Abstract
In this study, a highly specific ratiometric two-photon fluorescent probe GP-BAN was developed and well-characterized to monitor dipeptidyl peptidase IV in plasma and living systems. GP-BAN was designed on the basis of the catalytic properties and substrate preference of DPP-IV, and it could be readily hydrolyzed upon addition of DPP-IV under physiological conditions. Both reaction phenotyping and inhibition assays demonstrated that GP-BAN displayed good reactivity and high selectivity towards DPP-IV over other human serine hydrolases including FAP, DPP-VIII, and DPP-IX. The probe was successfully used to monitor the real activities of DPP-IV in complex biological systems including diluted plasma, while it could be used for high throughput screening of DPP-IV inhibitors by using human plasma or tissue preparations as enzyme sources. As a two-photon fluorescent probe, GP-BAN was also successfully used for two-photon imaging of endogenous DPP-IV in living cells and tissues, and showed high ratiometric imaging resolution and deep-tissue penetration ability. Taken together, a ratiometric two-photon fluorescent probe GP-BAN was developed and well-characterized for highly selective and sensitive detection of DPP-IV in complex biological systems, which could serve as a promising imaging tool to explore the biological functions and physiological roles of this key enzyme in living systems.
Keywords: Dipeptidyl peptidase IV, Two-photon fluorescent probe, Cells and tissues imaging, Inhibitor screening
Highlights
-
•
A highly specific two-photon fluorescent probe GP-BAN has been developed.
-
•
The probe was successfully applied to detect DPP-IV in complex biological samples.
-
•
The probe could be used for high throughput screening of DPP-IV inhibitors.
-
•
The probe was successfully used for two-photon imaging of DPP-IV in living cells and tissues.
1. Introduction
Dipeptidyl peptidase IV (DPP-IV, EC 3.4.14.5, CD26) is a multifunctional serine protease enzyme with key roles in the control of endocrine and immune function, cell metabolism, growth and adhesion (Hildebrandt et al., 2000). As an enzyme, DPP-IV cleaves the N-terminal dipeptide of many biologically active peptides (such as incretin hormones glucagon-like peptide-1 and glucose-dependent insulinotropic polypeptide) from the oligopeptide chain with proline or alanine at the second position (Mentlein, 1999). DPP-IV is widely expressed in various tissues or cells (high mRNA levels and abundant protein levels in the kidney and small intestine) as a membrane-anchored cell surface protein or as a soluble form in the plasma and other body fluids (Kameoka et al., 1993, Kim et al., 2014, Uhlén et al., 2015). The ubiquitous expression and the enzymatic actions of DPP-IV make this enzyme to be involved in many biological or pathological processes, including the regulation of glucose metabolism (Drucker and Nauck, 2006, Egan et al., 2014). Besides of being a famous therapeutic target for the treatment of type 2 diabetes, DPP-IV also exhibits co-stimulatory role in T-cell activation, adhesion to extracellular matrix proteins, and serves as the functional receptor for Middle East respiratory syndrome coronavirus infection (Ohnuma et al., 2008, Raj et al., 2013). Recently, DPP-IV has come back into the centre of attention as a novel molecular marker or a potential therapeutic target for cancer (Cordero et al., 2009, Javidroozi et al., 2012, Dahan et al., 2014, Arrebola et al., 2014, Davies et al., 2015). Furthermore, increasing evidence has demonstrated that the abnormal presence or altered level of DPP-IV in plasma can serve as a potential biomarker for early diagnosis and prognosis evaluation of many diseases, including cholestasis, non-alcoholic fatty liver disease and lung diseases (Perner et al., 1999, Firneisz et al., 2010, Sánchez-Otero et al., 2014). Thus, the accurate measurement of DPP-IV in complex biological samples, especially in plasma, is of great importance for disease diagnosis, drug discovery and clinical practice.
Recent years have witnessed a rapid expansion in the use of small molecule fluorescent probes to detect and monitor the key enzymes in human biological processes, due to their inherent advantages including high selectivity and sensitivity, non-destructiveness, easy management and capability of being applicable to high-throughput detection (Ueno and Nagano, 2011, Vendrell et al., 2012). Notably, dipeptidyl peptidases (DPPs) and fibroblast activation protein (FAP) are two highly homologous prolyl-cleaving dipeptidases, and the substrate spectra of FAP and DPPs (such as DPP-IV, DPP8 and DPP9) are commonly overlapped (Waumans et al., 2015). Up to date, most of DPP-IV substrates were found to be cleaved by FAP, DPP8 and DPP9 as well (Ajami et al., 2008, Keane et al., 2011, Lu et al., 2011). Although several fluorescent probes for DPP-IV have been developed and used in vitro bioassays, these probes do not have enough isoform selectivity to precisely evaluate the real activities of DPP-IV in complex biological systems, such as living cells and body fluids (Grant et al., 2002, Ho et al., 2006, Kawaguchi et al., 2010, Wang et al., 2016, Wang et al., 2016, Gong et al., 2016). Furthermore, almost all reported fluorescent probes for DPP-IV work with one photo microscopy (OPM) and monitor DPP-IV activity by intensity-responsive fluorescence signal, which can be easily interfered with the excitation and emission efficiency, probe concentration, biological matrix and environmental conditions (Zipfel et al., 2003). In contrast to traditional single-photon excited fluorescent probes, two-photon excited (TPE) fluorescent probes are induced by two-photon excitation with near-infrared (NIR) photons, which can minimize auto-fluorescence background, reduce phototoxicity or photo-damage to biological samples and increase specimen penetration, and thus more suitable for deep-tissue bioimaging and 3D imaging in living systems (Helmchen and Denk, 2005; Kim and Cho, 2015). These advantages make the TPE fluorescent probes excellent tools to investigate biomolecules in living cells and living tissues (Miao et al., 2014, Dai et al., 2015, Wang et al., 2015, Li et al., 2017). However, to the best of our knowledge, the TPE fluorescent probes for highly selective detection of DPP-IV in complex biological systems have not been reported yet. Thus it is urgently desirable to develop an isoform specific TPE probe for sensing DPP-IV in body fluids and living systems.
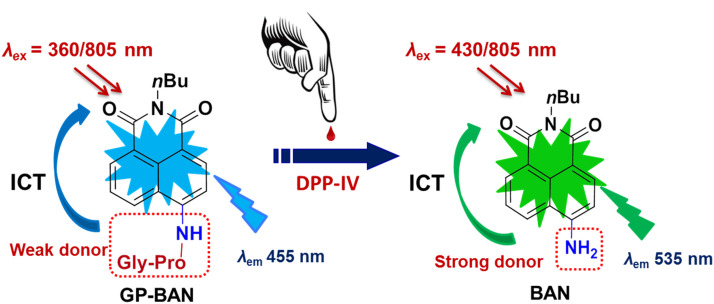
In this study, an isoform specific two-photon ratiometric fluorescent probe GP-BAN has been developed and well characterized for highly selective and sensitive sensing of DPP-IV in complex biological systems, on the basis of the catalytic properties and substrate preference of DPP-IV. Upon addition of DPP-IV, the probe can be readily hydrolyzed to release N-butyl-4-amino-1,8-naphthalimide (BAN), which brings remarkable changes in both colour and fluorescence intensity. The newly developed probe exhibits excellent specificity and ultrahigh sensitivity for DPP-IV over other hydrolases and common biological interferences in human body. Furthermore, GP-BAN has been successfully used for monitoring the real activities of DPP-IV in various biological samples, as well as for imaging endogenous DPP-IV in living cells and in deep-tissues with high ratiometric imaging resolution and deep-tissue penetration.
2. Experimental
2.1. Synthesis of GP-BAN
The detailed synthesis and structural characterization of GP-BAN are described in the Supplementary data (Scheme S1, Figs. S1–S3).
2.2. General procedure for the measurement of DPP-IV activities
All the measurements of DPP-IV activity were carried out in 100 mM phosphate buffer (PBS, pH =7.4). With a total incubation volume of 0.2 mL, culture mixtures that contained PBS and mentioned enzymes protein or microsomes were mixed gently. Then a proper amount of GP-BAN was added to start the reactions (contain 1% DMSO). After incubation at 37 °C in a shaking water bath for 60 min, an equal volume of ice-cold acetonitrile was added to terminate the reactions. The mixtures were then centrifuged for 5 min at 20,000g. Aliquots of supernatant were taken for further fluorescence analysis (Gain 60). To ensure that metabolites formation was enzyme dependent, control incubations were carried out without enzyme sources at the same time. All assays were performed in duplicates.
2.3. Sensing DPP-IV activities in HKM, HIM and HLM
In order to evaluate the applicability of GP-BAN as a selective indicator for DPP-IV activity in human biological samples, the formation rates of the GP-BAN metabolite were used to describe DPP-IV activities in a panel of three microsomes (HKM, HIM, HLM). The DPP-IV quantity was determined by ELISA and total protein ratio. In addition, the formation rates of BAN in HKM were compared with other microsomes. The concentrations of DPP-IV in HKM were also compared with others.
2.4. Sensing DPP-IV activities in human plasma and correlation studies
Plasma samples were collected from 13 healthy donors. Plasma sample was diluted 100-fold with a solution of GP-BAN (100 μM) in PBS. The fluorescence spectra were measured in PBS-acetonitrile (v:v =1:1, pH=7.4). The excitation wavelengths were set at 360 nm and 430 nm respectively. In addition, these plasma samples were selected to study the correlation relationship between the DPP-IV quantity determined by ELISA and the hydrolytic rate of GP-BAN hydrolyzed by DPP-IV.
2.5. Rapid screening of DPP-IV Inhibitors
The assay was performed in a 96-well microplate. A mixture of GP-BAN (100 μM), two DPP-IV selective inhibitors of sitagliptin (0–0.5 μM) and vildagliptin (0–2.5 μM), and recombinant DPP-IV (1 μg/mL) or human plasma (2 μL) in PBS (pH 7.4) containing less than 1% DMSO (total volume 200 μL), was incubated for 5 min at 37 °C. The IC50 values were determined by incubating GP-BAN with different concentrations of the inhibitor. The inhibitory effects were expressed as percent decrease in fluorescence intensities ratio (I 535/I 460). Data were fit to log (inhibitor) vs. normalized response -Variable slope equation in GraphPad Prism 6.0 (San Diego, CA).
The procedure was performed as described above. A mixture of GP-BAN (100 μM), two DPP-IV selective inhibitors of sitagliptin (0–2.5 μM) and vildagliptin (0–2.5 μM), and recombinant DPP-IV (1 μg/mL) or human intestine microsome (HIM, 20 μg/mL) in PBS (pH 7.4) containing less than 1% DMSO (total volume 200 μL), was incubated for 5 min at 37 °C. The IC50 values were determined by incubating GP-BAN with different concentrations of the inhibitor. The inhibitory effects were expressed as percent decrease in fluorescence intensities ratio (I 535/I 460). Data were fit to log (inhibitor) vs. normalized response-Variable slope equation in GraphPad Prism 6.0 (San Diego, CA).
2.6. Cell culture and live cell imaging
HepG2 or Caco-2 cells were grown in MEM/EBSS culture medium (contain 10% FBS). Cells were seeding at a density of 1×105 cells per dish (Φ 20 mm) and incubated in a humidified incubator containing 5% CO2 at 37 °C overnight. The adherent cells were washed twice with FBS-free culture medium and incubated with/without 50 μM vildagliptin (preparing in FBS free culture medium) for 30 min at 37 °C in 5% CO2 incubator. The stock solution of probe GP-BAN (50 mM) in DMSO was diluted into the cell culture media (FBS free) to a final concentration of 25 μM. The cells were then incubated at 37 °C for another 60 min, followed by rinsing with PBS (pH =7.4) for three times to remove the extracellular probe and imaged under confocal microscope (Olympus, FV1000). In one photon microscopy assay, excitation wavelength was set at 405 nm, blue emission was collected with a 430–480 nm window, and green emission was collected with a 520–570 nm window. In two-photon mode, images were acquired under 805 nm excitation and fluorescent emission windows of 420–460 nm (blue) and 495–540 nm (green).
2.7. Preparation of fresh mouse kidney slices and confocal fluorescence imaging
Slices were prepared from the kidney of 7-day old mice. Slices were cut to 1 mm thickness by a vibrating-blade microtome and placed into glass-bottomed dish. For the fluorescence imaging experiments, slices were incubated with 20 μM GP-BAN in PBS buffer bubbled with 95% O2 and 5% CO2 at 37 °C for 1 h. Slices were then washed three times with PBS and observed under a two-photon confocal microscope (Olympus FV1000).
3. Results and discussion
3.1. Design and synthesis of GP-BAN
It is well-known that DPP-IV can cleave the dipeptides from the N-terminal end of peptides or polypeptides with proline, alanine, serine, valine, glycine or leucine at the penultimate position. Particularly, DPP-IV prefers to hydrolyze the substrates with proline at the second position and the glycine at the terminal position (Grant et al., 2002, Ho et al., 2006). These facts inspired us that it is possible to develop a preferred fluorescent substrate for DPP-IV by introduction of a Gly-Pro substitute on a basic fluorophore. In this context, a ratiometric two-photon fluorescent probe (GP-BAN) was designed for sensing DPP-IV on the basis of the substrate preference of this key enzyme and the principle of the intramolecular charge transfer (ICT). Specifically, N-butyl-4-amino-1,8-naphthalimide was selected as the basic fluorophore due to its desirable photophysical properties including large Stokes shifts, high photostability and significant two-photon absorbability at near infrared wavelength (Zhang et al., 2011, Hettiarachchi et al., 2014, Jin et al., 2015). The probe GP-BAN was synthesized by the incorporation of a glycylprolyl (Gly-Pro) group into BAN via amidation, which served as a reserved metabolic site for DPP-IV ( Scheme 1). The detailed synthetic procedure for GP-BAN was provided in Scheme S1, while the chemical structure of GP-BAN was fully characterized by HRMS, 1H NMR and 13C NMR (Figs. S1–S3).
Scheme 1.
Proposed response mechanism of the GP-BAN/BAN system for DPP-IV detection. GP-BAN can be readily cleaved upon addition of DPP-IV and such biotransformation leads to the release of BAN, which strengthens intramolecular charge transfer and brings the redshift in both absorbance and fluorescence spectra.
3.2. Spectral properties of GP-BAN toward DPP-IV
As expected, GP-BAN could be readily hydrolyzed under physiological conditions and release a single metabolite upon addition of DPP-IV or DPP-IV containing biological samples, such as human kidney microsomes (HKM), human intestine microsomes (HIM) and human liver microsomes (HLM). The metabolite was identified as BAN by comparison of LC retention times, UV and MS spectra with the standard (Fig. S4). To demonstrate the ratiometric feature of the GP-BAN/BAN system, the UV–vis absorption and fluorescence emission spectra of GP-BAN with or without DPP-IV were depicted. As shown in Fig. S5a, GP-BAN exhibited an absorption band at 360 nm in phosphate buffer saline (PBS)-acetonitrile (v/v =1:1, pH 7.4 at 37 °C), and the solution was colorless due to the strong electron-withdrawal effect of the glycylprolyl moiety. In sharp contrast, upon addition of DPP-IV, the absorption at 360 nm decreased evidently, while a new absorption band at 430 nm emerged. Such a large red shift in the absorption behaviour changed the colour of the solution from colorless to yellow, indicating that GP-BAN can serve as a ‘‘naked-eye’’ colorimetric indicator for DPP-IV. Consistently, the maximum emission peak also underwent a desirable red-shift from 455 to 535 nm upon addition of DPP-IV (Fig. S5b). The desirable red-shifted emission after hydrolysis should be attributed to the stronger ICT efficiency of the released amino compound BAN. Further investigations showed that GP-BAN is quite stable in the pH range from 2.0 to 7.8, while the pH values do not affect the fluorescence intensity of both GP-BAN and BAN over the pH range of 2.0–7.8 (Fig. S6). The fluorescence quantum yields of GP-BAN and BAN in PBS-acetonitrile (v/v =1:1) were also determined as 0.45 and 0.67, respectively, by using fluorescein as the standard. These findings suggested that GP-BAN could function properly under physiological conditions (pH 7.4 at 37 °C).
3.3. Selectivity screening of GP-BAN
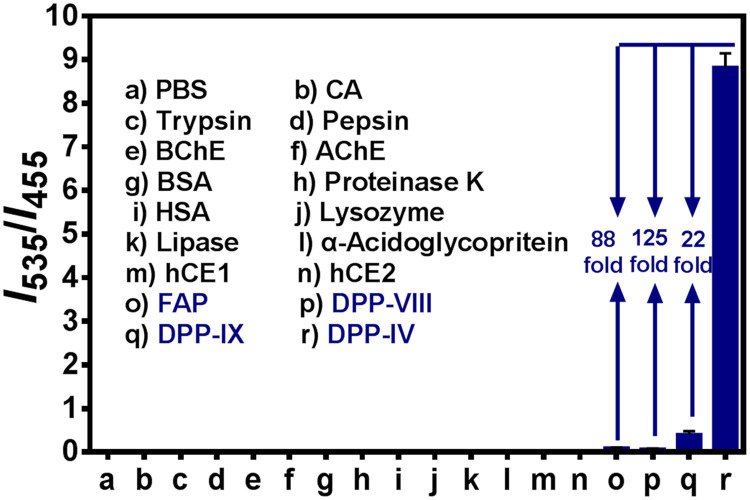
The specificity of GP-BAN hydrolysis was then investigated by a panel of human hydrolases including most known prolyl-cleaving dipeptidases. As shown in Fig. 1, only DPP-IV can elicit a marked increase in the fluorescence intensity at 535 nm, while the fluorescence intensity at 455 nm is decreased dramatically. In sharp contrast, other hydrolases including carbonic anhydrase (CA), trypsin, pepsin, butyryl-cholinesterase (BChE), acetylcholinesterase (AChE), proteinase K, human serum albumin (HSA), lysozyme, Lipase, α-acidoglycoprotein, human carboxylesterases (hCE1 and hCE2), and other human prolyl-cleaving dipeptidases including FAP, DPP-VIII, and DPP-IX led to negligible changes in fluorescence intensity. Notably, the rate of GP-BAN hydrolysis in DPP-IV is 22-fold higher than that in DPP-IX, and 125-fold higher than that in other DPP-VIII, suggesting that GP-BAN is an isoform specific probe for DPP-IV. Furthermore, the fluorescence performance of GP-BAN toward DPP-IV cannot be influenced by common biological metallic ions or amino acids in human tissues or fluids (Fig. S8). All these findings clearly demonstrate that GP-BAN displays an excellent specificity for DPP-IV over other human hydrolases and biologically relevant species in human tissues or fluids and thus holds great promise for highly selective detection of DPP-IV in complex biological samples.
Fig. 1.
Fluorescence intensities ratio (I535/I455) of GP-BAN (100 μM) upon addition of different hydrolases. The spectra were measured in PBS-acetonitrile (v/v =1:1, pH 7.4) at 37 °C for 60 min λex =360/430 nm.
3.4. Sensitivity and detection limit of GP-BAN based assay
For accurate measurement of the real activities of target enzyme in various biological samples, the linear fluorescent response with enzyme concentration is very essential. As shown in Fig. S9, the formation of BAN is time and DPP-IV dependent, the formation rates of BAN in DPP-IV are linear with the incubation time up to 60 min, while the ratio of fluorescence intensities (535 nm/455 nm) exhibits a good linearity (R2 >0.98) to the increasing concentrations of DPP-IV in the range from 0 to 80 ng/mL. The detection limit (3σ/slope) of GP-BAN for DPP-IV is also estimated as 0.78 ng/mL (7.43 pmol/L). It has been reported that the concentrations of DPP-IV in human serum have been determined ranging from 200 ng/mL to 700 ng/mL, while its expression levels in other tissues including kidney, liver and intestine are much higher than that in serum (Cordero et al., 2009). Thus, the sensitivity of GP-BAN is high enough to determine DPP-IV in human plasma or other complex biological samples. In addition, we compared the detection limits of GP-BAN and GP-AMC (a commercially available fluorescent probe for DPP-IV) using various concentrations of human DPP-IV. Under identical conditions, the sensitivity of the GP-BAN was about 4-fold more sensitive than that of GP-AMC. Such high sensitivity may be attributed to the very high catalytic efficacy of DPP-IV mediated GP-BAN hydrolysis, as well as to the high fluorescence intensity of BAN.
3.5. Enzymatic kinetics of DPP-IV mediated GP-BAN hydrolysis
It is well-known that the enzyme kinetic behaviour is very crucial for the quantitative applications of the activity-based fluorescent probes (Ge et al., 2013; Liu et al., 2014a, Liu et al., 2014b; Feng et al., 2015; Lv et al., 2015; Wang et al., 2016, Wang et al., 2016). Herein, the enzymatic kinetics of GP-BAN hydrolysis was well-characterized in different enzyme sources including DPP-IV, HKM and HIM. As depicted in Fig. S10, GP-BAN hydrolysis in DPP-IV, HKM and HIM followed the classic Michaelis-Menten kinetics, which was evidenced by the corresponding Eadie-Hofstee plots. The kinetic parameters for GP-BAN hydrolysis in different enzyme sources were listed in Table S1. It is evident from Table S1 that the K m value for GP-BAN hydrolysis in DPP-IV (42.93 μM) is very similar to that in HKM (48.04 μM) and HIM (41.46 μM), suggesting that DPP-IV plays predominant role in GP-BAN hydrolysis in human kidney and intestine. In order to compare the kinetic parameters for GP-BAN hydrolysis with other known probe reactions for DPP-IV, the kinetic parameters for GP-AMC hydrolysis in DPP-IV were determined under the same conditions (pH 7.4 at 37 °C). As shown in Fig. S11, DPP-IV mediated GP-AMC hydrolysis also followed classic Michaelis-Menten kinetic, with the apparent K m and V max values as 237.80 μM and 9.01 μmol/min/mg protein. It is evident from these results that DPP-IV displays a relatively high affinity towards GP-BAN (with the K m value of 42.93 μM) and a higher hydrolytic efficiency (with the V max value of 15.49 μmol/min/mg protein) (Grant et al., 2002). As a result, the catalytic efficacy (V max/K m) for GP-BAN hydrolysis in DPP-IV (360.82 mL/min/mg protein) is about 10-fold of that for GP-AMC hydrolysis in DPP-IV (37.89 mL/min/mg protein). These results demonstrated that GP-BAN was a good substrate for DPP-IV and displayed excellent kinetic behaviours, which encourage us to apply this probe reaction as an efficient tool to quantify trace DPP-IV in complex biological samples.
3.6. Quantification of DPP-IV in human tissues
To assess the applicability of this newly developed DPP-IV sensor, the GP-BAN-based assay was further applied to measure DPP-IV activities in microsomes from different tissues. These tissue preparations showed extremely different DPP-IV activities, while HKM showed the highest DPP-IV activity (Fig. S12a). These results were further validated by ELISA (Fig. S12b), and a good relevance was observed between the rate of GP-BAN hydrolysis and DPP-IV protein levels among various human tissues. These results demonstrated that GP-BAN could be used to sensitively determine the real activities of DPP-IV in complex biological samples, and the quantification was highly reliable.
3.7. Quantification of DPP-IV in diluted human plasma
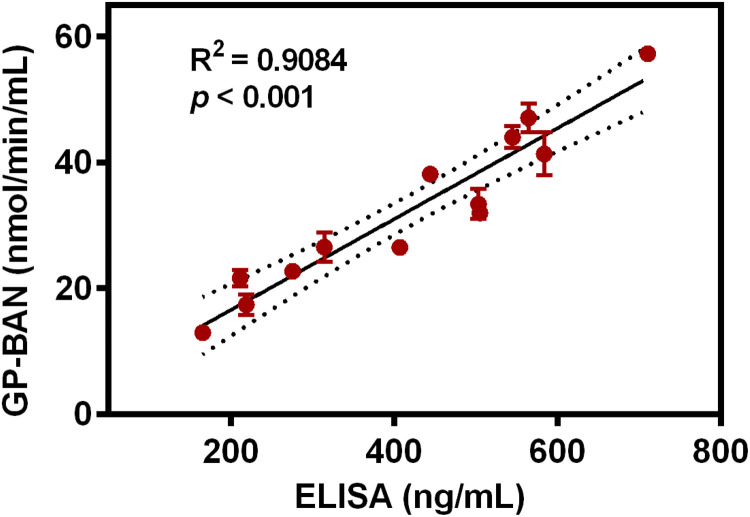
To further explore whether GP-BAN could be used to detect trace DPP-IV in complex biological systems, the applicability of GP-BAN for the detection of trace DPP-IV in human body fluids (such as plasma) was investigated. As shown in Fig. S13, the hydrolytic activities of DPP-IV in plasma samples from 13 health volunteers were carefully measured by using GP-BAN as a probe substrate. As shown in Fig. S13, about 4-fold variations in the catalytic activity of DPP-IV in human plasma were observed, which is in good agreement with the previously reported results in individual differences in DPP-IV activity in human plasma (Durinx et al., 2001). Furthermore, an ELISA method was used to determine the DPP-IV levels in these plasma samples, and the results agreed well with the rates of GP-BAN hydrolysis in these samples ( Fig. 2). These findings strongly suggested that GP-BAN could serve as a novel tool to detect trace amounts of DPP-IV in complex biological samples including diluted plasma. Given that DPP-IV served as a potential biomarker or target for many diseases, GP-BAN hold great promise for disease diagnosis and clinical practice.
Fig. 2.
Correlation analysis between the hydrolytic rates of GP-BAN and expression levels of DPP-IV in a panel of individual plasma samples (n =13).
3.8. High-throughput screening of DPP-IV inhibitors
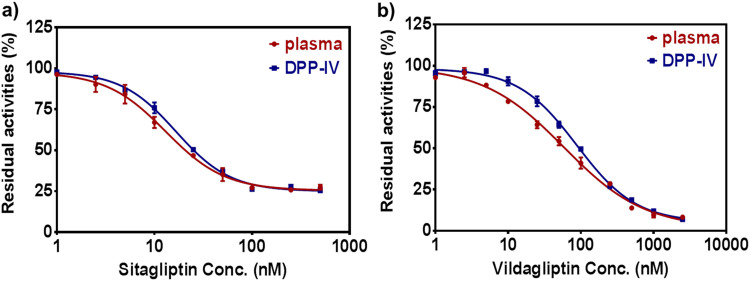
Encouraged by the above mentioned results, we subsequently employed GP-BAN hydrolysis as the probe reaction to rapidly screen DPP-IV inhibitors by using human plasma and HIM as enzyme sources. To this end, two known DPP-IV inhibitors including sitagliptin and vildagliptin were used, while their inhibitory effects on DPP-IV mediated GP-BAN hydrolysis in recombinant human DPP-IV, human plasma and HIM were assayed by a microplate reader with a 96-well microplate ( Fig. 3 and Fig. S14). As shown in Fig. 3a and b, the inhibitory tendency and the IC50 values of each inhibitor in both DPP-IV and human plasma were very similar. For instance, the IC50 values of sitagliptinon against DPP-IV in recombinant and human plasma were evaluated as 13.00 nM, and 16.59 nM, respectively. These results clearly shown that human plasma could be used as the enzyme source instead of expensive recombinant or purified DPP-IV for screening of DPP-IV inhibitors. Notably, the plasma needs to be 100-fold diluted by PBS for the GP-BAN-based assays, which means that only 2 μL of plasma is enough for DPP-IV related assays by using GP-BAN as probe substrate. Therefore, GP-BAN could serve as a promising tool for high-throughput screening (HTS) of DPP-IV inhibitors by using readily available human plasma as enzyme source in the field of drug discovery. Given that DPP-IV inhibitors are commonly used in clinical, it is urgently necessary to evaluate the sensitivity of a given DPP-IV inhibitor against DPP-IV among different individuals. Thus, GP-BAN holds a great promise to evaluate the efficacy of therapeutic drugs targeting DPP-IV and to guide the dosage adjustment for precision medicine in future.
Fig. 3.
a) Dose-inhibition curves of sitagliptinon (0–0.5 μM) on GP-BAN hydrolysis in both human plasma and DPP-IV. IC50 =13.00 nM, 16.59 nM, respectively. b) Dose-inhibition curves of vildagliptin (0–2.5 μM) on GP-BAN hydrolysis in both human plasma and DPP-IV. IC50 =58.65 nM, 85.12 nM, respectively.
3.9. Two-photon properties of GP-BAN
To further expand the potential applications of the newly developed probe in bioimaging related fields, especially cellular and tissue imaging, GP-BAN was intentionally applied for bioimaging of endogenous DPP-IV in living cells and deep tissues. Prior to bioimaging, the two-photon absorption cross sections of GP-BAN and BAN were determined in PBS-acetonitrile (v/v=1:1, pH 7.4) by the two-photon induced fluorescence measurement technique. As shown in Fig. S15, both GP-BAN and BAN could be excited with near-infrared light (two-photon absorption), while the two-photon action cross section (δ) values of both substrate and product were much close under excitation at 805 nm (about 600 g). Large two-photon action cross section of GP-BAN and BAN makes them attractive candidates as two-photon imaging agents.
3.10. Bioimaging of DPP-IV in living cells
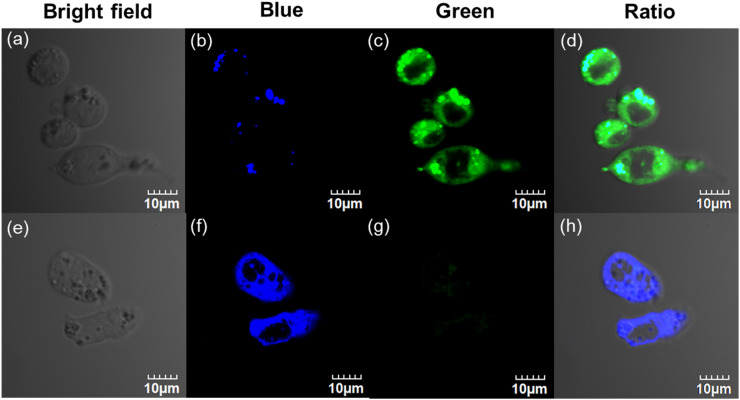
Prior to cell imaging, the cytotoxicity of GP-BAN and BAN was evaluated by a standard MTT assay. As shown in Figs. S16 and S17, both of GP-BAN and BAN exhibited relatively poor cytotoxicity to living hepatocellular carcinoma (HepG2) and colon adenocarcinoma (Caco-2) cells, as evidenced by the more than 80% cell viability of these two cell lines upon addition of GP-BAN (25 μM) or BAN (25 μM) at 37 °C for 48 h. In these cases, GP-BAN (25 μM) was co-incubated with HepG2 and Caco-2 cells, and the confocal fluorescence images were recorded both in one- and two-photon modes. As shown in Fig. 4 and Fig. S19, HepG2 and Caco-2 cells loaded with GP-BAN (25 μM) for 30 min at 37 °C showed an obvious strong green fluorescence (for BAN) and dim blue fluorescence (for GP-BAN) when excited at 805 nm. In contrast, following pretreatment of living cells with vildagliptin (a potent DPP-IV inhibitor, 50 μM), the fluorescence intensity decreased significantly in the green channel and increased evidently in the blue channel (Fig. 4e–h, Fig. S19e–h). It was evident that the changes in fluorescence response were DPP-IV dependent. The consistent changes in fluorescence were also observed in one photon microscopy (Figs. S18 and S20). To further validate the specific hydrolysis of GP-BAN by endogenous DPP-IV, chemical inhibition assays were also conducted in HepG2 and Caco-2 cells lysates. The results demonstrated that vildagliptin could remarkably reduce the formation of BAN in cells lysates from HepG2 and Caco-2 cells (Figs. S21 and S22). All these data clearly showed that GP-BAN could serve as a specific probe for both single-photon and two-photon-excited bioimaging and for sensing the real activities of DPP-IV in living cells.
Fig. 4.
Two-photon confocal fluorescence imaging of GP-BAN (25 μM) stained with HepG2 cells. (a-d) Cells were incubated with GP-BAN (25 μM) at 37 °C for 30 min (f–h) Cells were pretreated with vildagliptin (50 μM) and then added GP-BAN (25 μM) at 37 °C for 30 min. Images were acquired under excitation at 805 nm with two fluorescent emission windows: (b, f) blue =420–460 nm; (c, g) green =495–540 nm. Scale bars =10 µm.
3.11. Depth imaging of DPP-IV in mouse kidney tissues
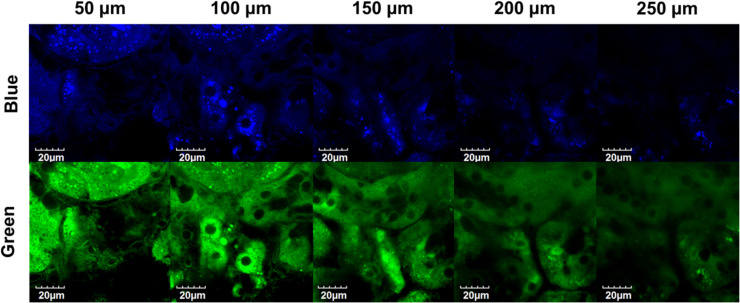
To further assess the application potential of GP-BAN as a two-photon fluorescent probe for bioimaging of DPP-IV in deep tissues, we next investigated the sensing ability of this probe for DPP-IV in fresh mouse kidney specimen. Prior to tissue imaging, the specificity of GP-BAN toward DPP-IV in mouse kidney was investigated. As shown in Fig. S23, vildagliptin could strongly inhibit the hydrolysis of GP-BAN in mouse kidney preparations, implying that GP-BAN hydrolysis in mouse kidney is mediated by DPP-IV. In this case, GP-BAN was co-incubated with mouse kidney slice in PBS (pH =7.4 at 37 °C) for 60 min and subsequently washed with PBS three times to remove the residual probe before confocal fluorescence imaging. As shown in Fig. 5, the application of GP-BAN in mouse kidney imaging was demonstrated by using a laser scanning confocal microscope under femtosecond laser excitation at 805 nm. Strong fluorescence was observed from green channel (495–540 nm) and the depth of tissue penetration was as far as 250 µm, indicating that GP-BAN displayed good capability of tissue penetration and was suitable for direct two-photon fluorescent depth imaging of trace DPP-IV in living tissues. These findings demonstrated that GP-BAN was capable of measuring endogenous DPP-IV in living tissues by TPM, which could be attributed to its good two photon fluorescence properties and the increased penetration depth by TPM.
Fig. 5.
Two-photon confocal fluorescent images of endogenous DPP-IV in a mouse kidney slice stained with GP-BAN (20 μM). Images (60× magnification) were acquired at depth within 250 µm with excitation at 805 nm and fluorescent emission windows of 420–460 nm and 495–540 nm. Scale bars =20 µm.
4. Conclusion
In summary, a novel ratiometric two-photon fluorescent probe GP-BAN has been designed and well characterized for highly selective detection of DPP-IV in complex biological systems including plasma, living cells and tissue slice. The newly developed probe displays excellent selectivity toward DPP-IV over other human hydrolases including FAP, DPP-VIII and DPP-IX. GP-BAN can be efficiently hydrolyzed by DPP-IV and release BAN which brings significant changes in both colour and fluorescence emission spectrum, allowing the naked-eye visible and fluorescence analysis. Furthermore, both GP-BAN and BAN exhibit excellent two-photon properties, which could be excited with near-infrared light to avoid interference of biological matrix and photodamage to living sample. Further biological applications demonstrate that GP-BAN can be used to measure DPP-IV in complex biological samples such as plasma and tissue preparations, as well as to monitor endogenous DPP-IV in living systems with high ratiometric imaging resolution and deep-tissue imaging depth. All these findings suggest that GP-BAN can serve as a highly specific fluorescent probe to image DPP-IV in complex biological systems and to explore the biological functions and medicinal roles of endogenous DPP-IV in living systems.
Acknowledgments
We gratefully acknowledge Prof. Baomin Wang of Dalian University of Technology for language proofreading. This work was supported by the National Key Research and Development Program of China (2016YFC1303900), National Basic Research Program of China (2013CB531805), NSF of China (21602219, 81473181, 81402822, 81573501, 81403003), the State Key Laboratory of Fine Chemicals (KF1504 & KF1408) and Beijing National Laboratory for Molecular Sciences (BNLMS 20150124).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bios.2016.11.068.
Contributor Information
Guang-Bo Ge, Email: geguangbo@dicp.ac.cn.
Ling Yang, Email: ylingdicp@gmail.com.
Appendix A. Supplementary data
Supplementary material
.
References
- Ajami K., Pitman M.R., Wilson C.H., Park J., Menz R.I., Starr A.E., Cox J.H., Abbott C.A., Overall C.M., Gorrell M.D. FEBS Lett. 2008;582:819–825. doi: 10.1016/j.febslet.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Arrebola Y.M., Gómez H., Valiente P.A., Chávez M. de los A., Pascual I. Biotecnol. Apl. 2014;31:102–110. [Google Scholar]
- Cordero O.J., Salgado F.J., Nogueira M. Cancer Immunol. Immun. 2009;58:1723–1747. doi: 10.1007/s00262-009-0728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan A., Wolk O., Yang P., Mittal S., Wu Z., Landowski C.P., Amidon G.L. Mol. Pharm. 2014;11:4385–4394. doi: 10.1021/mp500483v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z.R., Ge G.B., Feng L., Ning J., Hu L.H., Jin Q., Wang D.D., Lv X., Dou T.Y., Cui J.N., Yang L. J. Am. Chem. Soc. 2015;137:14488–14495. doi: 10.1021/jacs.5b09854. [DOI] [PubMed] [Google Scholar]
- Davies S., Beckenkamp A., Buffon A. Biomed. Pharmacother. 2015;71:135–138. doi: 10.1016/j.biopha.2015.02.031. [DOI] [PubMed] [Google Scholar]
- Drucker D.J., Nauck M.A. Lancet. 2006;368:1696–1705. doi: 10.1016/S0140-6736(06)69705-5. [DOI] [PubMed] [Google Scholar]
- Durinx C., Neels H., Van der Auwera J.C., Naelaerts K., Scharpé S., Meester I. Clin. Chem. Lab. Med. 2001;39:155–159. doi: 10.1515/CCLM.2001.026. [DOI] [PubMed] [Google Scholar]
- Egan A.G., Blind E., Dunder K., Graeff P.A., Hummer B.T., Bourcier T., Rosebraugh C.N. N. Eng. J. Med. 2014;370:794–797. doi: 10.1056/NEJMp1314078. [DOI] [PubMed] [Google Scholar]
- Feng L., Liu Z.M., Hou J., Lv X., Ning J., Ge G.B., Cui J.N., Yang L. Biosens. Bioelectron. 2015;65:9–15. doi: 10.1016/j.bios.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Firneisz G., Varga T., Lengyel G., Fehér J., Ghyczy D., Wichmann B., Selmeci L., Tulassay Z., Rácz K., So-mogyi A. PLoS One. 2010;18:e12226. doi: 10.1371/journal.pone.0012226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge G.B., Ning J., Hu L.H., Dai Z.R., Hou J., Cao Y.F., Yu Z.W., Ai C.Z., Gu J.K., Ma X.C., Yang L. Chem. Commun. 2013;49:9779–9781. doi: 10.1039/c3cc45250f. [DOI] [PubMed] [Google Scholar]
- Gong Q., Shi W., Li L., Wu X., Ma H. Anal. Chem. 2016;88:8309–8314. doi: 10.1021/acs.analchem.6b02231. [DOI] [PubMed] [Google Scholar]
- Grant S.K., Sklar J.G., Cummings R.T. J. Biomol. Screen. 2002;7:531–540. doi: 10.1177/1087057102238627. [DOI] [PubMed] [Google Scholar]
- Helmchen F., Denk W. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Hettiarachchi S.U., Prasai B., McCarley R.L. J. Am. Chem. Soc. 2014;136:7575–7578. doi: 10.1021/ja5030707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt M., Reutter W., Arck P., Rose M., Klapp B.F. Clin. Sci. 2000;99:93–104. [PubMed] [Google Scholar]
- Ho N.H., Weissleder R., Tung C.H. Bioorg. Med. Chem. Lett. 2006;16:2599–2602. doi: 10.1016/j.bmcl.2006.02.045. [DOI] [PubMed] [Google Scholar]
- Javidroozi M., Zucker S., Chen W.T. Dis. Markers. 2012;32:309–320. doi: 10.3233/DMA-2011-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Feng L., Wang D.D., Dai Z.R., Wang P., Zou L.W., Liu Z.H., Wang J.Y., Yu Y., Ge G.B., Cui J.N., Yang L. ACS Appl. Mater. Interfaces. 2015;7:28474–28481. doi: 10.1021/acsami.5b09573. [DOI] [PubMed] [Google Scholar]
- Kameoka J., Tanaka T., Nojima Y., Schlossman S.F., Morimoto C. Science. 1993;261:466–469. doi: 10.1126/science.8101391. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M., Okabe T., Terai T., Hanaoka K., Kojima H., Minegishi I., Nagano T.A. Chem. Eur. J. 2010;16:13479–13486. doi: 10.1002/chem.201001077. [DOI] [PubMed] [Google Scholar]
- Keane F.M., Nadvi N.A., Yao T.W., Gorrell M.D., Neuropeptide Y. FEBS J. 2011;278:1316–1332. doi: 10.1111/j.1742-4658.2011.08051.x. [DOI] [PubMed] [Google Scholar]
- Kim H.M., Cho B.R. Chem. Rev. 2015;115:5014–5055. doi: 10.1021/cr5004425. [DOI] [PubMed] [Google Scholar]
- Kim M.S., Pinto S.M., Getnet D. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.Q., Mao Z.Q., Rong L., Wu N., Lei Q., Zhu J.Y., Zhuang L., Zhang X.Z., Liu Z.H. Biosens. Bioelectron. 2017;87:73–80. doi: 10.1016/j.bios.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Liu Z.M., Feng L., Ge G.B., Lv X., Hou J., Cao Y.F., Cui J.N., Yang L. Biosens. Bioelectron. 2014;57:30–35. doi: 10.1016/j.bios.2014.01.049. [DOI] [PubMed] [Google Scholar]
- Liu Z.M., Feng L., Hou J., Lv X., Ning J., Ge G.B., Wang K.W., Cui J.N., Yang L. Sens. Actuators B: Chem. 2014;205:151–157. [Google Scholar]
- Lu C., Tilan J.U., Everhart L., Czarnecka M., Soldin S.J., Mendu D.R., Jeha D., Hanafy J., Lee C.K., Sun J., Izycka-Swieszewska E., Toretsky J.A., Kitlinska J. J. Biol. Chem. 2011;286:27494–27505. doi: 10.1074/jbc.M111.224089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X., Ge G.B., Feng L., Troberg J., Hu L.H., Hou J., Cheng H.L., Wang P., Liu Z.M., Finel M., Cui J.N., Yang L. Biosens. Bioelectron. 2015;72:261–267. doi: 10.1016/j.bios.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Mentlein R. Regul. Pept. 1999;85:9–24. doi: 10.1016/s0167-0115(99)00089-0. [DOI] [PubMed] [Google Scholar]
- Miao F., Zhang W., Sun Y., Zhang R., Liu Y., Guo F., Song G., Tian M., Yu X. Biosens. Bioelectron. 2014;55:423–429. doi: 10.1016/j.bios.2013.12.044. [DOI] [PubMed] [Google Scholar]
- Ohnuma K., Dang N.H., Morimoto C. Trends Immunol. 2008;29:295–301. doi: 10.1016/j.it.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Perner F., Gyuris T., Rákóczy G., Sárváry E., Görög D., Szalay F., Kunos I., Szönyi L., Péterfy M., Takács L. J. Lab. Clin. Med. 1999;134:56–67. doi: 10.1016/s0022-2143(99)90054-9. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H.H., Smits S.L. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Otero N., Rodríguez-Berrocal F.J., Cadena M.P., Botana-Rial M.I., Cordero O.J. Sci. Rep. 2014;4:3999. doi: 10.1038/srep03999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T., Nagano T. Nat. Methods. 2011;8:642–645. doi: 10.1038/nmeth.1663. [DOI] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vendrell M., Zhai D., Cheng J., Chang Y.T. Chem. Rev. 2012;112:4391–4420. doi: 10.1021/cr200355j. [DOI] [PubMed] [Google Scholar]
- Wang D.D., Jin Q., Zou L.W., Hou J., Lv X., Lei W., Cheng H.L., Ge G.B., Yang L. Chem. Commun. 2016;52:3183–3186. doi: 10.1039/c5cc09874b. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang B., Shi Z., Tang X., Dou W., Han Q., Zhang Y., Liu W. Biosens. Bioelectron. 2015;65:91–96. doi: 10.1016/j.bios.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wu X., Cheng Y., Zhao X. Chem. Commun. 2016;52:3478–3481. doi: 10.1039/c5cc08921b. [DOI] [PubMed] [Google Scholar]
- Waumans Y., Baerts L., Kehoe K., Lambeir A.M., Meester I.D. Front. Immunol. 2015;6:387. doi: 10.3389/fimmu.2015.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.F., Lim C.S., Bhuniya S., Cho B.R., Kim J.S. Org. Lett. 2011;13:1190–1193. doi: 10.1021/ol200072e. [DOI] [PubMed] [Google Scholar]
- Zipfel W.R., Williams R.M., Christie R., Nikitin A.Y., Hyman B.T., Webb W.W. Proc. Natl. Acad. Sci. USA. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material