Abstract
Programmed ribosomal frameshifting is used by many viruses to regulate the production of structural and enzymatic proteins. Altering the frameshifting efficiencies disrupts the virus life cycle and eliminates or reduces virus production. Ribosomal frameshifting therefore provides a unique target on which antiviral agents can function. This article describes a series of rapid assay strategies that have been developed and used to identify potential antiviral agents that target programmed −1 ribosomal frameshifting.
Keywords: virus, drug, assay, frameshifting, Saccharomyces cerevisiae, translation, ribosome
The ability of ribosomes to maintain the correct translational reading frame is fundamental to the integrity of protein translation and, ultimately, to cell growth and viability. Thus, the protein translational machinery has evolved to ensure that the intrinsic error rate of reading-frame maintenance is extremely low (<5×10−5 per codon[1]). However, a number of cases in which elongating ribosomes are programmed to shift their translational reading frame one base in the 5′ direction (−1 ribosomal frameshifting) have been identified (Table 1 ). Programmed −1 ribosomal frameshifting is most commonly observed in double-stranded RNA (dsRNA) and nonsegmented (+) strand RNA viruses; programmed +1 ribosomal frameshifting, which shifts the ribosome one base in the 3′ direction, has also been characterized in at least two viral systems. A few examples of programmed ribosomal frameshifting are known to occur in bacterial genes, and one example of programmed +1 ribosomal frameshifting has been documented in a eukaryotic gene; there are no reported examples of eukaryotic cellular mRNAs that use programmed −1 ribosomal frameshifting. These different ribosomal frameshift systems have been extensively reviewed elsewhere2, 3, 4, 5, and so this article will focus exclusively on programmed −1 ribosomal frameshifting as a target for antiviral intervention. Specifically, we will discuss assay strategies that have been developed for drug screening and recent work in which peptidyl-transferase inhibitors were found to have antiviral activities by altering the efficiency of viral programmed −1 ribosomal frameshifting[6].
Table 1.
Viruses that are known or suspected to use ribosomal frameshifting
| Animal viruses | |
| Retroviruses | |
| (almost all retroviruses use programmed −1 ribosomal frameshifting) | |
| Lentiviruses (immunodeficiency viruses, IVs) | Human HIV1 and HIV2, simian SIV (including many species-specific viruses), feline IV, bovine IV, Visna virus (sheep), arthritis–encephalitis virus of goats, equine infectious-anaemia virus. |
| T-cell lymphotrophic viruses (xTLVs) | Human HTLV I and II, simian STLVs, bovine LV |
| Avian leukosis viruses | Leukaemia and sarcoma viruses of many birds, e.g. Rous sarcoma virus |
| Type-B retroviruses | Includes mouse-mammary-tumour virus |
| Type-D Retroviruses | Mostly characterized in monkeys and sheep; includes Mason–Pfizer monkey virus and ovine pulmonary adenocarcinoma virus |
| Nidoviruses | (Genera Coronavirus, Torovirus and Arterivirus) |
| Human coronaviruses | 229-E, OC43, etc. Common cold, upper-respiratory-tract infections, pneumonia, gastroenteritis |
| Human toroviruses | Enteric and respiratory diseases |
| Animal coronaviruses | Calf coronavirus |
| Animal toroviruses | Breda virus (calves); bovine respiratory virus, Berne virus (horses), porcine torovirus, feline torovirus |
| Animal arteriviruses | Simian haemorrhagic-fever virus, equine arteritis virus, lelystad virus (porcine reproductive and respiratory syndrome virus); VR2332 virus (pigs), lactate-dehydrogenase-elevating virus (rodents) |
| Paramyxoviruses | −1 ribosomal frameshifting reported in measles |
| Astroviruses | Human astroviruses 1–5; bovine; ovine; porcine; canine; duck |
| Plant viruses | |
| Tetraviruses | |
| Sobemoviruses | e.g. Southern-bean mosaic virus; cocksfoot mottle virus |
| Leuteoviruses | e.g. Barley yellowdwarf virus; beet western yellows virus; potato leaf roll virus |
| Enamoviruses | e.g. Pea enation mosaic virus |
| Umbraviruses | e.g. Carrot mottle virus |
| Tombusviruses | |
| Tombusvirus | Tomato bushy stunt virus |
| Carmovirus | Carnation mottle virus |
| Necrovirus | Tobacco necrosis virus |
| Dianthoviruses | Red-clover necrotic mosaic virus |
| Machlomovirus | Maize chlorotic mottle virus |
| Totiviruses | L-A and L-BC (yeast); related viruses of other fungi? |
| Giardia lamblia virus (intestinal parasite) | |
| Triconella vaginella virus (human parasite) | |
| Leishmania brasiliensis virus (human parasite) | |
| Other viruses of protozoa? | |
| Bacteriophages | |
| Podoviruses | T7 phage |
| Siphoviruses | λ-Phage group |
1. The importance of ribosomal frameshifting
In viruses that utilize programmed −1 frameshifting, the open reading frame (ORF) encoding the major viral structural protein (typically the Gag protein) is located at the 5′ end of the mRNA, whereas the ORFs encoding proteins with enzymatic functions (typically Pro and Pol) are located at the 3′ end of the transcript and out of frame with the Gag ORF (Fig. 1 ). The enzymatic proteins are only translated as a result of a programmed ribosomal frameshift event, which occurs with an efficiency of 1–40%, depending on the specific virus and assay system employed[3]. Thus, the majority of translational events result in the production of the Gag protein, while only a minority yield viral enzymatic proteins (Fig. 1 ). The efficiency with which the frameshift occurs therefore determines the ratio of structural to enzymatic proteins available for virus-particle assembly (Fig. 2). This gene arrangement is important for virus-particle morphogenesis, because viruses require a large excess of structural components over the proteins with enzymatic activities. The importance of maintaining the appropriate ratio of these factors has been demonstrated using two endogenous viruses of the yeast Saccharomyces cerevisiae. The results demonstrate that altering the efficiency of programmed ribosomal frameshifting in either the −1 or +1 direction changes the ratio of Gag to Gag–Pol proteins synthesized, and thus the virus-particle assembly and RNA packaging, resulting in reduced viral titres6, 7, 8, 9, 10, 11, 12, 13, 14, 15(Fig. 2). In addition, it has been shown that gross changes in the ratio of Gag to Gag–Pol proteins in retroviruses like HIV or Moloney Murine Leukaemia Virus interferes with virus-particle formation16, 17. These studies suggest that programmed −1 ribosomal frameshifting presents a novel target for antiviral therapies.
Fig. 1.
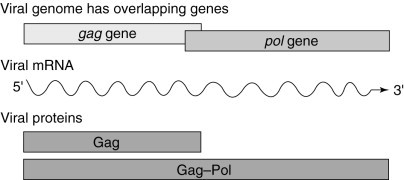
The genomes of viruses that use programmed ribosomal frameshifting are generally organized so that the open reading frames (ORFs) encoding structural proteins (e.g. gag) are 5′ of, and translated prior to, the ORFs encoding proteins with enzymatic functions (e.g. pol). Furthermore, pol is out of frame with respect to gag. The majority (90–98%) of host ribosomes translating the single viral mRNA terminate translation at the gag stop codon, resulting in the synthesis of Gag protein. A minority of ribosomes (2–10%) are induced to shift reading frame at the viral frameshift signal, resulting in the synthesis of the Gag–Pol fusion protein.
Fig. 2.
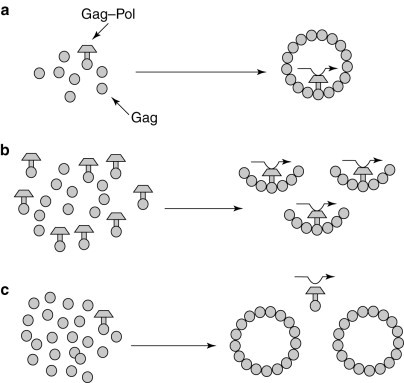
The efficiency of the frameshift determines the ratio of Gag to Gag–Pol proteins available for viral particle morphogenesis. (a) The normal frameshifting efficiency provides the correct ratio of Gag to Gag–Pol. (b) Increased frameshifting leads to the formation of incomplete viral particles. (c) Decreased frameshifting efficiency leads to the formation of Gag–Pol-deficient viral particles that cannot package the viral (+) strand.
2. The sequence elements that promote programmed −1 ribosomal frameshifting
The cis−acting sequences that promote efficient −1 ribosomal frameshifting have been well characterized in several viral systems2, 3, 4, 5. Two basic sequence elements are required to promote efficient levels of programmed −1 ribosomal frameshifting. The first sequence element is called the `slippery site' and consists of a heptamer sequence X XXY YYZ [the incoming 0-frame (e.g. the gag reading frame) is indicated by the positioning of the spaces], where XXX can be any three identical nucleotides, YYY can be AAA or UUU, and Z is A, U or C (Fig. 3 )10, 18, 19, 20. The second promoting element is usually a sequence that forms a defined RNA secondary structure, such as an RNA pseudoknot, approximately six nucleotides 3′ of the slippery site and is thought to increase the probability that the ribosome will slip reading frame in the −1 direction (Fig. 3)21, 22. The simultaneous slippage of both ribosome-bound tRNAs by one base in the 5′ direction still leaves their non-wobble bases correctly paired with the mRNA in the new reading frame (Fig. 3). Thus, the number of ribosomes that shift frame is affected by a number of parameters, including the ability of the ribosome-bound tRNAs to unpair from the 0-frame, the ability of these tRNAs to rebind to the −1 frame, the relative position of the RNA pseudoknot from the slippery site and the pseudoknot's thermodynamic stability10, 19, 20, 23, 24, 25, 26, 27, 28.
Fig. 3.
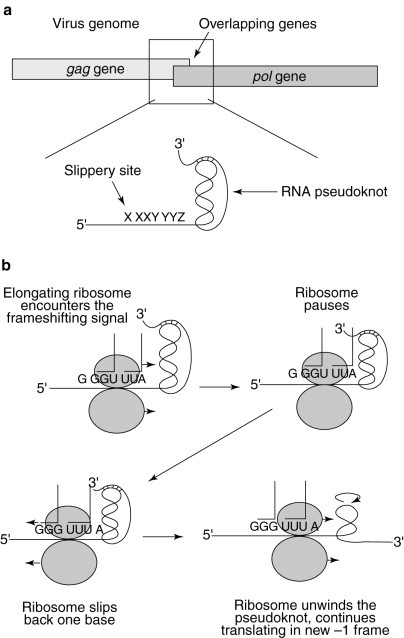
The simultaneous-slippage model of programmed ribosomal −1 frameshifting. (a) The programmed −1 ribosomal frameshift signal is located in the region where the gag and pol ORFs overlap. (b) An elongating ribosome encounters the RNA pseudoknot and pauses over the heptameric slippery site. The tRNAs in the ribosomal A and P sites unpair from the 0-frame codons and slip back one base so that their non-wobble bases rebind with the −1-frame codons. The ribosome unwinds the RNA pseudoknot and continues to translate the mRNA in the new −1 reading frame.
3. Advantages of targeting programmed frameshifting
The fact that programmed −1 ribosomal frameshifting appears to be virus specific makes it an attractive target to identify agents that affect the efficiency of this process and, consequently, of virus maintenance. We can envisage three major advantages to programmed −1 ribosomal frameshifting as a therapeutic target for antiviral agents.
-
1.
Small changes in frameshifting efficiencies can have large effects on virus production; for example, increasing or decreasing the efficiency of programmed −1 ribosomal frameshifting by the yeast L-A virus by as little as a factor of two interfered with the ability of yeast cells to maintain the M1 `killer' satellite virus of L-A6, 19.
-
2.
Compounds that change the efficiency of programmed ribosomal frameshifting function at concentrations that do not drastically inhibit the translational machinery (see below). Thus, these compounds should function as therapeutic agents at low drug concentrations, minimizing potential toxicity to the host. As described in greater detail below, it has been demonstrated that the peptidyl-transferase inhibitors anisomycin and sparsomycin efficiently inhibit viral propagation of the yeast killer virus at concentrations below those that inhibit cell growth and protein synthesis[6].
-
3.
The fact that this strategy targets a host-cellular process rather than a viral gene minimizes the ability of viruses to evolve drug-resistant mutants (Fig. 4 ). Most conventional antiviral strategies target a virus-specific protein—for example, nucleoside analogues and protease inhibitors, the most commonly used classes of antiviral agents, both target gene products encoded by the viral pathogen (Fig. 4). However, this therapeutic strategy generates a selective pressure for mutations in the viral genes that causes resistance to the actions of these drugs; drug-resistant mutants will therefore arise and, because virus populations evolve on the time scale of weeks to months, drug resistance emerges rapidly.
Fig. 4.
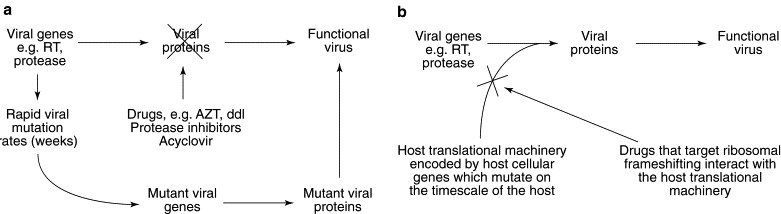
Advantages in targeting programmed −1 ribosomal frameshifting for antiviral therapies. (a) Conventional antiviral drugs target virus-encoded proteins. The rapid viral mutation rates ensure the selection of mutant viral genes encoding viral proteins that can bypass the actions of the drugs, resulting in drug-resistant functional virus. (b) Drugs that target programmed ribosomal frameshifting affect the host translational machinery, which is independent of the viral mutational capacity.
Compounds that target the host-cellular gene products that are involved in regulating programmed −1 ribosomal frameshifting have several advantages. First, any selective pressure on the host-cellular translational machinery to adapt to the drugs would have to occur at the host evolutionary time scale. Second, viral variants that might overcome the effects of the drugs by changing the efficiency of programmed −1 ribosomal frameshifting back to wild-type levels would theoretically have to involve multiple mutations in the slippery site and/or in the RNA pseudoknot according to the rules governing this process10, 19, 20, 24, 25, 26, 29, making it more difficult for drug-resistant mutants to arise. Thus, by shifting the selective pressure away from the virus' evolutionary strength, targeting programmed ribosomal frameshifting minimizes the ability of viruses to evolve drug-resistant mutants (Fig. 4b), and so agents that affect ribosomal frameshifting without deleteriously inhibiting the host-cellular translation apparatus would be excellent candidates for antiviral therapies.
4. Antiviral agents that alter programmed −1 ribosomal frameshifting
A series of yeast-based assay systems have been developed to identify compounds that modulate the translation machinery and alter programmed −1 ribosomal frameshifting efficiencies[6]. The advantages of using the yeast-based assay systems are threefold: (1) it has been amply demonstrated that the basic molecular mechanisms governing programmed −1 ribosomal frameshifting in yeast and humans are almost identical10, 19, 30, 31; (2) unlike mammalian-cell-culture systems or whole-animal models, yeast cells grow rapidly, providing rapid results with low turnaround times; (3) it is less expensive to culture yeast than to maintain mammalian cells or whole animals. For these reasons, yeast-based assay systems can be used for the identification of compounds that affect programmed ribosomal frameshifting. Below is a brief description of different assay strategies that have been used to monitor programmed −1 ribosomal frameshifting and virus maintenance.
4.1. In vivo and in vitro yeast-based frameshift-assay strategies
Three different reporter-construct strategies that allow the monitoring of programmed −1 frameshifting in intact yeast cells and translationally competent cell extracts are shown in Fig. 5 . The first strategy (Fig. 5a) has been used by many laboratories6, 19, 22, 30, 32, and is typically used to directly measure in vivo or in vitro programmed −1 ribosomal frameshift efficiencies and relies on the use of two different enzymatic reporter constructs that are assayed in parallel. The transcription-initiation and -termination signals, and the reporter gene in the two constructs are the same in both plasmids. The differences lie in the fact that the 0-frame control plasmid contains the reporter gene in the same reading frame as the translation start site with no intervening viral sequence, resulting in high-level expression of the reporter protein. The −1 frameshift reporter construct contains a virus-derived programmed −1 ribosomal frameshift signal inserted between the start codon and the reporter gene, in such a way that the reporter gene is in the −1 frame with respect to the translational start site. Thus, the reporter protein can only be translated if the ribosome shifts frame in the −1 direction. The efficiency of −1 ribosomal frameshifting is calculated by determining the ratio of reporter-protein activities measured in cells or cell extracts harbouring the −1 frameshift reporters to those harbouring the 0-frame controls. Because the efficiency of programmed ribosomal frameshifting is based on the ratio of frameshift-reporter to 0-frame-control activities, the efficiencies of programmed ribosomal frameshifting are always normalized to the effects that drugs have on overall translation by monitoring the reporter-protein activities of the 0-frame controls in the presence of the indicated drug. The changes in programmed frameshifting are determined by calculating the ratio of frameshifting efficiency in the presence of the drugs to that in cells grown in the absence of any drug.
Fig. 5.
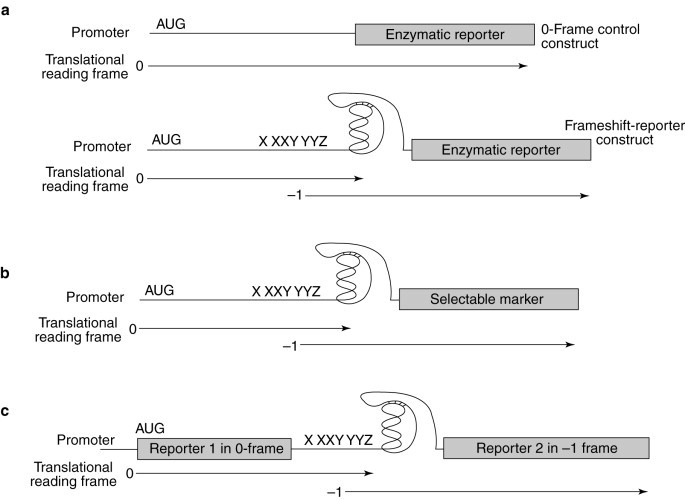
Programmed −1 ribosomal frameshift assay strategies. (a) General strategy for directly measuring the efficiency of programmed −1 ribosomal frameshifting. Pictured are the 0-frame control reporter and −1 ribosomal frameshift constructs. Both systems utilize a reporter protein (the lacZ-encoded β-galactosidase, in vivo, or luciferase, in vitro). In the −1 ribosomal frameshift construct, the reporter is cloned downstream of programmed −1 ribosomal frameshift signals and is in the −1 frame with a translational start site. Synthesis of the reporter protein thus requires a programmed ribosomal frameshift event. The 0-frame controls have the reporter genes cloned in frame with the start site and lack the intervening viral frameshift signals. The programmed ribosomal frameshift efficiency is determined by dividing the level of reporter-protein activity produced from the −1 reporters by that produced from the 0-frame control and multiplying by 100%. Translation of the reporters can occur in intact yeast cells or in translationally competent cell extracts. The effects of a candidate compound on both overall translation and on programmed −1 ribosomal frameshifting can be monitored. (b) General strategy for assays in intact yeast cells that detect changes in programmed −1 ribosomal frameshift efficiencies by using a selectable marker. A gene encoding a selectable marker is cloned downstream of, and in the −1 frame with respect to, a translational start site, such that synthesis of the gene product requires a programmed ribosomal frameshift event. Changes in programmed −1 ribosomal frameshift efficiencies are detected by monitoring the ability of cells to grow (positive selection) or not (negative selection) in the presence of a candidate compound. (c) Bicistronic constructs for measuring changes in programmed −1 ribosomal frameshifting efficiencies. Reporter-gene 1 constitutes the 0-frame control and can be used to monitor the effect of a compound on overall translation. Translation of reporter-gene 2 requires a programmed ribosomal frameshift event. Changes in programmed −1 ribosomal frameshift efficiencies are detected by monitoring for changes in the ratios between the activities of the two reporter proteins.
In the second general strategy (Fig. 5b), a programmed −1 ribosomal frameshift results in the synthesis of a selectable marker protein using intact cells. For example, increased expression of the CUP1 gene as a consequence of increased programmed −1 ribosomal frameshifting efficiency was used to isolate host chromosomal frameshifting mutants[29]. Although this strategy does not measure programmed −1 ribosomal frameshifting efficiencies directly, it could be employed in a high-throughput screen for agents that affect this process in intact cells.
The third general strategy (Fig. 5c) uses bicistronic reporter constructs in which translation of the first reporter protein serves as an internal control and synthesis of the second reporter protein requires a programmed −1 ribosomal frameshift27, 31. This strategy measures changes in the relative ratios of the two reporter-protein activities and is particularly well suited to cell-free translation systems.
Each of these three assay stategies can be used to provide rapid and reliable methods to measure the effects of agents on programmed ribosomal frameshifting, on overall protein translation and on cell growth and viability.
4.2. Yeast-based viral assays
One major caveat inherent in all of the in vivo strategies described above is that all of the frameshift-reporter genes contain 0-frame termination codons and thus encode unstable mRNAs that are substrates for rapid degradation via the nonsense-mediated mRNA-decay (NMD) pathway9, 33. Agents that inactivate the NMD pathway would stabilize these mRNAs, resulting in higher expression of the frameshift-reporter protein, appearing to increase the efficiency of programmed −1 ribosomal frameshifting. Thus, a second, independent assay system is required to confirm that the agent affects programmed −1 ribosomal frameshifting and that it has antiviral activity. The killer-viral system of the yeast S. cerevisiae has served as a model for investigations into the mechanisms governing −1 ribosomal frameshifting[2]. Typically, it is composed of the L-A helper virus and the M1 satellite virus. The dsRNA genome of L-A contains two ORFs, the 5′ gag gene encoding the major viral coat protein (Gag) and the 3′ pol gene encoding a multifunctional protein that includes the RNA-dependent RNA polymerase and a domain required for viral RNA packaging. A −1 ribosomal frameshift event is responsible for the production of the L-A encoded Gag–Pol fusion protein[19](Fig. 6 a). The M1 satellite dsRNA virus of L-A encodes a secreted `killer' toxin and its dsRNA genome is encapsidated and replicated inside the icosahedral 39-nm L-A viral particle. Changes in the efficiency of programmed −1 ribosomal frameshifting along the L-A mRNA result in rapid loss of M1, which can be monitored by replica-plating colonies of test cells on a lawn of cells that are sensitive to the killer toxin; cells maintaining the M1 virus secrete the killer toxin, creating a ring of growth inhibition[10](Fig. 6c).
Fig. 6.
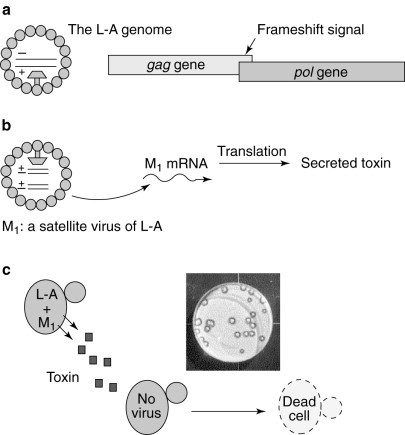
The yeast `killer-virus' system: an ideal assay system. The killer-virus system is composed of the L-A dsRNA helper virus and the M1 dsRNA satellite virus. (a) The gag and pol ORFs overlap in the L-A genome, and a programmed −1 ribosomal frameshift is used to synthesize the Gag and Gag–Pol fusion proteins to produce a functional viral particle containing the L-A dsRNA. (b) The dsRNA genome of the M1 satellite virus is encapsidated inside L-A-encoded viral particles; the M1 mRNA encodes a secreted toxin. (c) Cells harbouring both L-A and M1 secrete the toxin and are immune to its action, whereas virus-free cells are sensitive to the toxin. A picture of the yeast-killer assay is shown, in which cells harbouring L-A and M1 were replica-plated onto a lawn of virus-free cells. A ring of growth inhibition is indicative of the killer activity of the cells harbouring both L-A and M1.
5. Identification of compounds that affect frameshifting and promote killer-virus loss
Using the assay strategies described above, it has been shown that two peptidyl-transferase inhibitors (anisomycin and sparsomycin) specifically alter the efficiency of −1 ribosomal frameshifting in yeast cells when present at sublethal doses[6]. These drugs belong to a well-characterized class of small molecules that have been shown to affect the protein-synthetic machinery at the step at which the −1 ribosomal frameshift event is thought to occur[6]. For example, at anisomycin concentrations that do not affect the growth of yeast cells or the rates of protein translation, the efficiency of programmed −1 ribosomal frameshifting was altered. When cells harbouring the L-A and M1 viruses were grown at these concentrations of anisomycin, the rapid loss of both of these viruses was observed (Fig. 7 ).
Fig. 7.
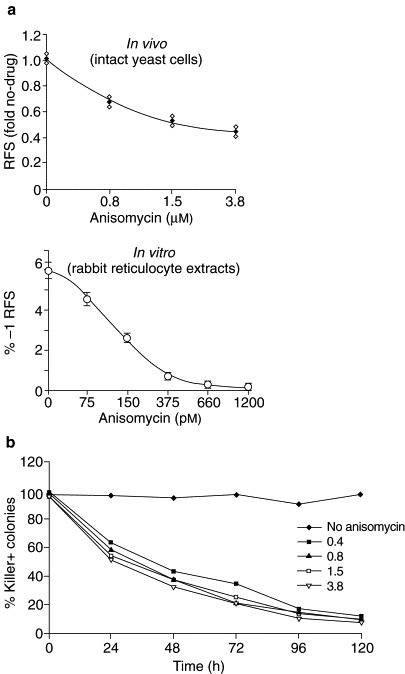
Anisomycin specifically decreases −1 ribosomal frameshifting efficiency and has antiviral activity. (a) The effects of anisomycin on programmed ribosomal frameshifting were assayed in intact yeast cells in vivo, and in translationally competent rabbit reticulocyte extracts in vitro. The indicated drug concentrations were subinhibitory for cell growth and division (in vivo), and for overall translation (both in vivo and in vitro) (not shown). Anisomycin specifically inhibits programmed −1 ribosomal frameshifting in both systems. (b) Yeast cells harbouring the L-A and M1 viruses were cultured in the presence of the indicated concentrations of anisomycin for the indicated times, after which the percentage of cells retaining killer activity was determined.
An alternative strategy involved the use of a series of 2′-methyl oligonucleotides designed to bind specifically to the sequences flanking the HIV frameshift signal[34]. It was shown that oligonucleotides that bound immediately 3′ of the HIV frameshift signal increased the efficiency of programmed −1 ribosomal frameshifting sixfold. Further investigations suggested that increasing RNA secondary structure downstream of the frameshift site increases the programmed −1 ribosomal frameshifting efficiencies[34]. Taken together, these results demonstrate that specific agents can be used to modulate the translation machinery, without inactivating translation, in order to alter the efficiency of programmed frameshifting to such a degree as to promote loss of the killer virus.
6. Future perspectives
Many viruses that cause significant human, animal and agricultural diseases utilize programmed −1 ribosomal frameshifting to regulate the production of their structural and enzymatic proteins (Table 1). Altering the frameshifting efficiencies disrupts the virus' life cycle, eliminating or reducing virus production. Programmed −1 ribosomal frameshifting should be an excellent target for compounds that function as antiviral agents, because it is predominantly utilized by these viruses to regulate gene expression. Small changes in ribosomal frameshift efficiencies can be achieved at sublethal concentrations of the chosen compounds, so that subtle modulations of the host-cellular translation machinery results in alteration of frameshifting efficiencies and virus loss. Importantly, because the correct efficiency of ribosomal frameshifting is determined in part by the host-cellular translational apparatus, and owing to the complex nature of the programmed −1 ribosomal frameshift signal, compensatory viral mutations will not be easily generated, greatly reducing the likelihood of drug-resistant variants.
A series of rapid and inexpensive yeast-based assay systems have been developed to identify compounds that alter the efficiency of programmed -1 ribosomal frameshifting and cure yeast cells of an endogenous virus. Clearly, more work is required to determine whether these compounds can be used as therapeutic agents for human viral diseases. The list of viruses that utilize programmed −1 ribosomal frameshifting includes HIV, the causative agent of AIDS; this demonstrates the value of identifying agents that function to affect programmed frameshifting and reduce viral titres. The results and assays presented here describe a new target to fight against viral disease and outline rapid, cost-effective approaches to identify potential antiviral agents that can be further characterized for their therapeutic value.
Interestingly, although the compounds described here have been previously demonstrated to have antibiotic function, there is also an alternative strategy for their use as potential antiviral agents. The key will be to use these agents to modulate the host-cellular translation machinery, rather than completely inhibit its function. Thus, these well-studied compounds that have been invaluable in bacterial infections may also now be utilized in a new arena in the fight against viral diseases.
Acknowledgements
The authors' work was supported in part by grants given to J. D. Dinman by the Foundation of the University of Medicine and Dentistry of New Jersey and the State of New Jersey Commission of Cancer Research (96-62-CCR-00), and by grants awarded to S. W. Peltz by the US National Institutes of Health (GM48631-01) and an American Heart Association Established Investigator Award. M. J. Ruiz-Echevarria has been supported by the New Jersey Affiliate of the American Heart Association.
References
- 1.Kurland C.G. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 2.Dinman J.D. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brierley I. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 4.Gesteland R.F., Atkins J.F. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 5.Farabaugh, P. J. (1997) Programmed Alternative Reading of the Genetic Code, R. G. Landes
- 6.Dinman J.D., Ruiz-Echevarria M.J., Czaplinski K., Peltz S.W. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasundaram D., Dinman J.D., Wickner R.B., Tabor C.W., Tabor H. Proc. Natl. Acad. Sci. U. S. A. 1994;91:172–176. doi: 10.1073/pnas.91.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng R.H. J. Mol. Biol. 1994;224:255–258. doi: 10.1006/jmbi.1994.1726. [DOI] [PubMed] [Google Scholar]
- 9.Cui Y., Dinman J.D., Peltz S.W. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinman J.D., Wickner R.B. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinman J.D., Wickner R.B. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinman J.D., Kinzy T.G. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- 13.Tumer N.E., Parikh B., Li P., Dinman J.D. J. Virol. 1998;72:1036–1042. doi: 10.1128/jvi.72.2.1036-1042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawakami K. Genetics. 1993;135:309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J., Boeke J.D. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein K.M., Goff S.P. J. Virol. 1988;62:2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park J., Morrow C.D. J. Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacks T., Varmus H.E. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 19.Dinman J.D., Icho T., Wickner R.B. Proc. Natl. Acad. Sci. U. S. A. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brierley I.A., Jenner A.J., Inglis S.C. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somogyi P., Jenner A.J., Brierley I.A., Inglis S.C. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu C., Tzeng T-H., Bruenn J.A. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brierley I.A., Dingard P., Inglis S.C. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brierley I.A., Rolley N.J., Jenner A.J., Inglis S.C. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacks T., Madhani H.D., Masiraz F.R., Varmus H.E. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa S., Bishop D.H.L. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kollmus H., Honigman A., Panet A., Hauser H. J. Virol. 1994;68:6087–6091. doi: 10.1128/jvi.68.9.6087-6091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda A., Nakamura T., Nishimura S. Biochem. Biophys. Res. Commun. 1995;213:575–582. doi: 10.1006/bbrc.1995.2170. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.I., Umen J.G., Varmus H.E. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6587–6591. doi: 10.1073/pnas.92.14.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson W., Braddock M., Adams S.E., Rathjen P.D., Kingsman S.M., Kingsman A.J. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 31.Stahl G., Bidou L., Rousset J-P., Cassan M. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belcourt M.F., Farabaugh P.J. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weng, Y. et al. (1997) in Modern Cell Biology. Post-transcriptional Gene Regulation: Todays RNA World (Harford, J. B. and Morris, D. R., eds), pp. 241–263, Wiley–Liss
- 34.Vickers T.A., Ecker D.J. Nucleic Acids Res. 1992;20:3945–3953. doi: 10.1093/nar/20.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]


