Abstract
We developed a series of oligonucleotide primers capable of detecting, typing, and subtyping influenza virus type A (H1 and H3) and type B and respiratory syncytial viruses types A and B. RNAs were isolated from culture fluid or clinical specimens, and cDNA synthesis and PCR were carried out with mixtures of primers specific to each virus. Amplified products were detected by ethidium bromide staining of amplified products after agarose gel electrophoresis. For each virus, five amplified products of different sizes could be distinguished on agarose gels. Multiplex RT-PCR can also be used to detect more than one viral template in the same reaction mixture, allowing identification of multiple strains in the same specimen.
Keywords: Multiplex PCR, Influenza virus, Respiratory syncytial virus, Respiratory infection
1. Introduction
Influenza A and B viruses and respiratory syncytial virus (RSV) are major pathogens involved in respiratory illness in the winter season, and these viruses are responsible for a large number of deaths each year. However, these pathogens have similar clinical symptoms, making accurate diagnosis difficult. Therefore, most cases with respiratory symptom are reported as ‘influenza-like illness,’ which may include diseases caused by other pathogens, such as parainfluenza virus, coronavirus, or human metapneumovirus (hMPV).
The standard method for diagnosis of influenza is the isolation of influenza virus particles using embryonated chicken eggs or MDCK cells. This system has the disadvantage that it requires upwards of 4–6 days to complete. An alternative approach for the rapid detection of influenza viruses, using reverse transcription-PCR (RT-PCR), has been reported.
Multiplex PCR is the inclusion of more than one primer set in an amplification reaction to allow the synthesis of one or more amplicons, indicating the presence of more than one gene or genome segment.
In this study, we developed a multiplex reverse transcription (RT)-PCR method that is capable of detecting and subtyping influenza A (H1 and H3), as well as B viruses and RSV types A and B.
2. Materials and methods
2.1. Clinical specimens
Clinical specimens from patients with respiratory symptoms were obtained from the Prince of Wales Hospital, Hong Kong. As clinical specimens, nasopharyngeal aspirates or throat swabs in 3 ml of virus transport medium were used.
2.2. Cells and virus stock
Influenza viruses and RSV were prepared from tissue culture fluid harvested from MDCK cells and HEp-2 cells, respectively.
Viruses were harvested when cytopathic effects were observed in 70% of the cell monolayer. After mechanically disrupting cells, the supernatants were used as virus stocks and were stored at −70 °C until use.
2.3. RNA
Viral RNA was extracted from 100 μl samples (culture fluid, clinical specimens) using a Roche high RNA kit and eluted with 50 μl of water PCR. For cDNA synthesis, aliquots of 10 μl of RNA were added to RT reaction mixtures.
2.4. PCR amplification
For both single primer pair and multiplex PCR, each primer set was used at 5 pmol. Amplicons were visualized by ethidium bromide staining following electrophoresis on 1.5% agarose gels.
3. Results
3.1. Design of primers
The primers described by Shimizu [1] and Stockton [2] were used for H1 and RSV, respectively.
Primers for H3 and B viruses were designed in our laboratory (Table 1) .
Table 1.
Oligonucleotide primers designed for typing and subtyping influenza and respiratory syncytial viruses
| Primer |
Sequence (5′ to 3′) |
Gene position |
Size (bp) |
|---|---|---|---|
| HA1-1NC | |||
| HA1-1N: | TGAGGGAGCAATTGAGTTCA | HA | |
| HA1-1C: | TGCCTCAAATATTATTGTGT | HA | 431 |
| HA3-al | |||
| HA3-5al: | GAGCTGGTTCAGAGTTCCTC | HA | |
| HA3-3al: | GTGACCTAAGGGAGGCATAATC | HA | 210 |
| NP-bl | |||
| NP-5bl: | GAAGTAGGTGGAGACGGAGGG | NP | |
| NP-3bl: | GTAAACACCCACATTCCAAACG | NP | 390 |
| RSVA | |||
| RSVA F: | GATGTTACGGTGGGGAGTCT | N | |
| RSVA R: | GTACACTGTAGTTAATCACA | N | 334 |
| RSVB (183 bp) | |||
| RSVB F: | AATGCTAAGATGGGGAGTTC | N | |
| RSVB R: | GAAATTGAGTTAATGACAGC | N | 183 |
For influenza A viruses, the gene for hemagglutinin (HA) was chosen as the target. These primers were designed to amplify a conserved fragment of the hemagglutinin (HA) gene. However, no homology was detected between subtypes H1 and H3.
The type B influenza, virus-specific primer was designed to amplify a fragment of 390 bp from the nucleoprotein gene (NP) using segments of the gene showing no homology to sequences of type A influenza viruses.
The primers were checked to ensure that the Tm was within the range of 50–60 °C, the GC content was less than 55%, and primer duplex formation, either within or between pairs, would not exceed five internal base pairs. Primers were also designed to allow five amplicons to be distinguishable based on their size after multiplex PCR (Fig. 1) .
Fig. 1.

Primers designed to allow five amplicons to be distinguishable based on their size after multiplex PCR.
3.2. Optimization of PCR
Annealing temperature for PCR was optimized by examining the yield over the range of 50–56 °C, and for the final reaction, annealing at 54 °C was found to give the optimum product yield (data not shown).
Biochemical optimization of the PCR conditions was performed using a range of buffer, salt, and pH conditions; final reaction conditions of pH 8.8 and 1.2 mM MgCl2 were found to give optimum product yield (data not shown).
Finally, we applied the methods as follows:
Viral RNA was extracted from 100-μl samples (egg fluid, culture fluid) using a Roche high RNA kit and eluted with 50 μl of water. Aliquots of 10 μl of RNA were added to the RT reaction mixtures. cDNA synthesis and PCR were performed using Superscript One step RT-PCR with Platinum Taq (Invitrogen, Carlsbad, CA).
After incubation for 30 min at 50 °C, cDNA was used for PCR amplification.
Amplification was carried out for 40 cycles under the following conditions: denaturation at 94 °C for 1 min, hybridization at 55 °C for 1 min, and elongation at 72 °C for 1 min.
The influenza AH1N1, AH3N2, B RSV (type A), and RSV (type B) virus primer sets, when used together in the multiplex reaction, amplified only the specific products of the theoretically correct size (431, 210, 390, 334, and 183 bp, respectively), which could be distinguished by agarose gel electrophoresis (Fig. 2) .
Fig. 2.
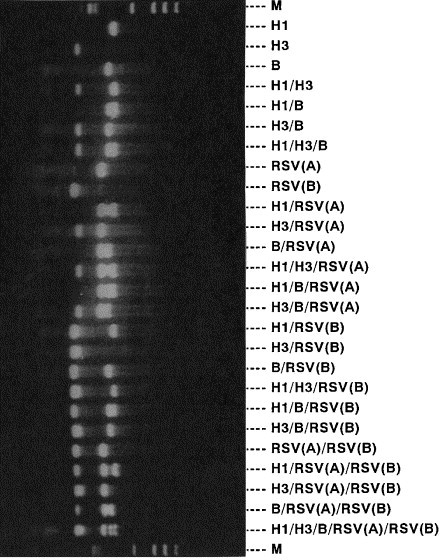
Typing and subtyping of influenza virus and RSV by multiplex PCR.
4. Specificity
Specimens from patients with respiratory infection were examined for the presence of other viruses by this multiplex system as follows: herpes simplex virus 1, parainfluenza 1, parainfluenza 2, parainfluenza 3, enterovirus, adenovirus 3, cytomegalovirus, and metapneumovirus. No amplified products from these virus RNAs were detected (data not shown).
5. Discussion
Multiplex RT-PCR involves the simultaneous amplification of more than one target sequence in a single reaction tube, using mixtures of primer sets.
Originally, multiplex PCR was applied for the diagnosis of genetic disorders using chromosomal DNA. Application of PCR for diagnosis of infectious diseases is more difficult than for identification of genetic disorders because of the low copy numbers of the DNA of interest in clinical specimens. Especially for diagnosis of RNA virus infection, cDNA synthesis is required before amplification by PCR.
Despite these difficulties, multiplex PCR has been introduced for the diagnosis of infectious diseases.
In our system, the detection of each different pathogen was dependent on distinguishing five amplification products of different sizes on agarose gels following RT-PCR with multiplex primer sets.
Multiplex RT-PCR can also be used to detect more than one viral template in the same reaction mixture, allowing identification of viral co-infection using the same respiratory specimen. The major advantage of this RT-PCR system is the rapidity with which diagnosis and characterization can be completed; in the same process, it allows typing and subtyping of respiratory viruses in a single RT-PCR step.
For simplicity, we used a commercial kit for RNA extraction and RT-PCR.
Rapid commercial methods for identification of influenza virus antigens by enzyme immunoassay have been introduced. Although such systems require less time than RT-PCR, they still have problems with regard to sensitivity.
The results of the present study indicated that five of the most common pathogens involved in respiratory infection could be detected quickly and easily using multiplex RT-PCR.
The emergence of severe acute respiratory syndrome (SARS) has prompted the need to be able to make a rapid differential diagnosis between influenza and SARS.
Further modifications of this protocol will be performed to allow detection of more pathogens (e.g., metapneumovirus, SARS-coronavirus). Our results suggest that multiplex RT-PCR is a simple, cost-efficient, and rapid method for the combined screening/typing of respiratory infection.
References
- 1.Shimizu H. The Journal of the Japanese Association for Infectious Disease. 1997;72:827–833. doi: 10.11150/kansenshogakuzasshi1970.72.827. [DOI] [PubMed] [Google Scholar]
- 2.Stockton J. Journal of Clinical Microbiology. 1998;36:2990–2995. doi: 10.1128/jcm.36.10.2990-2995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


