Abstract
Baculoviruses are a group of enveloped, double-stranded DNA insect viruses with budded (BV) and occlusion-derived (ODV) virions produced during their infection cycle. BVs are commonly described as rod shaped particles with a high apical density of protein extensions (spikes) on the lipid envelope surface. However, due to the fragility of BVs the conventional purification and electron microscopy (EM) staining methods considerably distort the native viral structure. Here, we use cryo-EM analysis to reveal the near-native morphology of two intensively studied baculoviruses, Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) and Spodoptera exigua MNPV (SeMNPV), as models for BVs carrying GP64 and F as envelope fusion protein on the surface. The now well-preserved AcMNPV and SeMNPV BV particles have a remarkable elongated, ovoid shape leaving a large, lateral space between nucleocapsid (NC) and envelope. Consistent with previous findings the NC has a distinctive cap and base structure interacting tightly with the envelope. This tight interaction may explain the partial retaining of the envelope on both ends of the NC and the disappearance of the remainder of the BV envelope in the negative-staining EM images. Cryo-EM also reveals that the viral envelope contains two layers with a total thickness of ≈6–7 nm, which is significantly thicker than a usual biological membrane (<4 nm) as measured by X-ray scanning. Most spikes are densely clustered at the two apical ends of the virion although some envelope proteins are also found more sparsely on the lateral regions. The spikes on the surface of AcMNPV BVs appear distinctly different from those of SeMNPV. Based on our observations we propose a new near-native structural model of baculovirus BVs.
Keywords: Baculovirus, Budded virus, Ultrastructure, Cryo-EM, Spike structure
1. Introduction
Baculoviruses are a family of enveloped, large double-stranded DNA viruses that predominantly infect insects. The genome of baculoviruses ranges from 80 to 180 kb in size and contains between 90 and 180 genes (Herniou et al., 2012). The viral genome is packed into rod-shaped nucleocapsids (NC) of 30–70 nm in diameter and 200–400 nm in length (Boucias and Pendland, 1998, Jehle et al., 2006). Two phenotypes of infectious enveloped virions are produced during the infection cycle: the occlusion-derived viruses (ODV) that initiate infection of the midgut of the host upon oral ingestion of occlusion bodies (OBs) and budded viruses (BV) that are responsible for cell-to-cell spread and further systemic infection. ODVs obtain their de novo assembled envelopes in the nucleus, while BVs acquire their envelope from the plasma membrane upon budding into the extracellular space or neighboring cells (Okano et al., 2006). Although the two phenotypes contain identical viral genomes and nucleocapsids, they differ in the lipid and protein composition and structure of their envelopes (Slack and Arif, 2007). The ODV envelope is more rigid than the BV envelope due to the presence of more saturated fatty acids (Braunagel and Summers, 1994, Slack and Arif, 2007). Several envelope-associated proteins of BVs (GP64, F), but no envelope-associated proteins of ODVs were found to be N-glycosylated (Hou et al., 2013). Unlike BVs, ODVs have tegument proteins, which package nucleocapsids and create a matrix layer between the nucleocapsid and the envelope (Slack and Arif, 2007). One or multiple ODV virions are further embedded into a proteinaceous OB. Due to the highly compact structure and OB protection the intact ODVs are more easily isolated in their native form than BVs.
The Baculoviridae family is divided into four genera: Alphabaculovirus (encompassing viruses with lepidopteran hosts), Betabaculovirus (lepidopteran hosts), Gammabaculovirus (hymenopteran hosts) and Deltabaculovirus (dipteran hosts) on the basis of the genome phylogeny (Herniou et al., 2012). The alphabaculoviruses are divided into two subgroups, group I and group II according to the sequence and phylogenetic analysis of conserved genes (Zanotto et al., 1993, Jehle et al., 2006). The subdivision between these two groups is correlated with employment of two different BV envelope glycoproteins, i.e. GP64 (group I) and F (group II), for virus-cell fusion and receptor binding (Lung et al., 2002). These two envelope glycoprotein types are highly distinct in sequence and structural features (Kadlec et al., 2008, Westenberg, 2004). In group I alphabaculoviruses both GP64 and F-like proteins are present; only GP64 mediates membrane fusion and the F-like protein appears to have lost its fusion function (Jehle et al., 2006, Lung et al., 2003, Wang et al., 2008). In members of group II alphabaculoviruses, the betabaculoviruses and the deltabaculoviruses the major envelope protein is the F protein, which mediates membrane fusion (Jehle et al., 2006, Lung et al., 2002).
The baculovirus F proteins differ from GP64 proteins not only in amino acid sequence, but also in their biochemical and structural properties, except that both proteins are activated at acidic pH (Blissard and Wenz, 1992, IJkel et al., 2000). F proteins require proteolytic cleavage to prime the membrane fusion (IJkel et al., 2000). Based on the structural elements and organization, F protein and GP64 proteins were characterized as class I and class III viral fusion proteins, respectively (Bosch et al., 2003, Kadlec et al., 2008). Both classes of fusion proteins are composed of trimers with their ectodomains oriented perpendicular to the viral membrane. Two trimers assemble into spikes on the surface of BV envelope. The fusogenic forms of class I and class III viral fusion proteins are of mainly α-helical structure or a mixture of α-helices and β-sheets, respectively [reviewed in (White et al., 2008)].
Previously, the baculovirus BV ultrastructure was studied using negative-staining electron microscopy EM (Adams et al., 1977, Beaton and Filshie, 1976, Fraser, 1986, Harrap, 1972). The BV envelopes of several group I and II alphabaculoviruses in baculovirus-infected cells and tissue of hosts were reported bulbous at one end. The surface of the envelope is serrated with notches, which are concentrated at the bulbous end (Adams et al., 1977). These BV notches, called spikes, on the surface were demonstrated to be composed of envelope proteins including GP64 and F protein (Volkman et al., 1984, Pearson et al., 2001).
BV and ODV structures have been studied by negative-staining EM (Adams et al., 1977, Beaton and Filshie, 1976, Fraser, 1986, Harrap, 1972). However, due to the fragility and flexibility of BV particles, the procedure of negative-staining and subsequent drying severely impairs the structural integrity of the viral envelope and causes virions to collapse. Intact BV particles are therefore rare and usually have lost part of their envelope. In the current study we have used cryo-EM of vitrified BV suspensions to re-visit the morphology of virions of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) and Spodoptera exigua multiple nucleopolyhedrovirus (SeMNPV), as representatives of group I (GP64) and group II (F), alphabaculoviruses to provide new insights on the ultrastructural organization and assembly of BV virions.
2. Materials and methods
2.1. Virus production and purification of budded baculovirus
Hemolymph-derived SeMNPV BVs and cell culture-derived AcMNPV BVs were obtained from Els Roode (Wageningen University) and subsequently used for virus production in cell culture. The titers of the SeMNPV and AcMNPV BV suspensions were first determined on Se301 and Sf9 cells, respectively, using end point dilution assays. Se301 cells were then infected with SeMNPV BVs at an MOI of 5 tissue culture infection dose 50% (TCID50) units per cell in HyClone CCM3 insect culture medium (Thermo Scientific). Sf9 cells were infected with AcMNPV BVs at an MOI of 5 TCID50 units per cell in Insect-XPRESS insect culture medium with l-glutamine (Lonza). At 48 h post-infection (p.i.) cell culture supernatants, where BVs were expected to be present, were clarified at 3000g for 15 min at 4 °C to remove cells and cellular debris. This supernatant was directly used for cryo-EM.
2.2. Fractionation of NCs from budded baculoviruses
The clarified cell culture supernatant from the procedure above was incubated with 1% Nonidet P-40 (NP-40) in TE buffer (10 mM Tris–HCl at pH 7.4, 137 mM NaCl, 2 mM EDTA) for 30 min at room temperature to remove the viral envelope. Six ml of NP-40 treated BV suspension were layered onto 3 ml of 25% and 50% iodixanol solution in TE buffer and centrifuged at 16,000g for 3 h at 4 °C in SW41Ti rotor (Beckman Coulter). The fractions were collected every 3 ml from the top to bottom. The NCs were most abundant in the third fraction from the top of the centrifuged tube upon examination with negative-staining EM as described below. The NC fraction was diluted in 30 ml phosphate-buffered saline (PBS) at pH7.4 (Lonza) and centrifuged at 100,000g for 1 h at 4 °C in an SW32Ti rotor (Beckman Coulter). The pellet was suspended in PBS.
2.3. Electron microscopy of budded baculovirus
For negative-staining EM, formvar/carbon coated 400-mesh copper grids were exposed to a glow discharge in air for 20 s. Ten μl of a BV or NC suspension was placed on the grids and incubated for 2 min. Negative-staining was performed with 1% phosphotungstic acid (PTA, pH 7.2) for 1 min. The specimens were examined in a JEOL 1011 transmission EM equipped with an Olympus Keenview (1K × 1K) and Veleta (2K × 2K) digital camera.
For cryo-EM, specimens were vitrified using a Vitrobot (FEI Company). Four μl of a suspension containing virions or NCs was placed on a quantifoil carbon or lacey carbon grid and allowed to absorb for 30 s. After blotting the grid was plunged into liquid ethane. The frozen specimen was examined at −180 °C in a JEOL 2100 TEM equipped with a Gatan CT3500 cryo-holder and a Gatan US4000 (4K × 4K) digital camera. Images were recorded at low dose with DigitalMicrograph software (Gatan) and analyzed with iTEM platform (Olympus).
3. Results
3.1. General architecture of baculovirus BVs
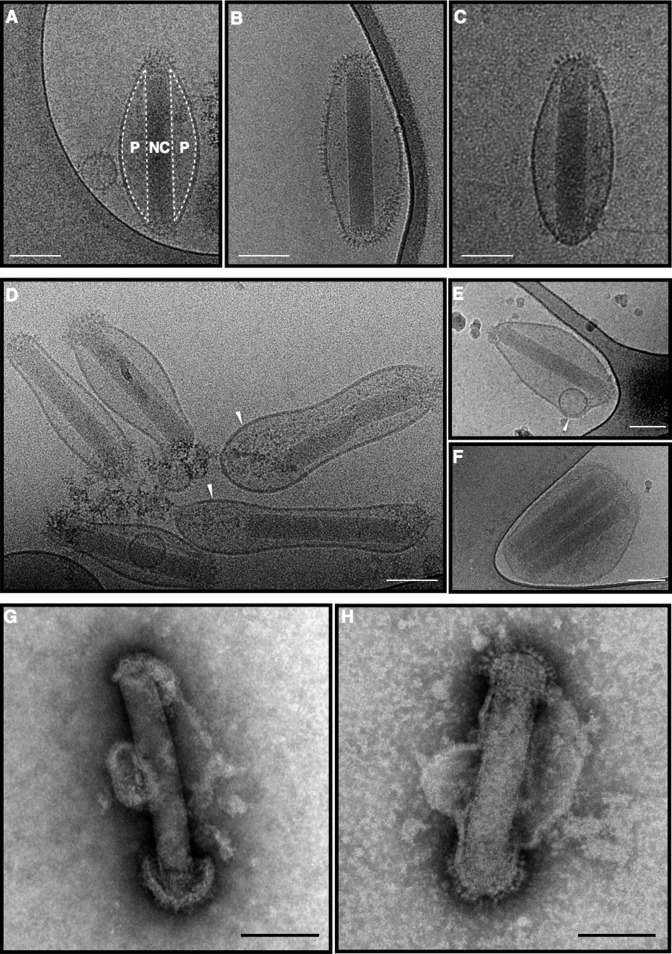
To study the near-native morphology of AcMNPV and SeMNPV BVs, virions were produced in Sf9 and Se301 cells respectively for 48 h, isolated from the supernatant and subjected to cryo-fixed by plunge freezing. Fig. 1 shows images of these BV particles, which exhibit remarkably different features as compared to the current knowledge on BV morphology. The majority (≈95%) of virions of both AcMNPV and SeMNPV with intact envelope show an extended ‘ovoid’ shape (Fig.1A–C) instead of a ‘rod’ shape as described previously (Adams et al., 1977). The ‘pocket’ between NC and lateral envelope appears to be electron-lucent, yet not empty because the electron density is darker than the ambient surrounding the BV particles. Small vesicles are apparently included in the lateral space of some virions (Fig.1E), indicating that the pockets might be filled with soluble content. The interactions between NC and envelope appeared to be limited to the two ends of NC and therefore the virions tend to have heterogeneous shapes around the lateral area even with cryo-fixation (Fig.1D). The virions marked with an arrowhead in Fig.1D appear to get compressed at the inner lateral space, which generated high tension at one end of NC. As a result the envelope–NC interaction at this end was disrupted. In this case the distance between envelope and NC is increased as compared to the other end of virion (indicated as white arrows in Fig.1D). This envelope–NC interaction is much more pronounced in the negative-staining EM micrographs of virions. The viral envelopes of negatively stained virions were disrupted to a great extent yet leaving the envelope part retained on the two ends of NCs (Fig.1G and H). Furthermore, imaging of intact BVs revealed that the envelope proteins, which appear as spikes perpendicular to the viral envelope, are present at both ends of most AcMNPV and SeMNPV BVs (Fig.1A and C). A small fraction (<1%) of virions is also distributed over the lateral region (Fig.1B). In addition, a fraction of the BVs (≈10%) possess multiple NCs (Fig.1F). The peculiar cases of lateral envelope protein distribution and multiple BV NCs per virion may be considered as outcomes of abundant virus infection with overexpression of envelope proteins and excessive number of NCs available for envelopment. By and large though the overall structure of AcMNPV (group I alphabaculovirus) and SeMNPV (group II alphabaculovirus) as seen via cryo-EM is very similar.
Fig. 1.
Virus morphology of group I and group II alphabaculovirus BVs. (A–F) Cryo-EM images of AcMNPV and SeMNPV BVs. (A–C) Ovoid-shaped of an AcMNPV BVs (A and B) and a SeMNPV BV (C). NC, nucleocapsid; P, pocket at lateral side. (D) AcMNPV virions with stretched sites of viral envelopes of AcMNPV around NC ends indicated with arrows. (E) AcMNPV BV with vesicle-like inclusion. The inclusion is indicated with an arrow. (F) AcMNPV BV with multiple nucleocapsids. (G and H) Negative-staining EM images of an AcMNPV BV (G) and a SeMNPV BV (H), showing fragments of the envelope attached to both ends of the NC. Scale bars represent 100 nm.
3.2. Budded virus envelope morphology
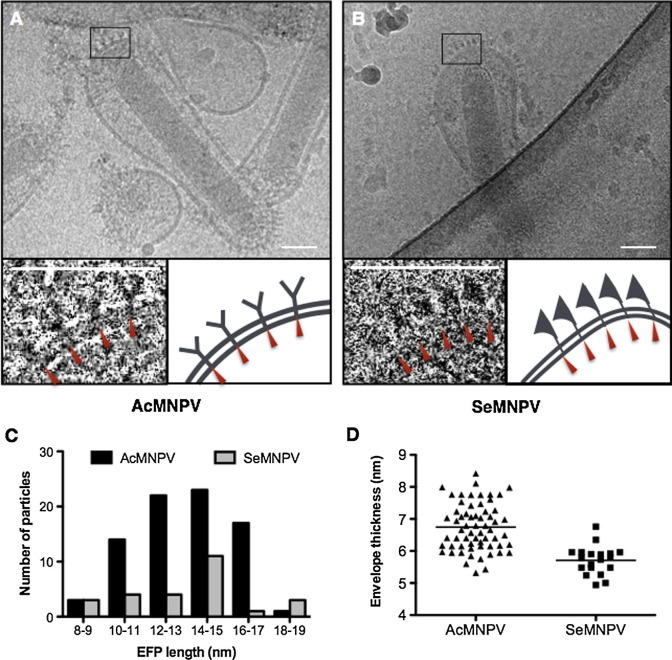
The BVs of AcMNPV and SeMNPV, representing the two groups of alphabaculoviruses, bear different envelope fusion proteins, GP64 and F respectively. In AcMNPV BVs an F-like protein is present. However, according to a proteomics study, the GP64 protein is the major envelope protein and is one of the most abundant proteins of the AcMNPV BV (Wang et al., 2010c). In a closely related virus, OpMNPV, this F-like protein remains associated with the nucleocapsid of the BV, when the envelope is removed (Pearson et al., 2001). So, it is safe to assume that the extended spikes of AcMNPV BVs observed in the cryo-EM are made up of GP64. In contrast, F is the only major envelope protein in SeMNPV (IJkel et al., 2000) as well as in other group II alphabaculoviruses. Cryo-EM images show that the spikes on AcMNPV and SeMNPV BVs appear to be ‘Y’ and ’burgee’ -shaped, respectively (Fig.2 A and B). The two open arms of Y shaped GP64 spikes appear to occupy more space than the ‘banner’ of burgee-shaped F spike. This is in good agreement with the distribution of the spikes, which are present more densely on SeMNPV than on AcMNPV BV envelopes (Fig.2A and B). However, the lengths of the majority of the spikes are similar on both baculoviruses (Fig.2C). The envelopes of BVs are derived from the plasma membrane of host cells (Blissard and Rohrmann, 1990) and the lipid composition of the viral envelope is therefore similar to the plasma membrane (Braunagel and Summers, 1994, Marheineke et al., 1998). Both AcMNPV and SeMNPV BV possess an envelope with a typical bilayer membrane structure (Fig. 2). These viral envelopes, with a thickness of approximately 6–7 nm (Fig.2D), are wider than general lipid bilayer thickness of up to 4 nm (Mitra et al., 2004). It is unknown if the lipid bilayers of Se301 or Sf9 cells from which these BVs are derived show a similar smaller thickness.
Fig. 2.
Ultrastructural characterization of baculovirus envelope proteins. (A and B) Cryo-EM images of envelope proteins on AcMNPV (A) and SeMNPV (B) BVs. The squared regions on the viral envelope were further focused in the lower panels showing more detailed structural patterns. Scale bars represent 50 nm. (C) Comparative analysis of length distributions on envelope fusion proteins from AcMNPV (n = 79; mean = 15 nm, SD = 2.1 nm) and SeMNPV (n = 30; mean = 14 nm, SD = 3.8 nm) BVs. (D) Thickness distributions of viral envelopes from AcMNPV (n = 62; mean = 7 nm, SD = 0.7 nm) and SeMNPV (n = 30; mean = 6 nm, SD = 0.4 nm). Spikes were excluded from membrane measurement.
3.3. NC structural organization
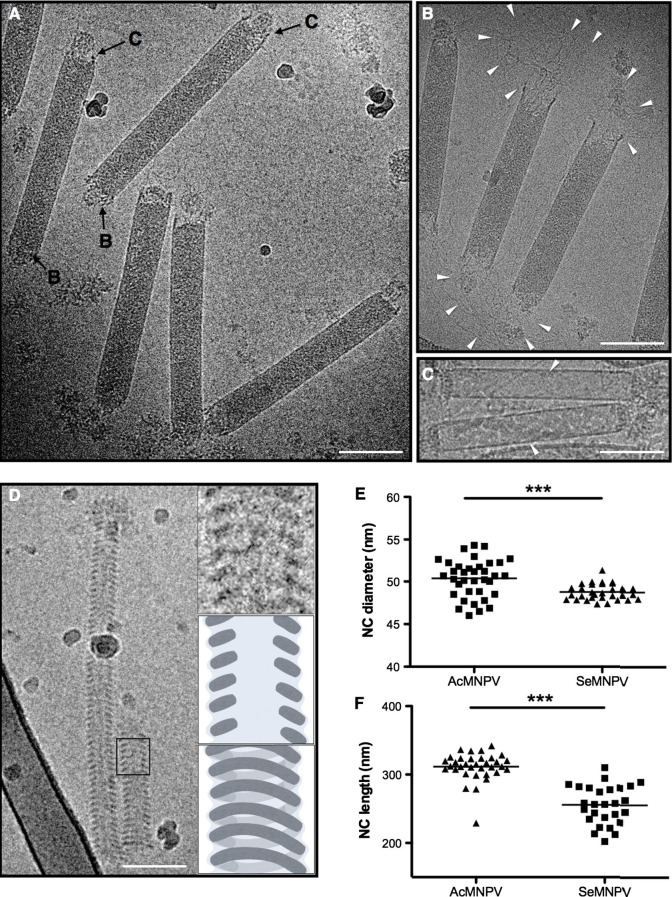
The ultrastructure of baculovirus NCs has only been studied from alkaline-dissolved ODVs with respect to organization of capsid and NC core (Beaton and Filshie, 1976, Bud and Kelly, 1980, Burley et al., 1982). We therefore investigated the structure of NC present in BVs in detail by cryo-EM. After cryo-fixation four distinct forms of NCs were observed from AcMNPV BVs: (i) NCs with a high electron density core and polar ends of apical nipple-like and basal claw-like structures (Fig. 3 A); (ii) NC partially disassembled from both ends (Fig.3B); (iii) empty capsids, in which NC cores are absent (Fig.3C) and (iv) disassembled NCs with a markedly spiral NC core (helical spring) structure (Fig.3D). The relaxed NCs and empty capsids were also observed within intact envelopes (data not shown). Compared to native NCs the relaxed NCs show a more prominent structural organization. The diameter of the relaxed NCs (Fig.3D) decreases approximately 10 nm as their length at the same time increases as compared with intact NCs (Fig.3A), resembling a helical spring in the expanded and compressed states. The empty capsids did not collapse but instead appear to be a rigid shell of tubular shape and identical size as the native NCs (Fig.3C), suggesting that the assembly of capsid sheath is independent of viral DNA packaging. Once the two ends of the capsid were open the viral DNA was extruded from capsid sheath as a non-supercoiled form (Fig.3B).
Fig. 3.
Characterization of nucleocapsid structure of BVs. (A–D) Cryo-EM images of nucleocapsids of AcMNPV. (A) Native nucleocapsids. (B) Partially uncoated nucleocapsids. The extruded fibrillar viral DNA is indicated with arrows. (C) Empty capsids without packaged viral DNA indicated with arrows. (D) “Relaxed” forms of nucleocapsids. The left panel shows the overall image of a relaxed NC. The panels at the right side show the detail of the spiral shapes of NC. (E) Distribution of the diameter of AcMNPV NC (n = 36, mean = 50.37 nm, SD = 2.24 nm and SeMNPV NC (n = 31, mean = 48.78 nm, SD = 0.93 nm). (F) Distribution of the length of AcMNPV NC (n = 34, mean = 311.5 nm, SD = 20.39 nm) and SeMNPV NC (n = 27, mean = 255.7 nm, SD = 28.31 nm). Significant differences between AcMNPV and SeMNPV are indicated (∗∗∗P < 0.0004 for NC diameter; P < 0.0004 for NC length). Scale bars represent 100 nm.
The genome sizes of SeMNPV (135,611 bp) (IJkel et al., 1999) and AcMNPV (133,894 bp) (Ayres et al., 1994) are nearly identical. However, the diameter and length of intact NCs (Fig.3A) showed significant differences (Fig.3E and F), indicating that the DNA packaging e.g. in terms of level of DNA condensation, or other steps in the assembly of these two types of BVs might differ in details. For example, the sequence divergence of DNA-binding protein P6.9 may contribute in the different levels of DNA condensation (Wang et al., 2010b).
4. Discussion
In cryo-EM, baculovirus BV particles exhibit an ovoid shape with nearly symmetrical curvature at the apical and basal regions. This is different from previous reports on the BV structure where the envelope was loosely wrapped around the nucleocapsid (Volkman et al., 1984). Interestingly, the envelope did not tightly surround the rod shaped NC at all but puffed up at the lateral region forming two large pockets (Fig.1A–C), which appear to be electron lucent. The negative-staining EM micrographs of virions showed retained envelope parts on the two ends of NCs and disrupted envelope at lateral region (Fig.1G and H) suggesting that there is an intimate interaction between envelope and capsids at the polar ends, but not at the lateral regions. Furthermore, the envelope proteins, visible as spikes perpendicular to the viral envelope, are present at both ends of most virions rather than only at the apical end of the virion as has been described previously (Adams et al., 1977, Harrison and Hoover, 2012) (Fig.1A and C). The polarized distribution of envelope proteins at apical and basal regions is a unique feature of baculovirus BVs. It is not known why baculovirus BVs have their envelope proteins polarized in contrast to other elliptic enveloped viruses, for instance poxviruses including vaccinia virus (Cyrklaff et al., 2005), myxoma virus (Duteyrat et al., 2006), orf virus (Tan et al., 2009) and Melolontha melolontha entomopoxvirus (Bergoin et al., 1969). Possibly, the polarized distribution of the envelope proteins advantages BV entry since orienting the BV perpendicular to the cellular membrane minimizes the membrane fusion area. Moreover, the position of envelope proteins in a curved membrane could be favorable for membrane fusion, since membrane curvature causes membrane tension, which could make the membrane less stable and hence lower the energy barrier for fusion.
Once the envelope proteins are embedded in the plasma membrane of host cells they are not likely to undergo dramatic rearrangement of their positions due to their confinement by the membrane skeleton (Ritchie and Kusumi, 2004). Instead, the localization of envelope proteins on the plasma membrane of infected cells might be pre-programmed before the onset of virus budding by sorting signals. The newly synthesized envelope proteins form clusters on the plasma membrane. The segregation distance between the protein clusters may correlate to the length of NCs so that two clusters would be localized on the two polar ends of a BV particle. On the other hand, the NCs might anchor at the envelope protein clustering sites through direct interaction with the envelope proteins. Deletion of part of the cytoplasmic tail of the envelope proteins was shown to reduce the production of infectious BVs (Long et al., 2006, Oomens and Blissard, 1999) possibly due to improper envelopment. The NC–envelope protein interaction has also been found crucial for virus assembly of several other enveloped viruses such as coronavirus (Narayanan et al., 2000), ebola virus (Noda et al., 2006), hantavirus (Wang et al., 2010a) and alphavirus (Lopez et al., 1994). Baculoviruses have been reported to utilize nuclear actin to transport from virogenic stroma in the nucleocapsids to the periphery of the nucleus through the interaction of the viral capsid protein VP80 (Marek et al., 2011). In addition these viruses translocate their NCs from the nucleus to the cell periphery along microtubules through interaction with the viral capsid proteins (VP39 and EXON0) and motor protein Kinesin-1 (Danquah et al., 2012). Thus, the NC may dictate actin and Kinesin-1 through its capsid proteins to travel to the plasma membrane where the NC interacts with envelope proteins starting the budding process.
The envelope of both SeMNPV and AcMNPV exhibits a typical bilayer structure. However, the thickness of the envelope was 6–7 nm, which exceeds the thickness of average biological membranes of about 4 nm (Mitra et al., 2004). The latter though were measured using a different methodology (X-ray scattering). Since plasma membranes and intact cells cannot be analyzed using cryo-EM, it would be of interest to apply X-ray scattering to BVs and to see if the difference in envelope thickness is real or an artifact of the respective technologies.
The unusual thickness of viral envelopes may suggest that some other envelope components such as proteins may contribute to the enhanced bilayer thickness. Although the current cryo-EM analysis was not able to resolve more details of the viral envelope, proteomic analysis of alphabaculovirus BVs revealed several other envelope proteins besides F and GP64, among which the most abundant one is ubiquitin (Hou et al., 2013, Wang et al., 2010c). Ubiquitin has been suggested to be attached to the inner membrane of the viral envelope by a type of phospholipid anchor (Guarino et al., 1995). Therefore, we propose ubiquitin as a BV structural protein that among others extends an inner layer on the viral envelope. This may explain a thicker appearance of the envelope in cryo-EM. High-resolution cryo-tomography could confirm the presence of this third layer of the baculovirus BV envelope.
The ‘Y’- and ‘bungee’-shaped appearance of the spikes for AcMNPV and SeMNPV BVs, respectively, in cryo-EM (Fig. 2) differs from what is expected on the basis of the symmetric, trimeric structure of envelope fusion proteins in general. It could be that what we see here is a fusion protein intermediate in its extended conformation. This conformation may also be different between F (SeMNPV) and GP64 (AcMNPV) explaining their unique appearance. Since information on the appearance of envelope fusion proteins is derived from vertebrate viruses, it is possible that such structures are different in invertebrate envelope fusion proteins. Further analysis using known vertebrate F proteins, such as those from parainfluenza virus (class I) or thogoto virus (class III), as well as other baculovirus F proteins using cryo-EM should provide further support for our observations.
In our cryo-EM micrographs of NCs the filled and empty capsids exhibit similar diameters and lengths (Fig.3A and C). This suggests that the capsid assembly is independent on the DNA packaging. Moreover, the two ends of NCs appear to be more fragile compared to the rest of the NC. These two ends are most likely composed with various structural proteins other than the major capsid protein VP39 [reviewed in (Slack and Arif, 2007)]. Once the ends are open, the viral DNA leaks out as an unstructured form (Fig.3B). These results support the NC assembly model proposed by Fraser (1986). In this model, the viral genome inserts into a pre-assembled capsid sheath through the cap structure (Fraser, 1986), which is consistent with the DNA packaging process for other large double-stranded (ds) DNA viruses such as bacteriophage lambda (Fuller et al., 2007b), T4 (Fuller et al., 2007a) and herpes virus (Mettenleiter et al., 2006). However, in case of the most extensively studied large dsDNA viruses including bacteriophage T4 and lambda, the terminase, an ATP-driven packaging motor protein, is responsible for DNA insertion into their pre-assembled NCs (Fuller et al., 2007a, Ortega et al., 2007). Although packaging the large circular genome into a capsid shell is an energy dependent process, up to now no protein associated with the NC or DNA has been demonstrated to be responsible for viral genome packaging into preformed BV capsids. The essential structural proteins at both ends of NC, which are related to NC morphogenesis and maturation, are not homologous to the bacteriophage terminase protein sequence. However, deletion of VP80 (Tang et al., 2008) or 38K (Wu et al., 2006) results in half filled or empty NCs, respectively without affecting DNA replication, indicating these two proteins may be involved in DNA encapsidation. Furthermore, yeast two-hybrid assays demonstrated that the 38K protein interacts with VP80 (Wu et al., 2008). The latter protein interacts with both F-actin and DNA (Marek et al., 2012). In addition, actin has been indicated to be involved in the DNA packaging since actin is transported from the cytoplasm to the nucleus (Volkman et al., 1992) and interacts with NC components (Lanier and Volkman, 1998) upon viral infection whereas inhibition of actin polymerization results in the empty NCs (Volkman, 1988). Taken together, VP80, 38K and actin cytoskeleton may function collectively on the DNA encapsidation rather than one specific motor protein directly interacting with viral DNA.
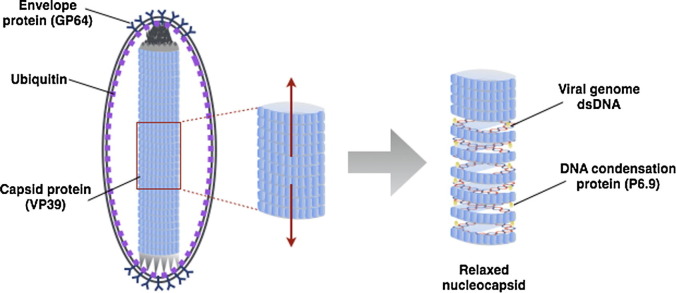
Based on our cryo-EM visualization we propose a new model for baculovirus BV structure (Fig. 4 ). The model emphasizes the strong interactions of envelopes at polar ends with the NCs and the space at the lateral region between NC and envelope of the BVs. The major envelope protein GP64 mediating fusion between viral envelope and cellular membrane, is predominantly distributed on the envelope around the both ends of the NC suggesting a vertical orientation to the cellular membrane during virus entry. The capsid sheath has a compact helical structure.
Fig. 4.
Structural model of the baculovirus BV represented by AcMNPV. Major envelope proteins GP64 are distributed at the polar ends of the ovoid shaped envelope. The DNA genome is condensed by P6.9 protein and organized as super coil in the nucleocapsid. The capsid sheath containing major protein VP39 forms a spiral structure visualized by its relaxed state.
The fragile nature of BVs compared to rigidity of ODVs (Slack and Arif, 2007) is the consequence of the baculovirus infection cycle. The BVs targets on cell-to-cell infection and therefore, are protected by the body fluid of host. The ODVs target per os infection of the host midgut epithelium; BVs do not have to survive the hostile conditions outside the host and hence this may permit a more fragile virion structure. Our understanding of baculovirus BV structure has long been hampered by the fragile nature of the virus and the limitations of conventional imaging techniques. Previous BV morphology has only been studied with negative-staining, which causes dramatic change of inner osmotic pressure of virions due to the air-drying procedure, which subsequently deform the viral envelope. In addition, high-speed centrifugal sedimentation and prolonged virus production time in cell culture introduce disruption of viral envelope and heterogeneity of viral morphology (Summers and Volkman, 1976, Zherebtsova et al., 1972). All these artifacts have led to a biased morphological view of BVs. Our cryo-EM analysis provides novel insight into the near-native structure and virus assembly of baculovirus BVs.
Acknowledgements
This work was supported by a Programme Strategic Scientific Alliances grant from the Ministry of Science and Technology of China and the Royal Academy of Arts and Sciences of the Netherlands (No. 2008DFB30220 to China and No. 08-PSA-BD-01 to the Netherlands).
References
- Adams J.R., Goodwin R.H., Wilcox T.A. Electron microscopic investigations on invasion and replication of insect baculoviruses in vivo and in vitro. Biol. Cellulaire. 1977;28:261–268. [Google Scholar]
- Ayres M.D., Howard S.C., Kuzio J., Lopez-Ferber M., Possee R.D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- Beaton C.D., Filshie B.K. Comparative ultrastructural studies of insect granulosis and nuclear polyhedrosis viruses. J. Gen. Virol. 1976;31:151–161. doi: 10.1099/0022-1317-31-2-151. [DOI] [PubMed] [Google Scholar]
- Bergoin M., Devauchelle G., Vago C. Electron microscopy study of the pox-like virus of Melolontha melolontha L (Coleoptera, Scarabeidae) Arch. Gesamte Virusforsch. 1969;28:285–302. doi: 10.1007/BF01240944. [DOI] [PubMed] [Google Scholar]
- Blissard G.W., Rohrmann G.F. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 1990;35:127–155. doi: 10.1146/annurev.en.35.010190.001015. [DOI] [PubMed] [Google Scholar]
- Blissard G.W., Wenz J.R. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucias D.G., Pendland J.C. Kluwer Academic Publishers; Norwell: 1998. Baculoviruses. Principles of Insect Pathology. pp. 111–146. [Google Scholar]
- Braunagel S.C., Summers M.D. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology. 1994;202:315–328. doi: 10.1006/viro.1994.1348. [DOI] [PubMed] [Google Scholar]
- Bud H.M., Kelly D.C. An electron microscope study of partially lysed baculovirus nucleocapsids: the intranucleocapsid packaging of viral DNA. J. Ultrastruct. Res. 1980;73:361–368. doi: 10.1016/s0022-5320(80)90095-7. [DOI] [PubMed] [Google Scholar]
- Burley S.K., Miller A., Harrap K.A., Kelly D.C. Structure of the baculovirus nucleocapsid. Virology. 1982;120:433–440. doi: 10.1016/0042-6822(82)90043-5. [DOI] [PubMed] [Google Scholar]
- Cyrklaff M., Risco C., Fernández J.J., Jiménez M.V., Estéban M., Baumeister W., Carrascosa J.L. Cryo-electron tomography of vaccinia virus. Proc. Natl. Acad. Sci. USA. 2005;102:2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danquah J.O., Botchway S., Jeshtadi A., King L.A. Direct interaction of baculovirus capsid proteins VP39 and EXON0 with Kinesin-1 in insect cells determined by fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy. J. Virol. 2012;86:844–853. doi: 10.1128/JVI.06109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duteyrat J.L., Gelfi J., Bertagnoli S. Ultrastructural study of myxoma virus morphogenesis. Arch. Virol. 2006;151:2161–2180. doi: 10.1007/s00705-006-0791-2. [DOI] [PubMed] [Google Scholar]
- Fraser M.J. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J. Ultra. Mol. Struct. Res. 1986;95:189–195. [Google Scholar]
- Fuller D.N., Raymer D.M., Kottadiel V.I., Rao V.B., Smith D.E. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. USA. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller D.N., Raymer D.M., Rickgauer J.P., Robertson R.M., Catalano C.E., Anderson D.L., Grimes S., Smith D.E. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L.A., Smith G., Dong W. Ubiquitin is attached to membranes of baculovirus particles by a novel type of phospholipid anchor. Cell. 1995;80:301–309. doi: 10.1016/0092-8674(95)90413-1. [DOI] [PubMed] [Google Scholar]
- Harrap K.A. The structure of nuclear polyhedrosis viruses: II. The virus particle. Virology. 1972;50:124–132. doi: 10.1016/0042-6822(72)90352-2. [DOI] [PubMed] [Google Scholar]
- Harrison R., Hoover K. In: Insect Pathology. Vega F.E., Kaya H.K., editors. Elsevier Inc.; 2012. Baculoviruses and other occluded insect viruses; pp. 73–131. [Google Scholar]
- Herniou E.A., Arif B.M., Becnel J.J., Blissard G.W., Bonning B., Harrison R., Jehle J.A., Theilmann D.A., Vlak J.M. In: Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. King A.M.Q., et al., editors. Elsevier Academic Press; 2012. Family Baculoviridae; pp. 163–173. [Google Scholar]
- Hou D., Zhang L., Deng F., Fang W., Wang R., Liu X., Guo L., Rayner S., Chen X., Wang H., Hu Z. Comparative proteomics reveal fundamental structural and functional differences between the two progeny phenotypes of a baculovirus. J. Virol. 2013;87:829–839. doi: 10.1128/JVI.02329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJkel W.F.J., van Strien E.A., Heldens J.G., Broer R., Zuidema D., Goldbach R.W., Vlak J.M. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J. Gen. Virol. 1999;80:3289–3304. doi: 10.1099/0022-1317-80-12-3289. [DOI] [PubMed] [Google Scholar]
- IJkel W.F.J., Westenberg M., Goldbach R.W., Blissard G.W., Vlak J.M., Zuidema D. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology. 2000;275:30–41. doi: 10.1006/viro.2000.0483. [DOI] [PubMed] [Google Scholar]
- Jehle J.A., Blissard G.W., Bonning B.C., Cory J.S., Herniou E.A., Rohrmann G.F., Theilmann D.A., Thiem S.M., Vlak J.M. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch. Virol. 2006;151:1257–1266. doi: 10.1007/s00705-006-0763-6. [DOI] [PubMed] [Google Scholar]
- Kadlec J., Loureiro S., Abrescia N.G.A., Stuart D.I., Jones I.M. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- Lanier L.M., Volkman L.E. Actin binding and nucleation by Autographa californica M nucleopolyhedrovirus. Virology. 1998;243:167–177. doi: 10.1006/viro.1998.9065. [DOI] [PubMed] [Google Scholar]
- Long G., Pan X., Westenberg M., Vlak J.M. Functional role of the cytoplasmic tail domain of the major envelope fusion protein of group II baculoviruses. J. Virol. 2006;80:11226–11234. doi: 10.1128/JVI.01178-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez S., Yao J.S., Kuhn R.J., Strauss E.G., Strauss J.H. Nucleocapsid–glycoprotein interactions required for assembly of alphaviruses. J. Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O., Westenberg M., Vlak J.M., Zuidema D., Blissard G.W. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 2002;76:5729–5736. doi: 10.1128/JVI.76.11.5729-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O.Y., Cruz-Alvarez M., Blissard G.W. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J. Virol. 2003;77:328–339. doi: 10.1128/JVI.77.1.328-339.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek M., Merten O.W., Galibert L., Vlak J.M., van Oers M.M. Baculovirus VP80 protein and the F-actin cytoskeleton interact and connect the viral replication factory with the nuclear periphery. J. Virol. 2011;85:5350–5362. doi: 10.1128/JVI.00035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek M., Merten O.W., Francis-Devaraj F., van Oers M.M. Essential C-terminal region of the baculovirus minor capsid protein VP80 binds DNA. J. Virol. 2012;86:1728–1738. doi: 10.1128/JVI.05600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marheineke K., Grunewald S., Christie W., Reilander H. Lipid composition of Spodoptera frugiperda (Sf9) and Trichoplusia ni (Tn) insect cells used for baculovirus infection. FEBS Lett. 1998;441:49–52. doi: 10.1016/s0014-5793(98)01523-3. [DOI] [PubMed] [Google Scholar]
- Mettenleiter T.C., Klupp B.G., Granzow H. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Mitra K., Ubarretxena-Belandia I., Taguchi T., Warren G., Engelman D.M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Ebihara H., Muramoto Y., Fujii K., Takada A., Sagara H., Kim J.H., Kida H., Feldmann H., Kawaoka Y. Assembly and budding of Ebolavirus. PLoS Pathog. 2006;2:e99. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano K., Vanarsdall A.L., Mikhailov V.S., Rohrmann G.F. Conserved molecular systems of the Baculoviridae. Virology. 2006;344:77–87. doi: 10.1016/j.virol.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Oomens A.G., Blissard G.W. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology. 1999;254:297–314. doi: 10.1006/viro.1998.9523. [DOI] [PubMed] [Google Scholar]
- Ortega M.E., Gaussier H., Catalano C.E. The DNA maturation domain of gpA, the DNA packaging motor protein of bacteriophage lambda, contains an ATPase site associated with endonuclease activity. J. Mol. Biol. 2007;373:851–865. doi: 10.1016/j.jmb.2007.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M.N., Russell R.L.Q., Rohrmann G.F. Characterization of a baculovirus-encoded protein that is associated with infected-cell membranes and budded virions. Virology. 2001;291:22–31. doi: 10.1006/viro.2001.1191. [DOI] [PubMed] [Google Scholar]
- Ritchie K., Kusumi A. Role of the membrane skeleton in creation of microdomains. Subcell. Biochem. 2004;37:233–245. doi: 10.1007/978-1-4757-5806-1_7. [DOI] [PubMed] [Google Scholar]
- Slack J., Arif B.M. The baculoviruses occlusion-derived virus: virion structure and function. Adv. Virus Res. 2007;69:99–165. doi: 10.1016/S0065-3527(06)69003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers M.D., Volkman L.E. Comparison of biophysical and morphological properties of occluded and extracellular nonoccluded baculovirus from in vivo and in vitro host systems. J. Virol. 1976;17:962–972. doi: 10.1128/jvi.17.3.962-972.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.L., Ueda N., Mercer A.A., Fleming S.B. Investigation of orf virus structure and morphogenesis using recombinants expressing FLAG-tagged envelope structural proteins: evidence for wrapped virus particles and egress from infected cells. J. Gen. Virol. 2009;90:614–625. doi: 10.1099/vir.0.005488-0. [DOI] [PubMed] [Google Scholar]
- Tang X.D., Xu Y.P., Yu L.L., Lang G.J., Tian C.H., Zhao J.F., Zhang C.X. Characterization of a Bombyx mori nucleopolyhedrovirus with Bmvp80 disruption. Virus Res. 2008;138:81–88. doi: 10.1016/j.virusres.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Volkman L.E. Autographa californica MNPV nucleocapsid assembly: inhibition by cytochalasin D. Virology. 1988;163:547–553. doi: 10.1016/0042-6822(88)90295-4. [DOI] [PubMed] [Google Scholar]
- Volkman L.E., Goldsmith P.A., Hess R.T., Faulkner P. Neutralization of budded Autographa californica NPV by a monoclonal antibody: identification of the target antigen. Virology. 1984;133:354–362. doi: 10.1016/0042-6822(84)90401-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman L.E., Talhouk S.N., Oppenheimer D.I., Charlton C.A. Nuclear F-actin: a functional component of baculovirus-infected lepidopteran cells? J. Cell Sci. 1992;103:15–22. [Google Scholar]
- Wang H., Alminaite A., Vaheri A., Plyusnin A. Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein Gn. Virus Res. 2010;151:205–212. doi: 10.1016/j.virusres.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Wang M., Tan Y., Yin F., Deng F., Vlak J.M., Hu Z., Wang H. The F-like protein Ac23 enhances the infectivity of the budded virus of gp64-null Autographa californica multinucleocapsid nucleopolyhedrovirus pseudotyped with baculovirus envelope fusion protein F. J. Virol. 2008;82:9800–9804. doi: 10.1128/JVI.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tuladhar E., Shen S., Wang H., Van Oers M.M., Vlak J.M., Westenberg M. Specificity of baculovirus P6.9 basic DNA-binding proteins and critical role of the C terminus in virion formation. J. Virol. 2010;84:8821–8828. doi: 10.1128/JVI.00072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Deng F., Hou D., Zhao Y., Guo L., Wang H., Hu Z. Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J. Virol. 2010;84:7233–7242. doi: 10.1128/JVI.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenberg M. Wageningen University; Wageningen, The Netherlands: 2004. Functional Analysis of a Novel Baculovirus Envelope Fusion Protein. ISBN 90-8504-010-8. [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Lin T., Pan L., Yu M., Li Z., Pang Y., Yang K. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. J. Virol. 2006;80:11475–11485. doi: 10.1128/JVI.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.B., Liang H.Q., Kan J.S., Liu C., Yuan M.J., Liang C., Yang K., Pang Y. Autographa californica multiple nucleopolyhedrovirus 38K is a novel nucleocapsid protein that interacts with VP1054, VP39, VP80, and itself. J. Virol. 2008;82:12356–12364. doi: 10.1128/JVI.00948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotto P.M., Kessing B.D., Maruniak J.E. Phylogenetic interrelationships among baculoviruses: evolutionary rates and host associations. J. Invertebr. Pathol. 1993;62:147–164. doi: 10.1006/jipa.1993.1090. [DOI] [PubMed] [Google Scholar]
- Zherebtsova E.N., Strokovskaya L.I., Gudz-Gorban A.P. Subviral infectivity in nuclear polyhedrosis of the great wax moth (Galleria mellonella L.) Acta Virol. 1972;16:427–431. [PubMed] [Google Scholar]