is probably because that there exists an inhibitory effect between the biotin-labeled reverse primer and reverse transcriptase, or between RT and RPA. This needs to try different strategies to make RT and RPA harmoniously run in a single tube. The other is that exists potential amplicons contamination. RPA product was pipetted to LFD for analysis by opening the reaction tube, which can cause amplicon contamination. This is a very prominent concern for quality control of these assays, especially which are used in a small hospital or rural areas with untrained staff. Previous studies indicate that Uracil-N-glycosylase (UNG) and dUTP can effectively eliminate amplicon contamination in PCR [31,32] or LAMP [33]. It is possible that the elimination of RPA amplicon contamination uses UNG and dUTP. Importantly, many sealed devices are developed for analysis of RPA product, and this can also avoid amplicon contamination [[34], [35], [36], [37]]. Therefore, the combination of RPA, UNG/dUTP, and/or a sealed LF device to reduce amplicon contamination will be performed in the future. Cost of the test per sample is approximately $2.5 (RT, $0.5; RPA reaction, $1.5; LFD, $0.5). A 25-μl reaction system and LFD assembled by ourselves greatly reduce the cost of test. With the improvement of availability and throughput, cost of RT-RPA-LFD will decrease in the further. For example, multiplexed recombinase polymerase amplification assay has been developed to detect intestinal protozoa reducing the cost of a lateral flow strip to $1 [26]. Furthermore, using LFDs can effectively save the cost of fluorescent detectors.
Overall, the RT-RPA-LFD assays developed in this study were advantageous for detection and subtyping of IAVs. They are suitable for low-resource settings because they are performed at a relatively low temperature of 39 °C and do not require expensive instruments. Moreover, they are sensitive and specific enough for routine clinical diagnosis and epidemiological investigations. In conclusion, RT-RPA-LFD assays were described here for detection and subtyping of IAVs, which offers relatively rapid, sensitive, and specific assays for diagnosis of IAVs and subtyping of H1 and H3.
Ethical approval
All procedures performed in this study involving human participants were in accordance with the ethical standards of Jinling Hospital of China and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This work is funded by the National Natural Science Foundation of China (No. 81601857), Science and Technology Program of Jiangsu Province (BL2014072), Medical and Health Research Major Project of Nanjing Military Region (14ZX17) and National Key Clinical Program of China (2014ZDZK003).
We acknowledge the Pediatrics Outpatient Department of Jinling Hospital, Nanjing, China, for collecting pharyngeal swab specimens. We also acknowledge Mr. Wang Zhixin for providing help and suggestions on the assembly of lateral flow dipsticks.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mcp.2018.10.004.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Fig. S1.
Fig. S2.
References
- 1.Hause B.M., Mariette D., Collin E.A., Ran Z., Liu R., Sheng Z., Anibal A., Bryan K., Suvobrata C., Hoppe A.D. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza c viruses. PLoS Pathog. 2013;9(2) doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S., Zhu X., Li Y., Shi M., Zhang J., Bourgeois M., Yang H., Chen X., Recuenco S., Gomez J. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10) doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garten R.J., Davis C.T., Russell C.A., Shu B., Lindstrom S., Balish A., Sessions W.M., Xu X., Skepner E., Deyde V. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannoun C. The evolving history of influenza viruses and influenza vaccines. Expert Rev. Vaccines. 2013;12(9):1085–1094. doi: 10.1586/14760584.2013.824709. [DOI] [PubMed] [Google Scholar]
- 5.Krammer F., Palese P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015;14(3):167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 6.Monto A.S. Implications of antiviral resistance of influenza viruses. Clin. Infect. Dis. 2009;48(4):397–399. doi: 10.1086/596312. [DOI] [PubMed] [Google Scholar]
- 7.Carrat F., Vergu E., Ferguson N.M., Lemaitre M., Cauchemez S., Leach S., Valleron A.J. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am. J. Epidemiol. 2008;167(7):775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 8.Uyeki T.M. Influenza diagnosis and treatment in children: a review of studies on clinically useful tests and antiviral treatment for influenza. Pediatr. Infect. Dis. J. 2003;22(22):164–177. doi: 10.1097/01.inf.0000050458.35010.b6. [DOI] [PubMed] [Google Scholar]
- 9.Smith K.J., Roberts M.S. Cost-effectiveness of newer treatment strategies for influenza. Am. J. Med. 2002;113(4):300–307. doi: 10.1016/s0002-9343(02)01222-6. [DOI] [PubMed] [Google Scholar]
- 10.Uyeki T.M., Prasad R., Vukotich C., Stebbins S., Rinaldo C.R., Ferng Y.H., Morse S.S., Larson E.L., Aiello A.E., Davis B. Low sensitivity of rapid diagnostic test for influenza. Clin. Infect. Dis. 2009;48(9):e89. doi: 10.1086/597828. [DOI] [PubMed] [Google Scholar]
- 11.Rothberg M.B., Fisher D., Kelly B., Rose D.N. Management of influenza symptoms in healthy children: cost-effectiveness of rapid testing and antiviral therapy. J. Gen. Intern. Med. 2010;18(10):808–815. [Google Scholar]
- 12.Fouchier R., Bestebroer T., Herfst S., Van der Kemp L., Rimmelzwaan G., Osterhaus A. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 2000;38(11):4096–4101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma X., Xu H., Shi L., Yang P., Zhang L., Sun X., Zhen W., Hu K. A multiplex PCR assay for the detection of five influenza viruses using a dual priming oligonucleotide system. BMC Infect. Dis. 2015;15:93. doi: 10.1186/s12879-015-0818-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payungporn S., Phakdeewirot P., Chutinimitkul S., Theamboonlers A., Keawcharoen J., Oraveerakul K., Amonsin A., Poovorawan Y. Single-step multiplex reverse transcription-polymerase chain reaction (RT-PCR) for influenza A virus subtype H5N1 detection. Viral Immunol. 2004;17(4):588–593. doi: 10.1089/vim.2004.17.588. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W., Evans D.H. Detection and identification of human influenza viruses by the polymerase chain reaction. J. Virol. Methods. 1991;33(1–2):165–189. doi: 10.1016/0166-0934(91)90017-t. [DOI] [PubMed] [Google Scholar]
- 16.Harmon K., Bower L., Kim W.-I., Pentella M., Yoon K.-J. A matrix gene-based multiplex real-time RT-PCR for detection and differentiation of 2009 pandemic H1N1 and other influenza A viruses in North America. Influenza Other Resp. 2010;4(6):405–410. doi: 10.1111/j.1750-2659.2010.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweiger B., Zadow I., Heckler R., Timm H., Pauli G. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J. Clin. Microbiol. 2000;38(4):1552–1558. doi: 10.1128/jcm.38.4.1552-1558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung J.H., Park B.H., Oh S.J., Choi G., Seo T.S. Integrated centrifugal reverse transcriptase loop-mediated isothermal amplification microdevice for influenza A virus detection. Biosens. Bioelectron. 2015;68:218–224. doi: 10.1016/j.bios.2014.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y., Yu X., Chen H., Diao Y. An immunoassay-based reverse-transcription loop-mediated isothermal amplification assay for the rapid detection of avian influenza H5N1 virus viremia. Biosens. Bioelectron. 2016;86:255–261. doi: 10.1016/j.bios.2016.06.063. [DOI] [PubMed] [Google Scholar]
- 20.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4(7):e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyle D.S., Lehman D.A., Lillis L. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio. 2013;4(2) doi: 10.1128/mBio.00135-13. e00135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teoh B.T., Sam S.S., Tan K.K., Danlami M.B., Shu M.H., Johari J., Hooi P.S., Brooks D., Piepenburg O., Nentwich O., Wilder-Smith A., Franco L., Tenorio A., Abu Bakar S. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification. J. Clin. Microbiol. 2015;53(3):830–837. doi: 10.1128/JCM.02648-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren H., Yang M., Zhang G., Liu S., Wang X., Ke Y., Du X., Wang Z., Huang L., Liu C., Chen Z. Development of a rapid recombinase polymerase amplification assay for detection of Brucella in blood samples. Mol. Cell. Probes. 2016;30(2):122–124. doi: 10.1016/j.mcp.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Wang W., Hu Y.A., Sun N., Yang B., Xia Z., Li X. Preliminary application of Tem-PCR combined with luminex for detection of four common respiratory viruses. J. Med. Postgra. 2016;29(9):958–963. [Google Scholar]
- 25.World Health Organization WHO information for molecular diagnosis of influenza virus - update. 2017. http://www.who.int/influenza/gisrs_laboratory/molecular_diagnosis/en/ 1-60.
- 26.Crannell Z., Castellanos-Gonzalez A., Nair G., Mejia R., White A.C., Richards-Kortum R. Multiplexed recombinase polymerase amplification assay to detect intestinal protozoa. Anal. Chem. 2016;88(3):1610–1616. doi: 10.1021/acs.analchem.5b03267. [DOI] [PubMed] [Google Scholar]
- 27.Toubanaki D.K., Christopoulos T.K., Ioannou P.C., Flordellis C.S. Identification of single-nucleotide polymorphisms by the oligonucleotide ligation reaction: a DNA biosensor for simultaneous visual detection of both alleles. Anal. Chem. 2009;81(1):218–224. doi: 10.1021/ac801870x. [DOI] [PubMed] [Google Scholar]
- 28.Nelson M.I., Simonsen L., Viboud C., Miller M.A., Holmes E.C. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 2007;3(9):e131. doi: 10.1371/journal.ppat.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garten R., Blanton L., Abd A.I., Alabi N., Barnes J., Biggerstaff M., Brammer L., Budd A.P., Burns E., Cummings C.N., Davis T. Update: influenza activity in the United States during the 2017-18 season and composition of the 2018-19 influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 2018;67:634–642. doi: 10.15585/mmwr.mm6722a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsushima Y., Uno N., Sasaki D., Morinaga Y., Hasegawa H., Yanagihara K. Quantitative RT-PCR evaluation of a rapid influenza antigen test for efficient diagnosis of influenza virus infection. J. Virol. Methods. 2015;212:76–79. doi: 10.1016/j.jviromet.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Branche A.R., Walsh E.E., Formica M.A., Falsey A.R. Detection of respiratory viruses in sputum from adults by use of automated multiplex PCR. J. Clin. Microbiol. 2014;52(10):3590–3596. doi: 10.1128/JCM.01523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward C. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J. Clin. Virol. 2004;29(3):179–188. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Liu D., Deng J., Wang Y., Xu J., Ye C. Loop-mediated isothermal amplification using self-avoiding molecular recognition systems and antarctic thermal sensitive uracil-DNA-glycosylase for detection of nucleic acid with prevention of carryover contamination. Anal. Chim. Acta. 2017;996:74–87. doi: 10.1016/j.aca.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 34.Chao C.C., Belinskaya T., Zhang Z., Ching W.M. Development of recombinase polymerase amplification assays for detection of Orientia tsutsugamushi or Rickettsia typhi. PLoS Neglect. Trop. D. 2015;9(7):1–21. doi: 10.1371/journal.pntd.0003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kappagantu M., Villamor D.E.V., Bullock J.M., Eastwell K.C. A rapid isothermal assay for the detection of Hop stunt viroid in hop plants (Humulus lupulus), and its application in disease surveys. J. Virol. Methods. 2017;245:81–85. doi: 10.1016/j.jviromet.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Karakkat B.B., Hockemeyer K., Franchett M., Olson M., Mullenberg C., Koch P.L. Detection of root-infecting fungi on cool-season turfgrasses using loop-mediated isothermal amplification and recombinase polymerase amplification. J. Microbiol. Methods. 2018;151:90–98. doi: 10.1016/j.mimet.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Liu Y., Wei Q., Liu Y., Liu W., Zhang X., Yu Y. Picoliter well array chip-based digital recombinase polymerase amplification for absolute quantification of nucleic acids. PloS One. 2016;11(4) doi: 10.1371/journal.pone.0153359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.