Abstract
Human enterovirus 71 (EV71) is the primary pathogen of hand, foot, and mouth disease (HFMD). EV71 infection may lead to neurologic damage, with higher incidence of fatality compared with other HFMD pathogens. An effective drug or vaccine against EV71 infection is currently unavailable. It is desirable to determine the pathogen of HFMD accurately and quickly for early treatment. In the current study, reverse-transcription and loop-mediated isothermal amplification (RT-LAMP) technology were developed to detect EV71. The efficacy of detecting EV71 was compared with regular nested reverse-transcription polymerase chain reaction (RT-PCR). After detecting 108 clinical specimens, results showed that RT-LAMP can specifically detect EV71, but not Coxsackie virus A16, and exhibited a specificity of 100% and a sensitivity of 97.1%, which was higher than regular RT-PCR. The findings indicate that RT-LAMP is a practical method for EV71 diagnostic applications, particularly in small county institutes of medical service. The detection ability of RT-LAMP was significantly affected by cryopreservation as the clinical specimens were repeatedly subject to freezing and thawing treatments.
Keywords: EV71, Reverse-transcription and loop-mediated isothermal amplification, Cryopreservation
1. Introduction
Enterovirus 71 (EV71), first isolated in 1969 in California, is a major public health issue across the Asia Pacific region, Europe, and other continents. The virus mostly affects young children and causes hand, foot, and mouth disease (HFMD), which is generally a mild illness characterized by blisters and ulcers (Cardosa et al., 2003, McMinn, 2002, Solomon et al., 2010, Wu et al., 2010). However, unlike other HFMD pathogens, such as the Coxsackie virus and echovirus, EV71 infection could result in severe neurologic complications, including aseptic meningitis, encephalitis, acute flaccid paralysis, myocarditis, pulmonary edema, and hemorrhage, with high incidence of fatality (Chang et al., 2007, Huang et al., 1999). EV71 was recently reported as the primary pathogen of HFMD among young children in China (Yang et al., 2009, Zhang et al., 2009), and over 1.77 million HFMD cases were reported in 2010, 905 of which were fatal. This number is a 53.6% increase from that reported in 2009, during which over 1.15 million cases were reported and 353 cases were fatal (Ministry of Health, PR China).
EV71 is a small, single-stranded, positive-sense RNA virus, belonging to the family of Picornaviridae. The EV71 genome, which is approximately 7.4 kb long, encodes a long polyprotein with a single open reading frame followed by a poly A tail. The single polyprotein is flanked by untranslated regions at both the 5′ and 3′ ends, and can be divided into 3 different genomic regions. The P1 genomic region encodes for structural proteins (VP1 to VP4), while the P2 and P3 genomic regions encode for nonstructural proteins 2A, 2B, 2C, 3A, 3B, 3C, and 3D (Cardosa et al., 2003, Solomon et al., 2010, Wu et al., 2010). Research on EV71 drugs and vaccines is ongoing and some progress has been achieved (reviewed by Lee and Chang, 2010, Wu et al., 2010), and the potential receptors for EV71 infection were also identified (Nishimura et al., 2009, Yamayoshi et al., 2009). However, an effective drug or vaccine against EV71 infection is currently unavailable. Thus, determination of the pathogen of HFMD for an early treatment is desirable.
Recently, a novel method of DNA amplification, known as loop-mediated isothermal amplification (LAMP) of a target nucleic acid, was reported by Notomi et al. (2000). LAMP is a 1-step amplification reaction that amplifies a target DNA sequence with high sensitivity and specificity under isothermal conditions by a DNA polymerase (Bst polymerase) and a set of primers. The addition of reverse transcriptase makes it possible to amplify DNA from RNA sequences (RT-LAMP). This method has already been applied to the detection of several RNA viruses, such as Norovirus (Iturriza-Gomara et al., 2008), avian influenza virus H5N1 (Imai et al., 2006, Imai et al., 2007), and severe acute respiratory syndrome coronavirus (Poon et al., 2005). Here we sought to determine whether or not RT-LAMP could be applied to EV71 amplification and determine its diagnostic performance in clinical specimens.
2. Material and methods
2.1. Viral RNA preparation and amplified by regular RT-PCR
Clinical stool specimens were collected from patients identified with HFMD and registered in Children's Hospital of Fudan University, Shanghai. A total of 0.2 g of stool from each patient was thoroughly suspended in 1.0 mL PBS buffer, after which viral RNA was extracted with Trizol (Invitrogen, USA) following the manufacturer's instructions. Five microliters of extracted RNA (a total of 20 μL) was reverse transcribed (RT) with M-MLV reverse transcriptase (Takara, Japan) in a volume of 20 μL consisting of 50 ng hexamer, RNAguard (Pharmacia, USA) at 400 U/mL, 200 U of M-MLV, 1 μL of 0.1 M DTT, and 2 μL of 10 mmol/L dNTP mixture. The RT reaction was incubated at 37 °C for 1 h and inactivated at 75 °C for 10 min. The RT products were then amplified by polymerase chain reaction (PCR). The PCR mixture, in a volume of 25 μL, consisted of 5 μL of RT products, 2.5 μL of 10 × PCR buffer, 2.5 μL of 25 mmol/L MgCl2, 2.5 μL of 2.5 mmol/L dNTP mixture, 1 μL of sense primer (10 μM), 1 μL of antisense primer (10 μM), and 0.25 μL of DNA Taq polymerase (5 U/μL). PCR was performed via a touch-down program: 94 °C for 4 min, after 10 cycles of 94 °C for 45 s, 60 °C for 45 s (decreasing at a rate of 0.5 °C/cycle for annealing in the next 9 cycles), 72 °C for 45 s, and then continued for another 20 cycles at 94 °C for 45 s, 55 °C for 45 s, 72 °C for 45 s, and, finally, at 72 °C extended for 7 min on a PCR machine (Eppendorf Mastercyler personnel 5333). Nested PCR was continued with 1 μL of the previous PCR products by inner sense and antisense primers. The amplified products were detected by running with 2% agarose gel and staining with 0.5% ethidium bromide (EB).
2.2. Viral RNA amplified by RT-LAMP
Five microliters of extracted RNA (the same amount for regular RT-PCR) was taken for RT-LAMP amplification in a 25-μL mixture consisting of Bst DNA polymerase (New England Biolab) 8 U, 10× thermal buffer 1.0 μL, AMV reverse transcriptase (Fermentas) 5 U, 5× AMV first-strand buffer 4.0 μL, 25 μM betaine (Sigma, USA) 1.0 μL, 2.5 mmol/L dNTP mixtures 5 μL, 0.1% Triton X-100 0.25 μL, and a primer mixture, including 0.2 μM F3 and B3, and 1.6 μM FIP and BIP. RT-LAMP primer sets were designed using the Primer Explorer V3.0 online software (http://primerexplorer.jp/v3_manual/index.html) and manually selected according to the principle for RT-LAMP (Table 1 and Fig. 1 ). After 60 °C for 1 h, the polymerases in the reaction mixture were inactivated at 80 °C for 5 min, and then 5 μL of RT-LAMP products was detected by running with 2% agarose gel and staining with EB.
Table 1.
Primers for RT-LAMP and regular nested RT-PCR amplification of EV71
| Detection methods | Primer name | Location (FJ713317) | Direction | Sequences |
|---|---|---|---|---|
| RT-LAMP | EV71-F3 | 2577–2597 | External sense | 5-act cca agc tgc tga aat tgg-3 |
| EV71-B3 | 2778–2797 | External antisense | 5-gca ttt gtg cgt aac ctg tt-3 | |
| EV71-FIP | 2650–2669/2610–2629 | Inner sense | 5-tca gct gtg ctg tgc gag ttt gct agt gac gag agc atg a-3 | |
| EV71-BIP | 2687–2710/2743–2763 | Inner antisense | 5-tct tca gca gag cgg gat tag ttg gtt ggc ata acc att tgg gtt-3 | |
| Nested RT-PCR | EV71-F | 2758–2781 | Sense | 5-gcc aac tgg gac ata gat ata ac-3 |
| EV71-R1 | 3312–3333 | External antisense | 5-ccc aag agt agt gat cgc tgt-3 | |
| EV71-R2 | 3137–3155 | Inner antisense | 5-ccc aca gtc cgc act gag-3 |
Fig. 1.
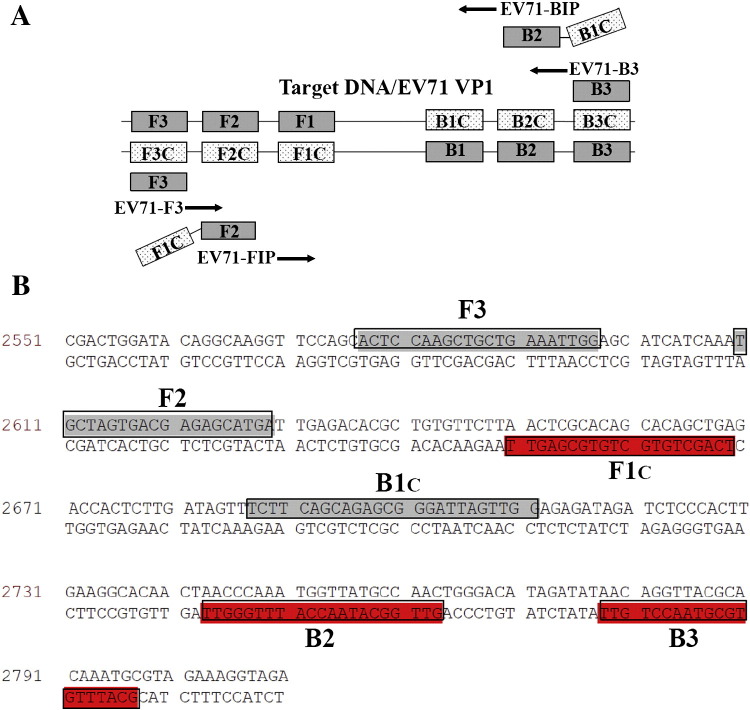
Oligonucleotide primers used for RT-LAMP assay of EV71 within the VP1 gene. (A) A schematic diagram showing the positions to which the primers attached for the target gene. The RT-LAMP primer sets were designed online by the Primer Explorer V3.0 software. The positions of the essential primers (F3, B3, FIP, and BIP) are shown here. The specially designed FIP and BIP inner primers contain the 2 distinct sequences F1c + F2 and B1c + B2, which respectively correspond to the sense and antisense segments of the target DNA. (B) The partial DNA sequence of the VP1 gene of EV71 (accession no. FJ713137) shows the target region for the LAMP assay. The inner and outer RT-LAMP primers designed from this region are indicated by closed boxes.
2.3. RT-LAMP products identified by Southern blot hybridization and sequencing
To confirm the facticity after amplification by RT-LAMP, 10 μL of RT-LAMP products was run with 1% agarose gel and then transferred onto a nylon membrane (Pharmacia Hybond N+) for Southern blot detection. The probe was prepared by a random primer-labeling method with digoxin using the EV71 VP1 gene as a DNA template, according to the manufacturer's instructions (Roche, SWI). The labeled probes were purified through a mini Quick Spin DNA Column (Roche). Hybridization was carried out in a volume of 10 mL hybridization solution (5× Denhart's, 6× SSC, 0.5% SDS, 100 μg/mL salmon sperm DNA) at 68 °C for 16 h, via a routine procedure as previously described (Long et al., 2005). Finally, the membrane was color developed by the NBT/BCIP system (Roche). The RT-LAMP amplification products were also purified after running with agarose gel, and 3 significant bands, as shown in Fig. 2A, were cloned into a pMT-18 (Takara) vector for sequencing.
Fig. 2.
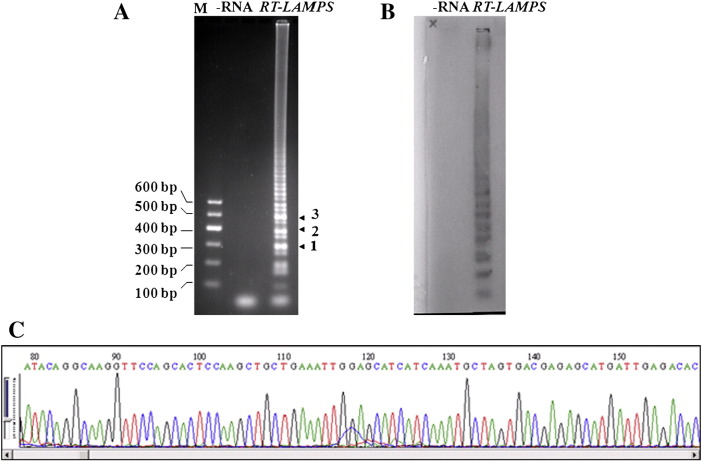
RT-LAMP amplification of EV71 RNA, confirmed by Southern blot and sequencing analysis. (A) RT-LAMP reaction solution detected by running with 2% agarose. M: DNA marker; −RNA: without EV71 RNA; RT-LAMP: complete EV71 RT-LAMP reaction. (B) Southern blot detection of EV71 RT-LAMP products. EV71 VP1 probe labeled by digoxin. (C) Representation of the partial sequencing results of bands 1–3 in (A).
2.4. Application of RT-LAMP to detect clinical specimens and comparison with regular RT-PCR
A total of 108 clinical stool specimens, collected from 100 EV71-infected patients, 4 Coxsackie virus A 16 (CA16)–infected patients, and 4 EV71 and CA16–double-infected patients (the pathogens of HFMD were identified by Shanghai Children's Hospital of Fudan University and Shanghai Center of Disease Control by real-time RT-PCR and virus isolation), were used to evaluate the RT-LAMP assay. RT-LAMP detection was carried out according to the above description, whereas RT-PCR was performed by half nesting inner primers with 1 μL of the first-round PCR products, which were initiated from the same amount of viral RNA for RT-LAMP after reverse transcription by M-MLV.
2.5. Effect of cryopreservation on the detection ability of RT-LAMP and RT-PCR
It is sometimes necessary to cryopreserve the clinical specimens, such as for research studies or clinical sample transfer. To determine the effect of cryopreservation on RT-LAMP, clinical stool specimens collected from HFMD patients in Shanghai Children's Hospital of Fudan University from March 2009 to December 2009 were classified into 3 groups, all of which underwent different treatments by freezing at −70 °C and thawing at room temperature 0−2, 3−5, or over 5 times before use. The specimens were then detected by RT-LAMP and RT-PCR as described above.
3. Results
3.1. RT-LAMP for EV71 detection
After the alignment of EV71 strain sequences performed by VECTOR NTI 8.0 based on the sequences of the VP1 gene, the primer set for EV71 amplification by RT-LAMP was designed online by the Primer Explorer V3.0 software and manually selected (Fig. 1). The extracted RNA was amplified by RT-LAMP, and the reaction solution was run with agarose gel. The products amplified by the RT-LAMP assay exhibited a typical ladder-like pattern on electrophoresis, indicating the formation of stem-loop DNA with inverted repeats (Fig. 2A). To confirm the amplification by RT-LAMP, the amplified products were detected by Southern blot with VP1 gene as the probe and the results showed a signal pattern similar to that obtained from electrophoresis (Fig. 2B). Three significant bands (marked in Fig. 2A) were purified and cloned into a T vector for sequencing; the results also showed that the sequences are conclusive for the VP1 gene (Fig. 2C).
3.2. End-point detection of RT-LAMP products by single staining
The RT-LAMP reaction produces insoluble magnesium pyrophosphate in the course of the amplification reaction. It has been reported that the turbidity analysis of RT-LAMP makes it possible to determine the results by visual end-point judgment of turbidity (Mori, et al., 2004, Saitou et al., 2010). Consequently, after amplification by RT-LAMP, the products were observed with the naked eye or by single staining with EB and then exposed to white light and ultraviolet (UV) light to detect amounts of amplified DNA. Results showed that, after amplification by RT-LAMP, the EV71-positive reaction solution gave a light milky disturbance, whereas the EV71-negative sample was clearly transparent as observed by the naked eye (Fig. 3A). After staining with EB, the EV71-positive reaction showed bright whiteness under UV light, whereas the EV71-negative sample showed only a slight whiteness on the tube wall and a grey darkness at the center of the liquid (Fig. 3B).
Fig. 3.
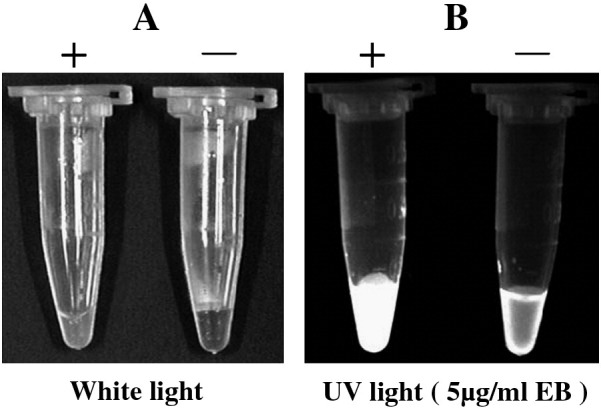
Detection of RT-LAMP products after amplification. (A) After RT-LAMP reaction, the EV71-positive sample showed turbidity (left), whereas the EV71-negative sample showed a clear solution, as determined by naked-eye observation. (B) After staining by EB (5 μg/mL), the EV71-positive sample became bright white (left), whereas the negative sample showed a dark liquid center under UV light.
3.3. Primary analysis of the sensitivity and specificity of RT-LAMP for detecting EV71
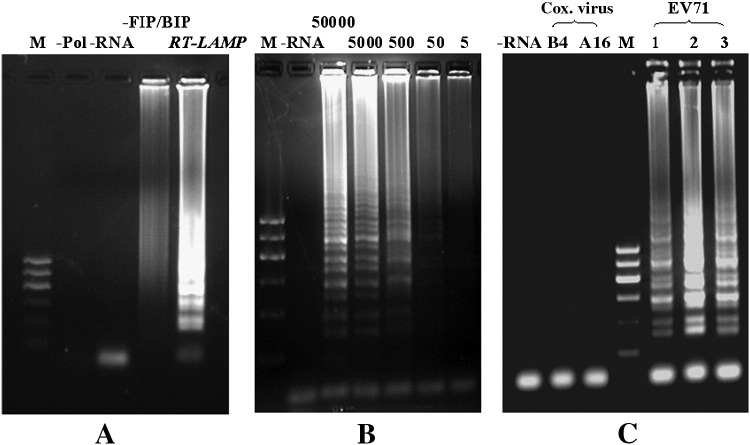
The RT-LAMP for amplification of EV71 RNA was significantly dependent on the inner primer pair; results showed that, without FIP/BIP, the efficiency of RT-LAMP amplification was very low. Without Bst DNA polymerase or RNA templates, RT-LAMP was unable to amplify RNA (Fig. 4A). To test the sensitivity of the detection limit of RT-LAMP for EV71, RNA genome copies from 5 to 50,000 were added for amplification. Results showed that the RT-LAMP amplification signal could be detected by electrophoresis when the number of RNA copies was equal to 5 (Fig. 4B). To confirm the specificity of amplification, RT-LAMP was used to determine the templates from phylogenic relative Coxsackie virus A16 and B4 RNA with initial copies at 105. The results revealed that only EV71 RNA can be amplified by RT-LAMP (Fig. 4C).
Fig. 4.
Sensitivity and specificity of RT-LAMP amplification of EV71. (A) RT-LAMP amplification of EV71 RNA. M: DNA marker; −Pol: without Bst DNA polymerase; −RNA: without RNA; −FIP/BIP: without inner pair primers of FIP and BIP; RT-LAMP: complete RT-LAMP. (B) The sensitivity of RT-LAMP amplification of EV71 RNA. The numerical value (5–50,000) over the electrophoresis land indicates the start copies of EV71 RNA. (C) The specificity of RT-LAMP amplification of EV71. Lanes marked with 1, 2, and 3 indicate the EV71 samples, Cox virus, and Coxsackie virus, respectively.
3.4. Evaluation of RT-LAMP for EV71 detection and a comparison with regular RT-PCR
To evaluate the assay of RT-LAMP for EV71 detection, a total of 108 clinical specimens, including 100 from patients infected with EV71, 4 from infected with CA16, and 4 from double infected with EV71 and CA16, were detected by RT-LAMP and RT-PCR (Fig. 5 ). Of the 104 EV71-infected samples (100 of EV71 samples and 4 of EV71 and CA16 double-infected samples), 101 were found to be positive for EV71 and 3 were negative by the RT-LAMP assay. Of 4 CA16-infected samples, none was found to be EV71 positive by RT-LAMP and RT-PCR (Table 2 ). Results revealed that the RT-LAMP for EV71 detection had a sensitivity of 97.1% (101/104) and a specificity of 100% (101/101), whereas RT-PCR yielded only a sensitivity of 83.7% (87/104) and a specificity of 100% (87/87). It indicated that RT-LAMP for EV71 detection was more sensitive than RT-PCR.
Fig. 5.
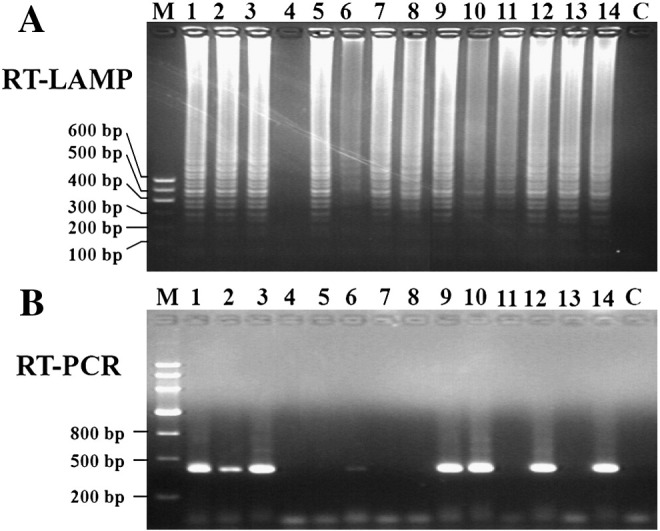
Representation of partial clinical samples from HFMD patients amplified by (A) RT-LAMP and (B) regular nested RT-PCR. M: DNA marker; 1–14: clinical samples, C: negative control.
Table 2.
RT-LAMP for detecting clinical specimens from HFMD patients and a comparison with RT-PCR
| Clinical patients identified with HFMD | Total number of detected specimens | Detection methods |
|||
|---|---|---|---|---|---|
| RT-LAMPa |
RT-PCR |
||||
| Positive | Negative | Positive | Negative | ||
| EV71 | 100 | 97 | 3 | 83 | 17 |
| EV71/CA16 double infected | 4 | 4 | 0 | 4 | 0 |
| CA16 | 4 | 0 | 4 | 0 | 4 |
In comparison with the RT-PCR assay, the sensitivity of RT-LAMP was 97.1% (101/104), whereas RT-PCR exhibited a sensitivity of 83.7% (87/104); both methods had a specificity of 100%.
3.5. Effect of cryopreservation on the detection ability of RT-LAMP and RT-PCR
In order to determine the effect of cryopreservation on the assay by RT-LAMP for EV71 detection, clinical stool specimens collected from HFMD patients from March 2009 to December 2009 were classified into 3 groups according to the number of times the samples were frozen and thawed, and then detected by RT-LAMP and RT-PCR (Table 3 ). The positive ratios for EV71 detected by RT-LAMP were 97.1% in group 1, 90% in group 2, and 58% in group 3, indicating that the number of times the specimens were frozen and thawed affected EV71 delectability. The same findings were also observed for RT-PCR when it was applied to detect EV71 in frozen and thawed specimens, with results showing positive ratios of 83.7% in group 1, 65% in group 2, and 54% in group 3. The results indicated that cryopreservation by freezing and thawing significantly affects the detection ability of RT-LAMP assay and with more severe impacts on the RT-PCR assay for EV71 detection.
Table 3.
Effect of cryopreservation on the detection ability of RT-LAMP and RT-PCR assay
| Specimen groups | Samples collected duration (number of freezing and thawing times)a | Detection methods |
|||
|---|---|---|---|---|---|
| RT-LAMP |
RT-PCR |
||||
| Positive | Negative | Positive | Negative | ||
| 1 | 2009.8–2009.10 (0–2) | 97.1% (101/104)b | 2.9% (3/104) | 83.7% (87/104) | 16.3% (17/104) |
| 2 | 2009.6–2009.7 (3–5) | 90% (36/40) | 10% (4/40) | 65% (26/40) | 35% (14/40) |
| 3 | 2009.3–2009.5 (>5) | 58% (29/50)b | 42% (21/50) | 54% (27/50) | 46% (23/50) |
Freezing and thawing mean that the samples after being collected from patients were stored at −70 °C and thawed to room temperature before use.
Number of positive or negative specimens/total detected specimens in this group.
4. Discussion
EV71 has reemerged in recent years as the main causative agent of HFMD outbreaks in China. A total of 488,955 HFMD cases were reported in 2008, over 1.15 million cases in 2009, and 1.77 million cases in 2010 (Ministry of Health, PR China). The significant increase in HFMD cases makes public prevention and therapy a critical issue. At present, no antiviral or vaccine is available for the disease, and most cases of HFMD are managed with symptomatic treatment. It has been reported that most of the deaths from HFMD caused by EV71 infection occurred within 24 h, when the children admitted exhibited neurologic complications associated with HFMD (Ho et al., 1999, Wang et al., 2006). A rapid and specific diagnosis method for EV71 from clinical specimens will enable clinicians to identify those who require closer monitoring or even hospitalization before deterioration of clinical conditions.
Classical clinical diagnosis of EV71 infection, such as virus isolation and neutralization assay, takes several days to complete, whose major drawback is the delay of treatment in patients at infirmaries. Methods based on RT-PCR are heavily dependent on the skill of the manipulator and cost machinery (Fujimoto et al., 2008, Singh et al., 2002, Tan et al., 2008). It is hard to be accepted by those small and condition-limited infirmaries in a developing country. An EV71 diagnostic kit based on viral specific IgM and IgG antibodies was recently approved by the Food and Drug Administration of China (2010, no. 3400411), thus providing significant benefits to the early diagnosis and epidemiologic survey of EV71 infection. It was reported that EV71 IgM antibodies could be detected 1–2 days after identification of HFMD, and the sensitivity of detection was up to 88–90.7%. However, up to 12.4% false-positive results were reported because of the cross-reactivity of EV71 IgM with other enteroviruses (Xu et al., 2010). Thus, the establishment of more rapid, convenient, sensitive, and accurate diagnostic method is particularly desirable.
In the current study, we developed a new method for EV71 detection based on RT-LAMP. LAMP is a novel method of DNA amplification that is dependent on the activity of DNA strand displacement, with the help of Bst polymerase and 4 highly specific primers targeting 6 distinct regions (Mori, and Notomi, 2009, Notomi et al., 2000). An advantage of LAMP-based diagnosis over regular RT-PCR is specificity because the LAMP reaction does not proceed without the hybridization of 6 distinct sequences in the target DNA by 4 highly specific primers. The ability of the method to amplify from fewer copies of initial target DNA than required in PCR was conclusively demonstrated (Gunimaladevi et al., 2005, Kono et al., 2004, Parida et al., 2004). The efficiency of LAMP does not seem to be affected by the presence of nontarget genomic DNA in the reaction mixture (Notomi et al., 2000), which is highly desirable for the development of a diagnostic system. Detection of target DNA by LAMP is significantly more convenient than detection by 2-step nested PCR because the former amplifies DNA within only 1 step at isothermal conditions. Real-time turbidity measurements and end-point visual judgment make LAMP very practical for the diagnosis of pathogens. Thus, many reports on LAMP or RT-LAMP for pathogen detection have been published (Cardoso et al., 2010, Imai et al., 2006, Le Roux et al., 2009, Mori et al., 2006, Parida et al., 2004, Yoshida et al., 2007).
In the current research, we established a method of RT-LAMP for EV71 detection. The VP1 structure protein is a serotype-dependent protein and capable of self-association and dimerization to participate in viral capsid formation; it is also a major immune determinant. Thus, a conserved part of the VP1 gene (2577–2797) was selected as the RT-LAMP target (Fig. 1) and the RT-LAMP primer set (Table 1) was designed using online software. The results showed that the primer set can be applied to specifically amplify EV71 RNA (Fig. 2), but not other phylogenetically related enteroviruses, such as Coxsackie virus A16 and B4 (Fig. 4C). Initial copies for RT-LAMP amplification of EV71 RNA were lowered to 5–50, and the amplification could be detected by agarose electrophoresis (Fig. 4B). The sensitivity of RT-LAMP for EV71 detection was significantly higher than that of regular 2-step RT-PCR in current research with positive results of up to 97.1% for RT-LAMP and 83.7% for RT-PCR (Table 2). When this article was in preparation, a similar RT-LAMP method for EV71 detection was reported and the sensitivity for EV71 detection was 92.9% by evaluation of 40 clinical samples by targeting the VP3 gene of EV71 (Jiang et al., 2011). It was noticeable that clinical specimens identified with HFMD were generally reported to exhibit EV71 infection with 40–90%, which was dependent on the origin of the collected specimens (Singh et al., 2002, Zhang et al., 2009).
RT-LAMP results can be determined by a single staining procedure, such as EB staining under UV light, or even by the naked eye (Fig. 3). Some staining chemicals, such as calcein and hydroxynaphthol blue dye, could be incubated with LAMP products, which then develop into a significant color (Cardoso et al., 2010, Tomita et al., 2008). The simplicity and clarity of the proposed method make it applicable for use even in resource-limited laboratories in rural areas of developing countries. Real-time turbidity measurements obtained by a cost-effective photometer with an incubation function enable kinetic analysis of the LAMP reaction without the need for any detection reagents, such as fluorescence, and allow quantification of the initial amount of template DNA in the samples (Mori, et al., 2001, Mori, et al., 2004, Saitou et al., 2010).
Where necessary laboratory equipment and time consumption are concerned, RT-LAMP for EV71 detection requires only an isothermal incubator, and the reaction takes place for only 1 h. RT-PCR requires a thermal cycler and 2-round PCR programs that, except for reverse transcription, could take anywhere from 5 to 8 h. Consequently, RT-LAMP is more convenient and easier to manipulate than RT-PCR.
It was noticeable that the positive ratio significantly decreased as the specimens were repeatedly subject to freezing and thawing treatments, and then detected by RT-LAMP or RT-PCR (Table 3); this result has been observed also in recent reports (Botling et al., 2009, Forster et al., 2008). Consequently, the clinical specimens should avoid unnecessary freezing and thawing, although the sensitivity provided by the RT-LAMP assay was very high, even higher than that of the RT-PCR assay.
In conclusion, RT-LAMP is a promising and practical method for EV71 detection. Its high sensitivity and specificity make it applicable for use even in resource-limited laboratories.
Footnotes
This work was supported by the National Science and Technology Major Project on Infectious Diseases (2009ZX10004-104, 2008ZX10002-011, and 2009ZX10004-505), the Shanghai Science and Technology Fund (No. 09411964500), and Fudan University Fund for Clinical and Basic Medical Science (2008).
Contributor Information
Hui Yu, Email: yuhui20@yahoo.com.
Jian-Er Long, Email: longjianer@fudan.edu.cn.
References
- Botling J., Edlund K., Segersten U., Tahmasebpoor S., Engström M., Sundström M., Malmström P.U., Micke P. Impact of thawing on RNA integrity and gene expression analysis in fresh frozen tissue. Diagn Mol Pathol. 2009;18:44–52. doi: 10.1097/PDM.0b013e3181857e92. [DOI] [PubMed] [Google Scholar]
- Cardosa M.J., Perera D., Brown B.A., Cheon D., Chan H.M., Chan K.P., Cho H., McMinn P. Molecular epidemiology of human enterovirus 71 strains and recent outbreaks in the Asia-Pacific region: comparative analysis of the VP1 and VP4 genes. Emerg Infect Dis. 2003;9:461–468. doi: 10.3201/eid0904.020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso T.C., Ferrari H.F., Bregano L.C., Silva-Frade C., Rosa A.C., Andrade A.L. Visual detection of turkey coronavirus RNA in tissues and feces by reverse-transcription loop-mediated isothermal amplification (RT-LAMP) with hydroxynaphthol blue dye. Mol Cell Probes. 2010;24:415–417. doi: 10.1016/j.mcp.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.Y., Huang L.M., Gau S.S., Wu Y.Y., Hsia S.H., Fan T.Y., Lin K.L., Huang Y.C., Lu C.Y., Lin T.Y. Neurodevelopment and cognition in children after enterovirus 71 infection. N Engl J Med. 2007;356:1226–1234. doi: 10.1056/NEJMoa065954. [DOI] [PubMed] [Google Scholar]
- Forster J.L., Harkin V.B., Graham D.A., McCullough S.J. The effect of sample type, temperature and RNA later on the stability of avian influenza virus RNA. J Virol Methods. 2008;149:190–194. doi: 10.1016/j.jviromet.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Fujimoto T., Yoshida S., Munemura T., Taniguchi K., Shinohara M., Nishio O., Chikahira M., Okabe N. Detection and quantification of enterovirus 71 genome from cerebrospinal fluid of an encephalitis patient by PCR applications. Jpn J Infect Dis. 2008;61:497–499. [PubMed] [Google Scholar]
- Gunimaladevi I., Kono T., Lapatra S.E., Sakai M. A loop mediated isothermal amplification (LAMP) method for detection of infectious hematopoietic necrosis virus (IHNV) in rainbow trout (Oncorhynchus mykiss) Arch Virol. 2005;150:899–909. doi: 10.1007/s00705-004-0468-7. [DOI] [PubMed] [Google Scholar]
- Ho M., Chen E.R., Hsu K.H., Twu S.J., Chen K.T., Tsai S.F., Wang J.R., Shih S.R. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med. 1999;341:929–935. doi: 10.1056/NEJM199909233411301. [DOI] [PubMed] [Google Scholar]
- Huang C.C., Liu C.C., Chang Y.C., Chen C.Y., Wang S.T., Yeh T.F. Neurologic complications in children with enterovirus 71 infection. N Engl J Med. 1999;341:936–942. doi: 10.1056/NEJM199909233411302. [DOI] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Tashiro M., Odagiri T. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24:6679–6682. doi: 10.1016/j.vaccine.2006.05.046. [DOI] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Van Tu P., Tien N.T., Tashiro M., Odagiri T. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene specific loop-mediated isothermal amplification method. J Virol Methods. 2007;141:173–180. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Iturriza-Gomara M., Xerry J., Gallimore C.I., Dockery C., Gray J. Evaluation of the Loopamp (loop-mediated isothermal amplification) kit for detecting Norovirus RNA in faecal samples. J Clin Virol. 2008;42:389–393. doi: 10.1016/j.jcv.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Jiang T., Liu J., Deng Y.Q., Xu L.J., Li X.F., Han J.F., Cao R.Y., Qin E.D., Qin C.F. Development and evaluation of a reverse transcription-loop-mediated isothermal amplification assay for rapid detection of enterovirus 71. J Clin Microbiol. 2011;49:870–874. doi: 10.1128/JCM.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T., Savan R., Sakai M., Itami T. Detection of white spot syndrome virus in shrimp by loop-mediated isothermal amplification. J Virol Methods. 2004;115:59–65. doi: 10.1016/j.jviromet.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Le Roux C.A., Kubo T., Grobbelaar A.A., van Vuren P.J., Weyer J., Nel L.H., Swanepoel R., Morita K., Paweska J.T. Development and evaluation of a real-time reverse transcription-loop-mediated isothermal amplification assay for rapid detection of Rift Valley fever virus in clinical specimens. J Clin Microbiol. 2009;47:645–651. doi: 10.1128/JCM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S., Chang L.Y. Development of enterovirus 71 vaccines. Expert Rev Vaccines. 2010;9:149–156. doi: 10.1586/erv.09.152. [DOI] [PubMed] [Google Scholar]
- Long J.E., Huang L.N., Qin Z.Q., Wang W.Y., Qu D. IFN-γ increases efficiency of DNA vaccine in protecting ducks against infection. World J Gastroenterol. 2005;11:4967–4973. doi: 10.3748/wjg.v11.i32.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn P.C. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol Rev. 2002;26:91–107. doi: 10.1111/j.1574-6976.2002.tb00601.x. [DOI] [PubMed] [Google Scholar]
- Mori N., Motegi Y., Shimamura Y., Ezaki T., Natsumeda T., Yonekawa T., Ota Y., Notomi T., Nakayama T. Development of a new method for diagnosis of rubella virus infection by reverse transcription-loop-mediated isothermal amplification. J Clin Microbiol. 2006;44:3268–3273. doi: 10.1128/JCM.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Mori, Y., Kitao M., Tomita N., Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004;59:145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Shimojima M., Tano Y., Miyamura T., Wakita T., Shimizu H. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat Med. 2009;15:794–797. doi: 10.1038/nm.1961. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Wong B.W., Chan K.H., Ng S.S., Yuen K.Y., Guan Y., Peiris J.S. Evaluation of real-time reverse transcriptase PCR and real time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J Clin Microbiol. 2005;43:3457–3459. doi: 10.1128/JCM.43.7.3457-3459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou Y., Kobayashi Y., Hirano S., Mochizuki N., Itou T., Ito F.H., Sakai T. A method for simultaneous detection and identification of Brazilian dog- and vampire bat-related rabies virus by reverse transcription loop mediated isothermal amplification assay. J Virol Methods. 2010;168:13–17. doi: 10.1016/j.jviromet.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Singh S., Chow V.T., Phoon M.C., Chan K.P., Poh C.L. Direct detection of enterovirus 71 (EV71) in clinical specimens from a hand, foot, and mouth disease outbreak in Singapore by reverse transcription-PCR with universal enterovirus and EV71-specific primers. J Clin Microbiol. 2002;40:2823–2827. doi: 10.1128/JCM.40.8.2823-2827.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T., Lewthwaite P., Perera D., Cardosa M.J., McMinn P., Ooi M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- Tan E.L., Yong L.L., Quak S.H., Yeo W.C., Chow V.T., Poh C.L. Rapid detection of enterovirus 71 by real-time TaqMan RT-PCR. J Clin Virol. 2008;42:203–206. doi: 10.1016/j.jcv.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Wang J.N., Yao C.T., Yeh C.N., Huang C.C., Wang S.M., Liu C.C., Wu J.M. Critical management in patients with severe enterovirus 71 infection. Pediatr Int. 2006;48:250–256. doi: 10.1111/j.1442-200X.2006.02198.x. [DOI] [PubMed] [Google Scholar]
- Wu K.X., Ng M.M., Chu J.J. Developments towards antiviral therapies against enterovirus 71. Drug Discov Today. 2010;15:1041–1051. doi: 10.1016/j.drudis.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Yan Q., Wang H., Niu J., Li L., Zhu F., He S., Zhang S., Weng Z., Cheng T., Cai Y., He D., Chen Y., Ge S., Yeo A.E., Zhang J., Ng M.H., Xia N. Performance of detecting IgM antibodies against enterovirus 71 for early diagnosis. PLoS One. 2010;5:e11388. doi: 10.1371/journal.pone.0011388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S., Yamashita Y., Li J., Hanagata N., Minowa T., Takemura T., Koike S. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med. 2009;15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- Yang F., Ren L., Xiong Z., Li J., Xiao Y., Zhao R., He Y., Bu G., Zhou S., Wang J., Qi J. Enterovirus 71 outbreak in the People's Republic of China in 2008. J Clin Microbiol. 2009;47:2351–2352. doi: 10.1128/JCM.00563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Fujino M., Ota Y., Notomi T., Nakayama T. Simple differentiation method of mumps Hoshino vaccine strain from wild strains by reverse transcription loop-mediated isothermal amplification (RT-LAMP) Vaccine. 2007;25:1281–1286. doi: 10.1016/j.vaccine.2006.09.093. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tan X.J., Wang H.Y., Yan D.M., Zhu S.L., Wang D.Y., Ji F., Wang X.J., Gao Y.J., Chen L., An H.Q., Li D.X., Wang S.W., Xu A.Q., Wang Z.J., Xu W.B. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J Clin Virol. 2009;44:262–267. doi: 10.1016/j.jcv.2009.02.002. [DOI] [PubMed] [Google Scholar]