Abstract
There are few reports describing diarrhea of adult cattle caused by group A rotaviruses. Here, we report the identification of a novel bovine group A rotavirus from diarrhea of adult cows. A group A rotavirus was detected from an epizootic outbreak of diarrhea in adult cows with a decrease in milk production in Japan in 2013. The comprehensive genomic analyses from fecal samples by viral metagenomics using a next-generation sequencer revealed that it had an unreported genotype combination G15P[14]. The genome constellation of this strain, namely, RVA/Cow-wt/JPN/Tottori-SG/2013/G15P[14] was G15-P[14]-I2-R2-C2-M2-A3-N2-T6-E2-H3 representing VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5, respectively. Each gene segment of Tottori-SG was most closely related to Japanese bovine group A rotaviruses suggesting that Tottori-SG might have derived from multiple reassortment events from group A rotavirus strains circulating among Japanese cattle. No other diarrhea pathogen of adult cattle was detected by routine diagnosis and metagenomics. Viral metagenomics, using a next-generation sequencer, is useful to characterize group A rotaviruses from fecal samples and offers unbiased comprehensive investigations of pathogen.
Keywords: Rotavirus, Novel genotype, Adult cow diarrhea, Viral metagenomics, Japan
1. Introduction
Group A rotavirus (RVA) is one of the most important pathogens of neonatal calf diarrhea (Dhama et al., 2009, Papp et al., 2013). RVAs are icosahedral, non-enveloped viruses and belong to the family Reoviridae. The rotavirus genome consists of 11 segments encoding six structural proteins (VP1, VP2, VP3, VP4, VP6 and VP7) and six nonstructural proteins (NSP1-NSP6) (Estes and Kapikian, 2007). RVAs are classified into multiple G and P types defined by the two outer capsid proteins, VP7 and VP4, respectively. To date, 27G and 35P genotypes have been determined, many of which have been identified in the last five years (Matthijnssens et al., 2011). The Rotavirus Classification Working Group proposed a new classification system using nucleotide sequences of all of the 11 genomic RNA segments, Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx representing VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5, respectively (Matthijnssens et al., 2011).
Co-infection of a cell by two or more different strains of a segmented virus can give rise to a “reassortant” with phenotypic characteristics that might differ from those of the parental strains. Reassortment has been observed in Reoviridae including rotavirus (Watanabe et al., 2001). Reassortment is an important mechanism which drives the genetic diversity observed in rotaviruses.
RVA is the most common cause of diarrhea in infants and young children (Estes and Kapikian, 2007). Although there are several reports of adult diarrhea caused by RVA including suspected zoonotic transmission from artiodactyl hosts (Griffin et al., 2002, Midgley et al., 2012), the role of RVA as a pathogen in adult humans has long been underappreciated (Anderson and Weber, 2004). There are few reports describing diarrhea in adult cattle caused by RVA (Fukai et al., 2007, Onuma et al., 2003, Sato et al., 1997). Similarly, the role of RVA as a pathogen in adult cattle is also unclear.
Next Generation Sequencing, non-Sanger-based sequencing technologies have delivered on a high-throughput sequencing methodology which generates millions of sequences simultaneously from one sample (Schuster, 2008). This method can detect and characterize pathogens without prior knowledge of their existence, cultured material, or the requirement of specific primers (Marston et al., 2013).
Here, we report a case that occurred in Japan, in which novel G15P[14] RVA was identified from adult cattle with watery diarrhea and a decrease in milk production.
2. Materials and methods
2.1. Routine diagnosis
The fecal samples from five cows were tested for RVA by commercial immunochromatographic assay kit (Dipstick ‘Eiken’ Rota (Eiken Chemical Co., Ltd, Tokyo, Japan)), Salmonella species and Escherichia coli K99 by using standard techniques, and Coccidium species and Cryptosporidium species by a sucrose floatation method. The samples were also examined by reverse transcription-polymerase chain reaction (RT-PCR) for bovine coronavirus (BCV) (Tsunemitsu et al., 1999), bovine torovirus (BToV) (Hoet et al., 2003), bovine group B rotavirus (RBV) (Chinsangaram et al., 1994), bovine group C rotavirus (RCV) (Tsunemitsu et al., 1996) and bovine viral diarrhea virus (BVDV) (Vilcek et al., 1994). Serum samples were collected from affected cows during the acute phase of the disease and again, three weeks later. These paired sera were tested for antibodies to BCV and bovine adenovirus type 7 (BAV7) by hemagglutination inhibition tests and to BVDV-1 and BVDV-2 by virus neutralization tests.
2.2. Total RNA extraction from fecal samples, building cDNA library and sequencing
Five fecal samples were diluted 1:9 (W/V) in sterile PBS and the viral RNAs were extracted from them using ISGEN LS (NipponGene, Tokyo, Japan), followed by DNase I treatment (TaKaRa Bio Inc., Shiga, Japan). RNA samples were normalized to 10–100 ng of RNA using Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) and libraries for Illumina sequencing were constructed using the NEBNext® Ultra RNA Library Prep Kit for Illumina Version 1.0 (New England Biolabs, Ipswich, MA, USA) according to the manufacture's guidelines with minor modifications: for selecting 400–500 bp fragments after A-Tailing and adaptor ligation, two beads clean-up steps (firstly, ×0.4 volume and secondly, the surpernatant from the first bind was taken for ×0.15 volume clean-up) were done using the Agencourt AMPure XP (Beckman Coulter, Pasadena, CA, USA). After assessing the library quality and quantity on a Bioanalyzer® (Agilent technologies, Santa Clara, CA, USA) and Qubit® 2.0 Fluorometer (Invitrogen), sequencing was carried out on a MiSeq bench-top sequencer (Illumina, San Diego, CA, USA) using 151 single-end reads.
2.3. Sequence data analysis
Data analysis was performed using the MiSeq reporter program (Illumina) to generate FASTQ formatted sequence data. Contigs were assembled from the obtained sequence reads using de novo assembly command in the CLC Genomics Workbench (CLC bio, Aarhus, Denmark). Using the assembled contigs as query sequences, the BLAST non-redundant nucleotide database was searched using the blastn program. Nucleotide sequences were aligned using ClustalW and phylogenetic analyses performed by the Neighbor-Joining method using MEGA5.22 (Tamura et al., 2011).
3. Results
3.1. Clinical history and routine detection of potential pathogens of cattle
In February 2013, two out of 42 lactating cows on a dairy farm in Tottori Prefecture, Japan, suddenly showed signs of anorexia and started having watery diarrhea. Subsequently, three more lactating cows neighboring the two infected cows, started showing similar symptoms, and within a week, all adult cows on the farm were affected. The diarrhea feces were liquid and greenish yellow, but not bloody. All animals recovered within a week; however, a decrease in milk production on the farm was observed the day after the first finding of diarrhea and continued for two weeks. Preliminary diagnosis using Dipstick ‘Eiken’ Rota for RVA revealed that all fecal samples from five cows with diarrhea were positive. All fecal samples in the acute-phase were negative for BCV, BToV, RBV, RCV and BVDV by RT-PCR, and Salmonella species, E. coli K99, Coccidium species and Cryptosporidium species by standard techniques. Examination of paired sera from affected cows revealed no significant increase in antibody titers to BCV, BAV7 and BVDV-1 and 2.
3.2. Full genome sequencing of RVA
The sequencing from Illumina MiSeq yielded a range of 800,298 to 1,180,474 sequence reads (about 150-mer length) from five fecal samples. First, convenient genotyping of RVA was carried out using the CLC Genomics Workbench. Mapping sequence reads from five samples to VP4 and VP7 reference sequences of representative strains (Minami-Fukuda et al., 2013) revealed poor alignment with G6, G8, G10, P[1], P[5] and P[11] – the most common bovine genotypes. Near complete sequences of the 11 segments of RVA could be obtained from the resultant contigs generated by reads assembled from five samples. The sequences of all 11 segments of RVA were detected from all five samples and confirmed to be identical. The genotypes for each of gene segments of this strain (RVA/Cow-wt/JPN/Tottori-SG/2013/G15P[14] (Tottori-SG)) were determined using the online RotaC genotyping tool (Maes et al., 2009). A very rare G/P combination, G15P[14] was found. Applying the nucleotide-sequence-based complete genome classification system, the constellation of Tottori-SG was determined as G15-P[14]-I2-R2-C2-M3-A3-N2-T6-E2-H3 representing VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5, respectively. The sequences were deposited in the DNA Data Bank of Japan (DDBJ/EMBL/GenBank accession numbers: AB853890 to AB853900 for the 11 segments).
3.3. Phylogenetic analyses and pairwise comparison
Phylogenetic analysis based on entire open reading frame nucleotide sequences of the VP7 gene revealed that though Tottori-SG clustered with RVA/Cow-wt/ARG/B383/1998/G15P[11] (Garaicoechea et al., 2006, Matthijnssens et al., 2009) and three Indian G15P[11] and G15P[21] strains (Rao et al., 2000, Ghosh et al., 2008), it formed a single G15 cluster, distinct from other G genotype strains (Fig. 1A). Nucleotide (nt) and amino acid (aa) of Tottori-SG showed 87.9–89.9% and 91.7–92.9% sequence identity to other G15 strains at nucleotide (nt) and amino acid (aa) levels, respectively. The VP4 gene of Tottori-SG formed a cluster with RVA/Cow-tc/JPN/Sun9/2000/G8P[14] (Fukai et al., 2004) and RVA/Human-tc/THA/Mc35/XXXX/G10P[14] (Urasawa et al., 1993) (Fig. 1B). Tottori-SG had the highest level of nt and aa identity to strain Sun9 (95.8%, 97.2%) and Mc35 (89.7%, 95.6%), respectively. With regard to the VP6 gene segment, Tottori-SG clustered with the unusual Japanese bovine RVA strains G21P[29] and G24P[33], RVA/Cow-wt/JPN/AzuK-1/2006/G21P[29] (Abe et al., 2009) and RVA/Cow-wt/Dai-10/2008/G24P[33] (Abe et al., 2011), and Simian G3P[2] RVA strains (Mlera et al., 2013). The VP6 gene of Tottori-SG exhibited high nucleotide identity with AzuK-1, Dai-10 and RVA/Simian-tc/ZAF/SA11-N5/1958/G3P[2] (96.0, 96.6 and 95.2%), respectively (Fig. 1C). Phylogenetic analysis of VP1, VP2, VP3, NSP2, NSP3, NSP4 and NSP5 showed that Tottori-SG clustered with genes of bovine or bovine-like rotavirus (Fig. 1D–F, H–K). Similarly, NSP1, which clustered with simian G8P[1] and human G6P[14] strains, was thought to be the result of transmission from ruminant hosts (Matthijnssens et al., 2009, Matthijnssens et al., 2010) (Fig. 1G). Overall, the nucleotide sequences of the 11 genome segments of Tottori-SG had relatively high nucleotide identities to bovine or bovine-like RVA strains (Table 1). Furthermore, apart from the VP7 gene, the gene segments of Tottori-SG were highly homologous with those of Japanese bovine RVA strains (Table 2).
Fig. 1.
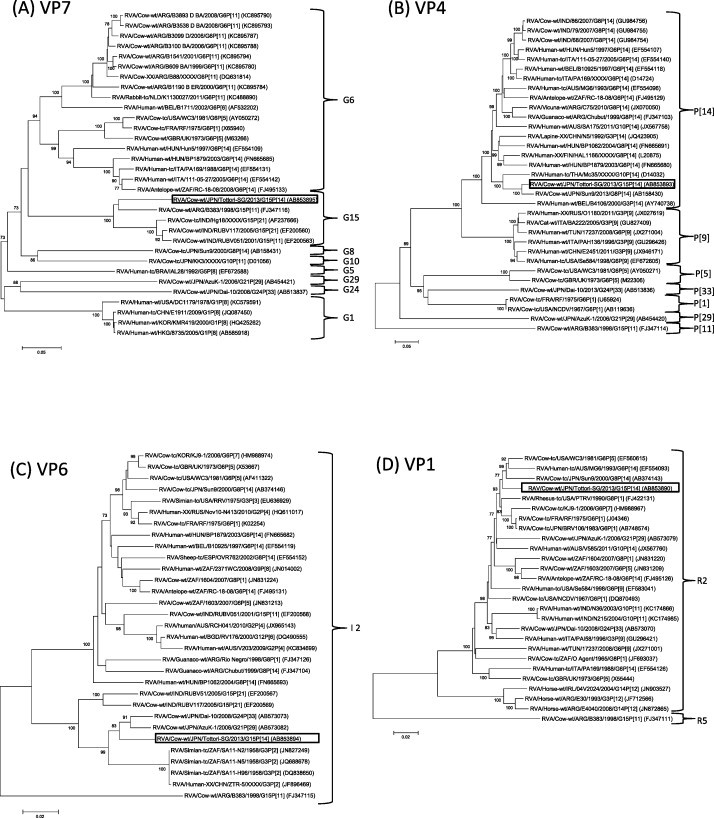
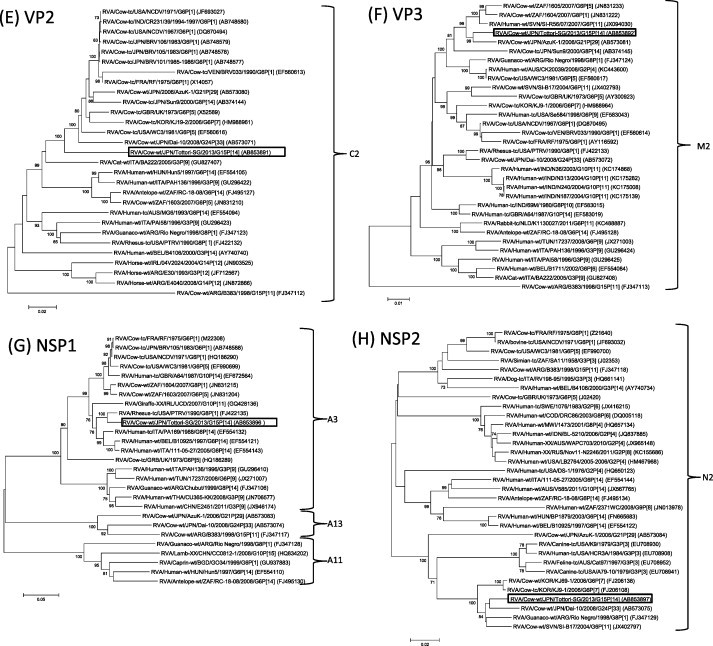
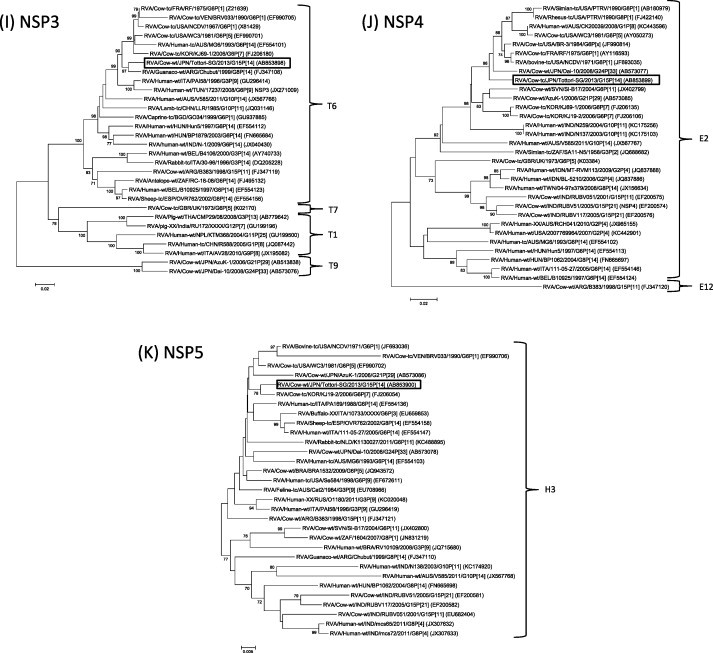
Phylogenetic analysis based on the nucleotide sequences of VP7 (A), VP4 (B), VP6 (C), VP1-VP3 (D-F) and NSP1-NSP5 (G-K) genes of Tottori-SG. Phylogenetic trees were constructed using the neighbor-joining methods in MEGA5.22 with bootstrap values (1000 replicates) above 70 are shown. The scale bar indicates nucleotide substitutions per site. The genotypes are indicated at the right-hand side. The rotavirus strains detected in this study is shown by a black open square.
Table 1.
Closest nucleotide identities of the 11 segments of RVA/Cow-wt/JPN/Tottori-SG/2013/G15P[14].
| Gene | Strains that had highest sequence homology in the database |
Genotype of Tottori-SG | ||
|---|---|---|---|---|
| Strain | Accession no. | Homology (%) | ||
| VP7 | RVA/Cow-wt/ARG/B383/1998/G15P[11] | FJ347116 | 89.9 | G15 |
| VP4 | RVA/Cow-tc/JPN/Sun9/2000/G8P[14] | AB158430 | 95.8 | P[14] |
| VP6 | RVA/Cow-wt/JPN/Dai-10/2008/G24P[33] | AB573073 | 96.6 | I2 |
| VP1 | RVA/Cow-tc/FRA/RF/1975/G6P[1] | J04346 | 97.2 | R2 |
| VP2 | RVA/Cow-tc/USA/NCDV/1967/G6P6[1] | DQ87049 | 93.2 | C2 |
| VP3 | RVA/Cow-wt/JPN/AzuK-1/2006/G21P[29] | AB573081 | 96.9 | M2 |
| NSP1 | RVA/Rhesus-tc/USA/PTRV/1990/G8P[1] | FJ422135 | 97.0 | A3 |
| NSP2 | RVA/Cow-wt/JPN/Dai-10/2008/G24P[33] | AB573075 | 96.7 | N2 |
| NSP3 | RVA/Guanaco-wt/ARG/Chubut/1999/G8P[14] | FJ347108 | 97.0 | T6 |
| NSP4 | RVA/Cow-tc/FRA/RF/1975/G6P[1] | AY116593 | 97.2 | E2 |
| NSP5 | RVA/Cow-tc/KOR/KJ19-2/2006/G6P[7] | FJ206054 | 98.9 | H3 |
Table 2.
Nucleotide identity of the 11 segments of RVA/Cow-wt/JPN/Tottori-SG/2013/G15P[14] to Japanese and non-Japanese bovine rotavirus strains from GenBank.
| Strain | Genotype (Homology to RVA/Cow-wt/Tottori-SG/2013/G15P[14] (%)) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | |
| RVA/Cow-wt/JPN/Tottori-SG/2013/G15P[14] | G15 (100) | P[14] (100) | I2 (100) | R2 (100) | C2 (100) | M2 (100) | A3 (100) | N2 (100) | T6 (100) | E2 (100) | H3 (100) |
| Japanese bovine strains | |||||||||||
| RVA/Cow-tc/JPN/Sun9/2000/G8P[14] | G8 (72.0) | P[14] (95.8) | I2 (88.4) | R2 (96.0) | C2 (92.2) | M2 (94.1) | |||||
| RVA/Cow-wt/JPN/AzuK-1/2006/G21P[29] | G21 (71.6) | P[29] (64.7) | I2 (96.0) | R2 (96.4) | C2 (93.1) | M2 (96.9) | A13 (74.4) | N2 (89.4) | T9 (77.6) | E2 (94.5) | H3 (97.1) |
| RVA/Cow-wt/JPN/Dai-10/2008/G24P[33] | G24 (70.6) | P[33] (67.6) | I2 (96.6) | R2 (94.9) | C2 (92.2) | M2 (92.7) | A13 (73.0) | N2 (96.7) | T9 (78.1) | E2 (96.2) | H3 (97.0) |
| RVA/Cow-tc/JPN/BRV105/1983/G6P[1] | G6 (71.8) | P[1] (67.7) | I2 (87.7) | R2 (97.1) | C2 (93.2) | M2 (92.6) | A3 (92.9) | N2 (86.4) | T6 (96.0) | E2 (96.8) | H3 (97.5) |
| non-Japanese bovine strains | |||||||||||
| RVA/Cow-tc/USA/NCDV/1967/G6P[1] | G6 (73.6) | P[1] (67.7) | I2 (88.3) | R2 (95.8) | C2 (93.2) | M2 (92.8) | A3 (93.2) | N2 (86.6) | T6 (95.7) | E2 (96.4) | H3 (98.3) |
| RVA/Cow-tc/FRA/RF/1975/G6P[1] | G6 (73.7) | P[1] (67.6) | I2 (88.2) | R2 (97.2) | C2 (93.1) | M2 (92.8) | A3 (93.1) | N2 (86.6) | T6 (96.1) | E2 (97.2) | H3 (97.4) |
| RVA/Cow-tc/UK/1973/G6P[5] | G6 (74.2) | P[5] (65.3) | I2 (88.8) | R2 (92.5) | C2 (92.6) | M2 (93.1) | A3 (87.8) | N2 (87.4) | T7 (83.0) | E2 (90.6) | H3 (93.1) |
| RVA/Cow-tc/USA/WC3/1981/G6P[5] | G6 (73.4) | P[5] (64.9) | I2 (88.3) | R2 (96.9) | C2 (93.0) | M2 (94.3) | A3 (92.8) | N2 (88.0) | T6 (95.7) | E2 (92.0) | H3 (97.4) |
| RVA/Cow-wt/ARG/B383/1998/G15P[11] | G15 (89.9) | P[11] (60.2) | I2 (85.5) | R5 (80.7) | C2 (84.1) | M2 (85.9) | A13 (73.8) | N2 (87.7) | T6 (88.1) | E12 (87.5) | H3 (97.7) |
An open space means the sequence data could not be available.
3.4. Analysis of resultant contigs using BLAST
The sequence reads from the FASTQ formatted sequence data were assembled and contigs generated using the CLC Genomics Workbench. The generated 728–2407 contigs containing 477,632–923,026 sequence reads from five samples were analyzed using a BLAST search against non-redundant databases. The BLAST search using contigs identified 73,039–464,263 (7.4–77.5%), 52,382–698,684 (11.0–70.7%), 2850–9734 (0.5–1.1%) and 20–10,345 (0.0–1.2%) reads as RVA, non-pathogenic bacteria, mammalian and plant, and plant virus, respectively. Apart from RVA, no other genomes of virus pathogens of bovine were detected from sequencing reads.
4. Discussion
In Japan, G6 and G10 genotypes are predominantly distributed (Ishizaki et al., 1996), and G15 genotype has never been isolated. G15 genotype is very rare in nature. To date, only five RVA strains possessing this genotype are available in database. These including three Indian G15P[21] strains – Hg18, RUBV51 and RUBV117 – isolated in 1999 and 2005 (Ghosh et al., 2008, Rao et al., 2000), and two G15P[11] strains – B383 and RUBV051 – isolated from Argentina and India in 1998 and 2001, respectively (Ghosh et al., 2008, Matthijnssens et al., 2009). Tottori-SG is the sixth G15 strain and the first report of a G15P[14] combination strain worldwide. All G15 strains were isolated from cattle. VP7 of Tottori-SG demonstrated modest nucleotide identity with other G15 strains, suggesting that VP7 of Tottori-SG is novel and might have an independent ancestor. On the other hand, P[14] genotype has been found in rabbits (Ciarlet et al., 1997), humans (Cowley et al., 2013, Gerna et al., 1994, Matthijnssens et al., 2006, Matthijnssens et al., 2009, Mphahlele et al., 1999, Urasawa et al., 1993), goats, antelope (Ghosh et al., 2007), sheep (Ciarlet et al., 2008), guanacos (Parreño et al., 2004) and cattle (Chitambar et al., 2011, Fukai et al., 2004). A recent study suggested that the human P[14] strains were related to RVA strains isolated from even-toed ungulates belonging to the mammalian order Artiodactyla (Matthijnssens et al., 2009). The P[14] VP8* protein has a high affinity to type A human histo-blood group antigens (Hu et al., 2012, Liu et al., 2012). The VP8* of P[14] also binds the A type antigen of bovine and porcine, which could be a reason that P[14] RVAs infect both bovine and human (Liu et al., 2012). In Japan, only one strain, Sun9, has been reported from calves (Fukai et al., 2004). However, the P[14] genotype has never been isolated from other species within Japan, including humans. Tottori-SG shared high nucleotide identity with VP4 gene of Sun9. Although Sun9 remains the only P[14] strain to have been isolated in Japan, it is of interest to examine whether the P[14] strains may be prevalent among Japanese cattle. Further analyses of genes other than VP7 and VP4 revealed that structural and nonstructural protein genes of Tottori-SG showed high identity with those of bovine or bovine-like RVA strains, suggesting Tottori-SG strain might adapt to cattle (Table 1). Furthermore, Tottori-SG shared a similar genetic background with Japanese bovine RVAs (Table 2). Interestingly, VP6 of Tottori-SG was closely related to unusual Japanese RVA strains, Dai-10 (G24P[33]) and AzuK-1 (G21P[29]), together with the Indian strain RUBV51 (G15P[21]), as well as Simian RVA strains. In regard to the NSP2 and NSP3 gene, Tottori-SG clustered with Dai-10 and P[14] guanaco RVA strain in NSP2, and with P[14] guanaco one in NSP3, suggesting relationship between Japanese bovine RVA and this guanaco strain. These data suggest that Tottori-SG might have derived from multiple reassortment events among RVA strains circulating among Japanese cattle.
Main causes of epizootic outbreak of viral diarrhea in adult cows including BCV and BTov, RBV and RCV have also been sporadically reported in Japan and worldwide. However, symptomatic infections of RVA in adult cattle are rare. In the present case, considering both the absence of viral, bacterial and protozoan pathogens of diarrhea in the acute-phase fecal samples, and the detection of RVA antigen only during the acute-phase of diarrhea but not after recovery, it is suggested that Tottori-SG associated with the epizootic outbreak of diarrhea in adult cows. At least three cases of symptomatic RVA infection in adult cattle have been reported in Japan (Fukai et al., 2007, Onuma et al., 2003, Sato et al., 1997). In two out of three cases, two rare G and P genotype strains of RVA, BRV16, which belongs to G8P[1]genotype, and Tak2, which probably belongs to G21P[29] genotype, were isolated from fecal samples in adult cows with diarrhea (Fukai et al., 2007, Sato et al., 1997). One of the possible factors responsible for symptomatic infections by RVA in adult cattle is a lack of immunity in cattle against such rare genotype RVAs. The G15P[14] strain may efficiently transmit from infected cattle to immune-naïve ones, causing unusual age pattern of RVA infection. To elucidate the mechanisms responsible for epizootic diarrhea of adult cattle caused by rare RVA genotypes, further serological and pathogenic studies are needed.
In this study, we employed the characterization of bovine RVA and detection of potential pathogens of RNA viruses using de novo sequencing by a next-generation sequencer. Exhaustive investigation is useful for directly detecting pathogenic viruses, and no step requiring sequence-specific primers for PCR amplification or bait-based enrichment is needed (Djikeng et al., 2008). This feature provides the opportunity of bioinformatically detecting any other known RNA virus at the same time (Batty et al., 2013). We could not only generate near whole genome sequences of RVA, but could also give the opportunity of bioinformatically detecting any other known RNA virus at the same time. The resultant contigs generated by reads assembly were investigated using BLAST search. However no other RNA virus genome of cattle pathogen was detected within the sequencing reads, suggesting that epizootic diarrhea of adult cows caused by RVA.
In conclusion, this study demonstrated that Tottori-SG, which was detected from fecal samples of adult cows with diarrhea had yet not reported genotype combination of G15P[14]. Full genome analyses of Tottori-SG suggested that Totttori-SG might be derived by multiple reassortment events from RVA strains circulating among Japanese cattle. Finally, comprehensive genomic analyses from fecal samples by de novo sequencing using a next-generation sequencer will contribute to easy determination of full genomes of RVA and increasing sequence data of RVAs from individual regions will shed light on RVA infection of livestock industry.
Acknowledgment
This work was supported by the Grants-in-Aid for Research on Emerging and Re-emerging Infectious Diseases by the Ministry of Health, Labour and Welfare, Japan (grant H24-shinkou-ippan-005).
References
- Abe M., Ito N., Morikawa S., Takasu M., Murase T., Kawashima T., Kawai Y., Kohara J., Sugiyama M. Molecular epidemiology of rotaviruses among healthy calves in Japan: isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus. Res. 2009;144:250–257. doi: 10.1016/j.virusres.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Abe M., Ito N., Masatani T., Nakagawa K., Yamaoka S., Kanamaru Y., Suzuki H., Shibano K., Arashi Y., Sugiyama M. Whole genome characterization of new bovine rotavirus G21P[29] and G24P[33] strains provides evidence for interspecies transmission. J. Gen. Virol. 2011;92:952–960. doi: 10.1099/vir.0.028175-0. [DOI] [PubMed] [Google Scholar]
- Anderson E.J., Weber S.G. Rotavirus infection in adults. Lancet. Infect. Dis. 2004;4:91–99. doi: 10.1016/S1473-3099(04)00928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty E.M., Wong T.H., Trebes A., Argoud K., Attar M., Buck D., Ip C.L., Golubchik T., Cule M., Bowden R., Manganis C., Klenerman P., Barnes E., Walker A.S., Wyllie D.H., Wilson D.J., Dingle K.E., Peto T.E., Crook D.W., Piazza P. A modified RNA-Seq approach for whole genome sequencing of RNA viruses from faecal and blood samples. PLoS One. 2013;8:e66129. doi: 10.1371/journal.pone.0066129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarlet M., Estes M.K., Conner M.E. Comparative amino acid sequence analysis of the outer capsid protein VP4 from four lapine rotavirus strains reveals identity with genotype P[14] human rotaviruses. Arch. Virol. 1997;142:1059–1069. doi: 10.1007/s007050050142. [DOI] [PubMed] [Google Scholar]
- Ciarlet M., Hoffmann C., Lorusso E., Baselga R., Cafiero M.A., Bányai K., Matthijnssens J., Parreño V., de Grazia S., Buonavoglia C., Martella V. Genomic characterization of a novel group A lamb rotavirus isolated in Zaragoza, Spain. Virus Genes. 2008;37:250–265. doi: 10.1007/s11262-008-0257-6. [DOI] [PubMed] [Google Scholar]
- Chinsangaram J., Akita G.Y., Osburn B.I. Detection of bovine group B rotaviruses in feces by polymerase chain reaction. J. Vet. Diagn. Invest. 1994;6:302–307. doi: 10.1177/104063879400600304. [DOI] [PubMed] [Google Scholar]
- Chitambar S.D., Arora R., Kolpe A.B., Yadav M.M., Raut C.G. Molecular characterization of unusual bovine group A rotavirus G8P[14] strains identified in western India: emergence of P[14] genotype. Vet. Microbiol. 2011;148:384–388. doi: 10.1016/j.vetmic.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Cowley D., Donato C.M., Roczo-Farkas S., Kirkwood C.D. Novel G10P[14] rotavirus strain, Northern Territory, Australia. Emerg. Infect. Dis. 2013;19:1324–1327. doi: 10.3201/eid.1908.121653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K., Chauhan R.S., Mahendran M., Malik S.V. Rotavirus diarrhea in bovines and other domestic animals. Vet. Res. Commun. 2009;33:1–33. doi: 10.1007/s11259-008-9070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Halpin R., Kuzmickas R., DePasse J., Feldblyum J., Sengamalay N., Afonso C., Zhang X., Anderson N.G., Ghedin E., Spiro D.J. Viral genome sequencing by random primer methods. BMC Genom. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M.K., Kapikian A.Z. Rotaviruses and their replication. In: Fields B.N., Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Fields Virol. 5th ed. Lippincott, Williams and Wilkins; Philadelphia: 2007. pp. 1917–1974. [Google Scholar]
- Fukai K., Saito T., Inoue K., Sato M. Molecular characterization of novel P[14], G8 bovine group A rotavirus, Sun9, isolated in Japan. Virus. Res. 2004;105:101–106. doi: 10.1016/j.virusres.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Fukai K., Takahashi T., Tajima K., Koike S., Iwane K., Inoue K. Molecular characterization of a novel bovine group A rotavirus. Vet. Microbiol. 2007;123:217–224. doi: 10.1016/j.vetmic.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaicoechea L., Bok K., Jones L.R., Combessies G., Odeón A., Fernandez F., Parreño V. Molecular characterization of bovine rotavirus circulating in beef and dairy herds in Argentina during a 10-year period (1994–2003) Vet. Microbiol. 2006;118:1–11. doi: 10.1016/j.vetmic.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Gerna G., Sears J., Hoshino Y., Steele A.D., Nakagomi O., Sarasini A., Flores J. Identification of a new VP4 serotype of human rotaviruses. Virology. 1994;200:66–71. doi: 10.1006/viro.1994.1163. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Varghese V., Samajdar S., Sinha M., Naik T.N., Kobayashi N. Evidence for bovine origin of VP4 and VP7 genes of human group A rotavirus G6P[14] and G10P[14] strains. J. Clin. Microbiol. 2007;452:751–2753. doi: 10.1128/JCM.00230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Samajdar S., Sinha M., Kobayashi N., Taniguchi K., Naik T.N. Molecular characterization of rare bovine group A rotavirus G15P[11] and G15P[21] strains from eastern India: identification of simian SA11-like VP6 genes in G15P[21] strains. Virus Genes. 2008;37:241–249. doi: 10.1007/s11262-008-0260-y. [DOI] [PubMed] [Google Scholar]
- Griffin D.D., Fletcher M., Levy M.E., Ching-Lee M., Nogami R., Edwards L., Peters H., Montague L., Gentsch J.R., Glass R.I. Outbreaks of adult gastroenteritis traced to a single genotype of rotavirus. J. Infect. Dis. 2002;185:1502–1505. doi: 10.1086/340218. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Nielsen P.R., Hasoksuz M., Thomas C., Wittum T.E., Saif L.J. Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J. Vet. Diagn. Invest. 2003;15:205–212. doi: 10.1177/104063870301500301. [DOI] [PubMed] [Google Scholar]
- Hu L., Crawford S.E., Czako R., Cortes-Penfield N.W., Smith D.F., Pendu J.L., Estes M.K., Prasad B.V.V. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki H., Sakai T., Shirahata T., Taniguchi K., Urasawa T., Urasawa S., Goto H. The distribution of G and P types within isolates of bovine rotavirus in Japan. Vet. Microbiol. 1996;48:367–372. doi: 10.1016/0378-1135(95)00168-9. [DOI] [PubMed] [Google Scholar]
- Liu Y., Huang P., Tan M., Liu Y., Biesiada J., Meller J., Castello A.A., Jiang B., Jiang X. Rotavirus VP8*: Phylogeny, host range, and interaction with histo-blood group antigens. J. Virol. 2012;86:9899–9910. doi: 10.1128/JVI.00979-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes P., Matthijnssens J., Rahman M., Van Ranst M. RotaC: a web-based tool for the complete gemone classification of group A rotaviruses. MBC Microbiol. 2009;9:238. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston D.A., McElhinney L.M., Ellis R.J., Horton D.L., Wise E.L., Leech S.L., David D., de Lamballerie X., Fooks A.R. Next generation sequencing of viral RNA genomes. BMC Genom. 2013;14:444. doi: 10.1186/1471-2164-14-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Rahman M., Martella V., Xuelei Y., De Vos S., De Leener K., Ciarlet M., Buonavoglia C., Van Ranst M. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J. Virol. 2006;80:3801–3810. doi: 10.1128/JVI.80.8.3801-3810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Potgieter C.A., Ciarlet M., Parreño V., Martella V., Bányai K., Garaicoechea L., Palombo E.A., Novo L., Zeller M., Arista S., Gerna G., Rahman M., Van Ranst M. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J. Virol. 2009;83:2917–2929. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Taraporewala Z.F., Yang H., Rao S., Yuan L., Cao D., Hoshino Y., Mertens P.P., Carner G.R., McNeal M., Sestak K., Van Ranst M., Patton J.T. Simian rotaviruses possess divergent gene constellations that originated from interspecies transmission and reassortment. J. Virol. 2010;84:2013–2026. doi: 10.1128/JVI.02081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Banyai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gomara M., Johne R., Rahman M., Ruggeri F.M., Saif L.J., Santos N., Steyer A., Taniguchi K., Patton J.T., Desselberger U., Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch. Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midgley S.E., Hjulsager C.K., Larsen L.E., Falkenhorst G., Böttiger B. Suspected zoonotic transmission of rotavirus group A in Danish adults. Epidemiol Infect. 2012;140:1013–1017. doi: 10.1017/S0950268811001981. [DOI] [PubMed] [Google Scholar]
- Minami-Fukuda F., Nagai M., Takai H., Murakami T., Ozawa T., Tsuchiaka S., Okazaki S., Katayama Y., Oba M., Nishiura N., Sassa Y., Omatsu T., Furuya T., Koyama S., Shirai J., Tsunemitsu H., Fujii Y., Katayama K., Mizutani T. Detection of Bovine Group A Rotavirus Using Rapid Antigen Detection Kits RT-PCR and Next-Generation DNA Sequencing. J. Vet. Med. Sci. 2013;75:1651–1655. doi: 10.1292/jvms.13-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlera L., O’Neill H.G., Jere K.C., van Dijk A.A. Whole-genome consensus sequence analysis of a South African rotavirus SA11 sample reveals a mixed infection with two close derivatives of the SA11-H96 strain. Arch Virol. 2013;158:1021–1030. doi: 10.1007/s00705-012-1559-5. [DOI] [PubMed] [Google Scholar]
- Mphahlele M.J., Peenze I., Steele A.D. Rotavirus strains bearing the VP4P[14] genotype recovered from South African children with diarrhoea. Arch. Virol. 1999;144:1027–1034. doi: 10.1007/s007050050565. [DOI] [PubMed] [Google Scholar]
- Onuma N., Kudo K., Ogawa S., Sakurada M., Sunahara E., Mawatari T., Tsunemitsu H. The role of the group A rotavirus in adult-cow diarrhea. J. Jpn. Vet. Assoc. 2003;56:245–248. (In Japanese) [Google Scholar]
- Papp H., László B., Jakab F., Ganesh B., De Grazia S., Matthijnssens J., Ciarlet M., Martella V., Bányai K. Review of group A rotavirus strains reported in swine and cattle. Vet. Microbiol. 2013;165:190–199. doi: 10.1016/j.vetmic.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreño V., Bok K., Fernandez F., Gomez J. Molecular characterization of the first isolation of rotavirus in guanacos (Lama guanicoe) Arch. Virol. 2004;149:2465–2471. doi: 10.1007/s00705-004-0371-2. [DOI] [PubMed] [Google Scholar]
- Rao C.D., Gowda K., Reddy B.S. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology. 2000;276:104–113. doi: 10.1006/viro.2000.0472. [DOI] [PubMed] [Google Scholar]
- Sato M., Nakagomi T., Tajima K., Ezura K., Akashi H., Nakagomi O. Isolation of serotype G8, P[1] bovine rotavirus from adult cattle with diarrhea. J. Clin. Microbiol. 1997;35:1266–1268. doi: 10.1128/jcm.35.5.1266-1268.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S.C. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2719–2731. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemitsu H., Jiang B., Saif L.J. Sequence comparison of the VP7 gene encoding the outer capsid glycoprotein among animal and human group C rotaviruses. Arch. Virol. 1996;141:705–713. doi: 10.1007/BF01718328. [DOI] [PubMed] [Google Scholar]
- Tsunemitsu H., Smith D.R., Saif L.J. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch. Virol. 1999;144:167–175. doi: 10.1007/s007050050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasawa T., Taniguchi K., Kobayashi N., Mise K., Hasegawa A., Yamazi Y., Urasawa S. Nucleotide sequence of VP4 and VP7 genes of a unique human rotavirus strain Mc35 with subgroup I and serotype 10 specificity. Virology. 1993;195:766–771. doi: 10.1006/viro.1993.1428. [DOI] [PubMed] [Google Scholar]
- Vilcek S., Herring A.J., Herring J.A., Nettleton P.F., Lowings J.P., Paton D.J. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch. Virol. 1994;136:309–323. doi: 10.1007/BF01321060. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Nakagomi T., Koshimura Y., Nakagomi O. Direct evidence for genome segment reassortment between concurrently-circulating human rotavirus strains. Arch. Virol. 2001;146:557–570. doi: 10.1007/s007050170162. [DOI] [PubMed] [Google Scholar]


