Abstract
Objective
The non-translation RNA-microRNA (miRNA) has been demonstrated to correlate to various disease occurrence in body. Serum miRNA was gradually considered as molecular markers for disease diagnosis. This study was designed to analyze differential serum miRNAs level in hormone-induced non-traumatic osteonecrosis of the femoral head (hormone-NOFH) patients.
Methods
We selected 30 patients with hormone-NOFH as case group, and 30 healthy volunteers were recruited as control group. miRCURYTM LNA miRNA chip and quantitative RT-PCR were used to examine differential miRNAs expression. Correlation assay was performed between miRNAs and NOFH trait.
Results
We found that 9 miRNAs were upregulated while 3 miRNAs were downregulated in hormone-TOFH patient serum by result of miRNA chip. QRT-PCR assay revealed that the level of miR-423-5p was significantly increased and miR-10a-5p was significantly decreased. Using Spearman correlation analysis, we observed that miR-423-5p serum level is positive association to FHC levels whereas miR-10a-5p has no association with FHC levels. Furthermore, miR-423-5p is negatively correlated to its downstream molecule-adiponectin.
Conclusion
We report a miRNA profile of hormone-NOFH and provide a new perspective to understand this intricate disease. This novel information suggests the potential roles of miR-423-5p in the diagnosis, prognosis biomarkers, or therapy targets of hormone-NOFH.
Keyword: MicroRNA, Hormone-induced non-traumatic osteonecrosis of the femoral head (hormone-NOFH), miR-423-5p, miRNA microchip
1. Introduction
Non-traumatic osteonecrosis of the femoral head (NOFH) is a common disease that regularly affects patients aged 20–50 years and is characterized by a secondary femoral head collapse (FHC), hip pain and dysfunction [1]. The main causes of NOFH is blood circulation obstacle of the femoral head, however incentives to the NOFH pathogenesis are complex. Some studies indicated that steroid, which extensively used in various disease such as severe acute respiratory syndrome (SARS), systemic lupus erythematosus, rheumatoid arthritis, leukemia, is an important inducer for occurrence of NOFH [2]. However, the mechanism underlying steroid-induced NOFH remains to be elaborated.
Micro-RNAs (miRNAs) are small noncoding regulatory RNAs of about 19–22 nucleotides. They function as translational inhibitor or mRNA degradation promoter by binding to the 3′untranslated region of their target mRNAs [3]. Recently, the biological roles of miRNA are under intense investigation, and their dysfunction has been considered to be extensively involved in the pathogenesis of numerous types of disease, including cancers, cardiovascular disease, lymphocytic leukemia as well as orthopaedics diseases [4], [5], [6]. Therefore, miRNA may play an important roles in the occurrence and development of various types of disease and may function as a novel biomarker and tool for clinical therapy. Currently, abnormal miRNA expression has been shown to play an important role in development of bone dysfunction. Currently, the best studied miRNAs that are involved in differentiation of osteoclasts are miR-21, miR-155 and miR-223 [7], [8], [9]. Studies by Zhang et al. established that at least 11 different miRNAs (i.e., miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-217, miR-218, and miR-338) contributed to control osteoblast differentiation [10] and are expressed to different degrees in both osteogenic and non-osteogenic cells. miR-210 is intensely expressed in osteonecrosis, and was speculated to play a role in osteonecrosis pathogenesis [11]. Dysfunction of miR-17-5p is demonstrated to contribute to NOFH pathogenesis [12]. Adiponectin, considering as a serum biomarker for NOFH [13], is significantly associated with the presence of NOFH and also has been proved to regulated by mi-378, miR-221 and miR-423-5p [14]. Yuan et al. have identified thousands of upregulations such as miR-21, miR-17, and miR-92a, whereas others tended to downregulation, such as miR-205 and miR-145, in reparative interface of the femoral head with osteonecrosis by using high-throughput techniques-gene chip [15]. Notably, evidence increasingly shows serum miRNAs differential expression might be a marker for disease diagnosis. Although about 1000 miRNAs have been estimated in human, previous studies have only screened few miRNA expression in NOFH patients. Given this, it is necessary to screen NOFH with larger collections of miRNAs by gene chip.
To explore more related miRNAs, we have used 6th generation of miRNA array that contains more than 1891 probes, which include nearly all human miRNAs and identified aberrant expression miRNAs between hormone-NOFH serum and healthy serum.
2. Material and methods
2.1. Study subject
A total of 30 patients with hormone non-traumatic osteonecrosis of the femoral head (case group; aged at 50.5 ± 2.7; 45% men) were recruited from our hospital. In addition, a total of 30 healthy volunteers (control group; aged at 49.5 ± 3.1; 55% men) enrolled at our hospital. The two groups were matching in age, gender, race and region. All subjects were obtained from March 2013 to January 2014. The inclusion criteria for the case group were: imaging in the diagnosis of stage in III duration (Femoral head is a bit deformation, has collapsed, subchondral fracture, but no significant change in acetabulum and joint); no intervention contraindicated; patients have taken steroidal hormones. The exclusion criteria for the case group were: femoral head necrosis has reached IV period; patients have obvious contraindicated intervention operation. The inclusion criteria for the control group were: healthy person is with no history of serious disease and never took hormone. All the relevant data were collected from clinical, baseline laboratory medicine, as well as the relevant demographic data.
The study was approved by Linyi People’s Hospital and the study-related procedures were initiated when informed consent were signed by participants.
2.2. Serum isolation and storage
An 20 mL aliquot of blood was collected from each of the 60 participants (30 patients in the ‘case group’ and 30 individuals in the ‘control group’) with serum collection tubes (BD vacutainer® rapid serum tube, No. 367981; USA). The whole blood was stand for 30 min at room temperature and then was centrifuged at 1800×g for 5 min for serum separation. The resultant serum was aliquoted into microtubes and immediately frozed with liquid nitrogen. All samples were finally stored at −80 °C for following miRNA chip or quantitative reverse transcription–polymerase chain reaction (qRT-PCR) assay.
2.3. miRNA chip assay
A total of 100 μL aliquot of serum was incubated at 56 °C overnight with 0.65 mg/mL proteinase K (Sigma–Aldrich, CA, USA). The RNAs were extracted by acid phenol chloroform and maintained in ethanol overnight at −20 °C. Following, RNAs were performed with a second acid phenol chloroform extraction, and re-suspended in 200 μL of double distilled water (DDW). Among this, 2 μL sample was took out and diluted with 198 μL DDW for purity assessment using spectrophotometer.
After isolated from the samples, the miRNAs were labeled by the miRCURY Hy3/Hy5 using power labeling kit (Exiqon, Vedbaek, Denmark) according to the manufacturer’s protocol. The microgram of each sample was labeled with Hy3 fluorescent label on its 3′-end catalyzed by T4 RNA ligase. In brief, RNA in 2.0 μL of DDW was combined with 1.0 μL of CIP buffer and CIP (Exiqon) and incubated at 37 °C for 30 min. This incubation was stopped by mixture staying for 5 min at 95 °C. Then the mixture was added with 3.0 μL of labeling buffer, 1.5 μL of fluorescent label (Hy3), 2.0 μL of DMSO, and 2.0 μL of labeling enzyme. The labeling reaction was performed at 16 °C for 1 h and stopped by incubation for 15 min at 65 °C. After terminating the labeling procedure, the Hy3-labeled samples were hybridized on the miRCURY LNA array (v.16.0) (Exiqon) according to array manual. Briefly, the total 25 μL mixture from Hy3-labeled samples was denatured for 2 min at 95 °C, incubated on ice for 2 min, and then hybridized to the microarray for 16–20 h at 56 °C in a 12-Bay Hybridization Systems (Hybridization System, Nimblegen Systems, Inc., Madison, WI, USA), which provides an active mixing action and constant incubation temperature to improve hybridization uniformity. Following hybridization, the slides were obtained after being washed several times with Wash buffer kit (Exiqon), and finally dried. Finally, the slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA).
2.4. RNA extraction and qRT-PCR
miRNA levels of samples were determined by qRT-PCR. In brief, the whole sample was mixed with the poly(A) and undergone cDNA procedure with poly(A) polymerase (NEB-M0276L) and Super-Script II reverse transcriptase (Invitrogen, USA) in a total volume of 20 mL for 1 h at 37 °C. The cDNA product was diluted 1:40 for following real time PCR, which is miRNA specific, using 0.125 mL of serum per quantification of one miRNA. The content in this reaction mixture includes miRNAs-specific forward primers, a TaqMan probe complementary to the 3′ end of the specific miNA sequence, as well as to part of the poly(A) adaptor sequence, and a universal reverse primer complementary to the consensus 3′ sequence of the oligo (dT tail). The negative controls studied along with the RNA samples served to detect potential contaminations and/or non-specific amplifications. The cycle number at which the fluorescence passes the threshold [cycle threshold (Ct)] was measured for each miRNA in each sample.
2.5. Data analysis and statistics
The experimental data and clinical diagnose data were statistically analyzed using software SPSS18.0. Data were represented as mean ± SE. Microarray data were statistically analyzed by fold change (FC). All statistical significance was defined as P < 0.05 or FC > 2.
To assess the correlation between them, iR-423-5p or miR-10a-5p and Index of FHC or Adiponectin, the Spearman correlation coefficient was used. The relationship was described as the correlation coefficient r and statistical significance was defined as P < 0.05. Serum miRNA were normalized by scaling with the mean Ct of the samples: for each sample the average Ct of all miRNAs measured in the sample was subtracted from the Ct of each miRNA.
3. Results
3.1. Characteristics of the subjects
The clinical demographic and clinical characteristics of the hormone-NOFH and control group are shown in Table 1 . The two groups were similar regarding age, gender, body mass index (BMI). Predicted significant changes, reflecting the different nature of the hormone-NOFH compared to the control group, were in the C-reactive protein (CRP), erythrocyte sedimentation (ESR), fibrinogen (FIB) and adiponectin. The clinical characterization of hormone-NOFH is III-IV phase by FICAT staging and appearance of femoral head collapse (FHC).
Table 1.
The demographic and clinical characteristics of patients.
| Parameters | Control | Patients |
|---|---|---|
| Age | 49.5 ± 3.1 | 50.5 ± 2.7 |
| Male (%) | 55% | 45% |
| BMI (kg/m2) | 23.9 ± 2.3 | 24.7 ± 4.3 |
| CRP (mg/L) | 2.14 ± 0.41 | 4.9 ± 0.63* |
| ESR (mm/h) | 4.12 ± 0.71 | 24 ± 6.9* |
| FIB (g/L) | 1.72 ± 0.63 | 4.2 ± 0.9* |
| WBC (109/L) | 5.4 ± 2.1 | 5.7 ± 2.7 |
| Adiponectin (μg/mL) | 11.23 ± 3.2 | 6.5 ± 1.4* |
| FICAT staging | NA | III–IV |
| Index of FHC | NA | 0.34 ± 0.12 |
BMI, body mass index; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; WBC, white blood cell counts; FIB, fibrinogen; FHC, femoral head collapse; NA, not applicable.
P < 0.05.
3.2. Clustering analysis of the significantly changed miRNA Genes
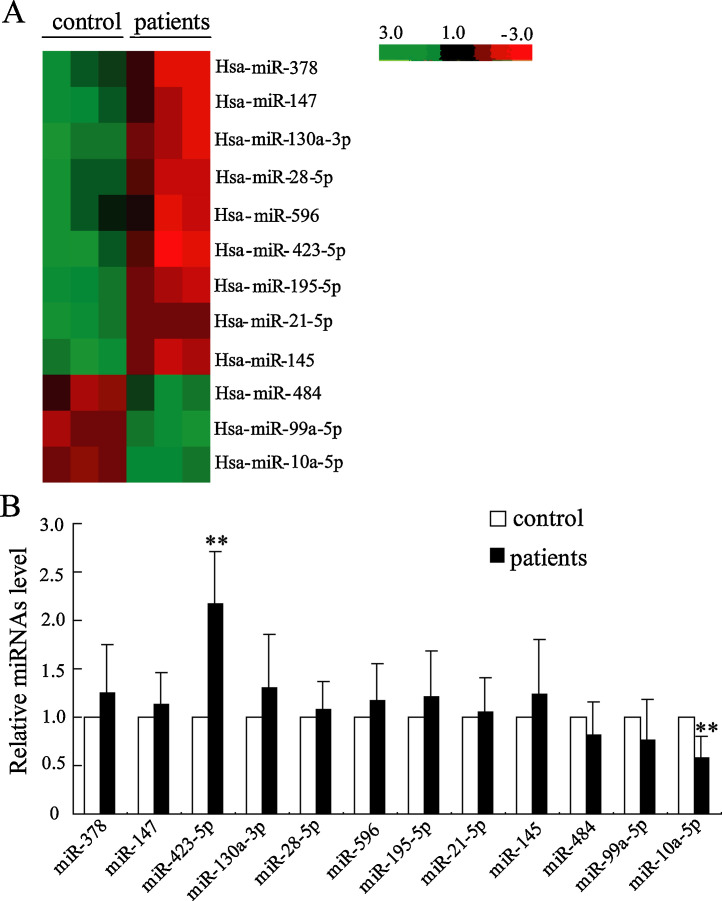
We used miRCURY™ LNA miRNA chip to evaluate miRNA expression profiles in all serum samples. The distinguishable miRNA expression profiling among NTFH and healthy control were presented by the result of hierarchical clustering. As shown in Fig. 1 A, compared to healthy control, 9 miRNAs are upregulated and 3 miRNAs are downregulated in hormone-NTFH. To confirm the differentially expressed miRNAs, expression levels of the 9 upregulated miRNAs genes and 3 downregulated miRNAs genes were analyzed by qRT-PCR. Our data showed that among these aberrant expression miRNAs, miR-423-5p was significantly upregulated and miR-10a-5p was significantly downregulated (Fig. 1B) in hormone-NTFH patients.
Fig. 1.
MiRNAs expression in plasma of non-traumatic osteonecrosis of the femoral head patient. (A) Hierarchical clustering for differentially expressed miRNA in NTFH patients versus healthy control; green, low relative expression; red, high relative expression; FC ≥ 2. (B) qRT-PCR was performed to verify miRNAs expression. Data were expressed as mean ± SD. **P < 0.01 compared with control.
3.3. Correlation analysis between miRNAs and index of FHC
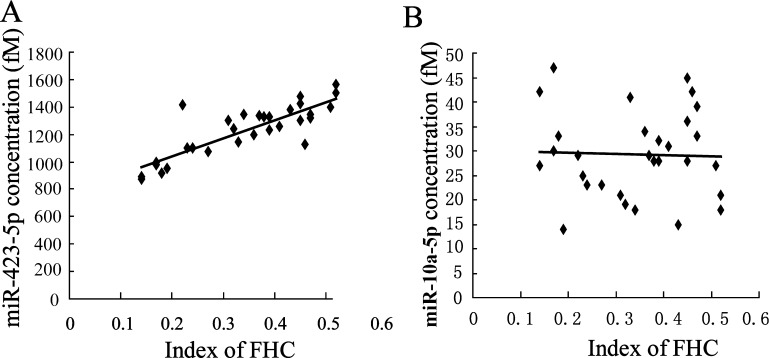
To identify association between differentially expressed miRNAs and FHC, Spearman’s rank correlation coefficient assay was performed. Our data in Fig. 2 A showed a strong correlation between miR-423-5p level and FHC (Spearman correlation is 0.84, P = 0.000). While miR-10a-5p is no correlation to FHC (Spearman correlation is −0.31, P = 0.869; Fig. 2B).
Fig. 2.
Significant correlation analysis between the serum miRNAs concentration and Index of FHC. Serum miRNAs levels were determined by using absolute quantitative detection. (A) miR-423-5p level is associated with Index of FHC. (B) miR-10a-5p is associated with Index of FHC.
3.4. Correlation analysis between miR-423-5p and index of FHC
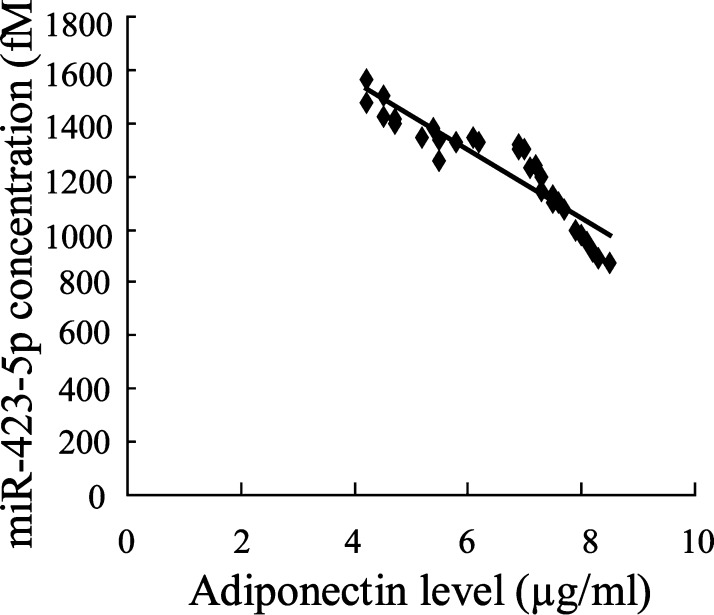
Next, we evaluated whether miR-423-5p expression is correlated with adiponectin level. The analysis data showed that there is a strong negative association between miR-423-5p concentration and Adiponectin level in NOFH patients (Spearman correlation is −0.92, P = 0.000; Fig. 3 ).
Fig. 3.
Significant correlation analysis between the serum miR-423-5p concentration and Adiponectin. miR-423-5p level is associated with Adiponectin level (P = 0.000).
4. Discussion
Hormone usage probably induced secondary action and is recognized as a common cause for NOFH pathogenesis. Due to complexity of pathogenesis, the therapeutic method has been always explored. Notably, the therapeutic trials to regulate the endogenous miRNAs related to diseases provide the potential of therapeutical targeting. Presently, the obstacle is the determination of the target miRNA. In this study, by screening with miRNA microchip, we found 9 prominent miRNAs (miR-378, miR-147, miR-130a-3p, miR-28-5p, miR-592, miR-423-5p, miR-195-5p, miR-21-5p, miR-145) with increased levels and three prominent miRNAs (miR-10a-5p, miR-484, miR-99a-5p) with decline levels in the serum of hormone-NOFH patients vs. healthy control. Based on this result, we test this differential miRNAs expression by qRT-PCR, and found out one upregulated miRNA (miR-423-5p) expression and one downregulated miRNA (miR-10a-5p) expression. Moreover, in NOFH patients, there is significant association between the serum miR-423-5p level and an important known diagnostic parameters (Index of FHC) as well as serum mark for NOFH (adiponectin). The diagnostic implications of adiponectin was demonstrated in Shuai et al. study performed in NOFH patients [13].
NOFH pathogenesis is characteristic of disorder of bone metabolism that reflects a delicate balance in anabolic activities of osteoblasts and the catabolic actions of osteoclasts [16]. As a key controller for epigenetic regulating genes, miRNAs are revealed to be important regulator for bone resorbing activity mediated by osteoclasts, as well as osteoblast proliferation and differentiation. For example, miR-378 was shown to directly upregulate bone morphogenetic protein 2 (BMP2) expression that is essential for osteogenic differentiation in mesenchymal progenitor cells [17]. Tumor necrosis factor (TNF)-alpha is a key cytokine regulator of bone and mediates inflammatory bone loss, however, it-induced osteoblasts apoptosis was attenuated by miR-23a [18]. miR-204 attenuated osteoblast differentiation by directly targeting Runx2 in MSCs [19]. we reveal miR-34a as a key osteoclast suppressor of osteoclastogenesis, bone resorption and the bone metastatic niche by inhibiting transforming growth factor-β-induced factor 2 (Tgif2) that is significant for bone resorption.
Previous studies which revealed altered serum levels of miRNAs in the osteoporotic fractures [20], osteosarcomas [21] as well as osteogenesis imperfect [22] combined with increasing understanding of their role in osteoblasts proliferation have led to an appreciation of the potential use of miRNAs as biomarkers of bone tissue disorder. Given this, we combined miRNA microchip assay and qRT-PCR in NOFH serum and attempt to find out changed levels of specific miRNAs in circulation of NOFH. The results of our study are consistent with this notion, showing that increased level of miR-423-5p and decreased level of miR-10a-5p in hormone-NOFH patients may be used as markers for NOFH diagnosis. Currently, although miRNAs has been demonstrated to be correlated with bone tissue homeostasis, the understanding of the clinical impact of miRNAs in NOFH patients is still in its infancy. In a recently published study, Jia et al. proved that dysfunction of miR-17-5p is contribute to NOFH pathogenesis [12]. Thus we using Spearman correlation assay to test whether exist a association between circulating miRNAs and hormone-NOFH clinical parameters. From the data, we confirmed that upregulated miR-423-5p has close relevance to diagnostic parameters (Index of FHC) that commonly appears in end-stage disease.
Next, we analyzed the correlation between miR-423-5p and serum adiponectin in hormone-NOFH. Adiponectin is a collagen-like cytokine and commonly found in human serum with a range of 5–30 μg/mL [23]. Adiponectin possesses anti-atherogenic and anti-inflammatory properties and can influence lipid metabolism [24]. Abnormal lipid metabolism and bone marrow fat cell hypertrophy/proliferation play important roles in NOFH. Moreover, recent studies have also implicated adiponectin in the regulation of bone metabolism [25]. Oshima et al. reported that adiponectin could increase bone mass by suppressing osteoclastogenesis and activating osteoblastogenesis [26]. Shinoda et al. suggested distinct adiponectin actions on bone formation [27]. With regard to the important role of adiponectin in NOFH pathogenesis, Shuai et al. proved that low adiponectin level is significantly associated with the presence of NOFH and adiponectin may act as a biomarker for NOFH diagnosis. Our data showed that there is a strong negative association between miR-423-5p concentration and adiponectin level in hormone-NTFH patients.
In summary, this study firstly defined a specific unregulated serum miRNAs-miR-423-5p that was highly correlated to important prognostic parameters (Index of FHC and adiponectin) in hormone-NOFH. This discrimination predicted miR-423-5p was potentially a biomarker for hormone-NOFH diagnosis in clinic. Although the role of miRNAs in the clinical management of NOFH disease has yet to be studied, our work shows that they are exciting potential candidates for this challenge.
Conflict of interest
The authors have no actual or potential conflicts of interest to declare.
Acknowledgment
This work was supported by Science and Technology Develop Project of Shandong province (No. 2014GSF119022).
References
- 1.Mouzas O.D., Zibis A.H., Bonotis K.S., Katsimagklis C.D., Hadjigeorgiou G.M., Papaliaga M.N. Psychological distress, personality traits and functional disability in patients with osteonecrosis of the femoral head. J. Clin. Med. Res. 2014;6:336–344. doi: 10.14740/jocmr1851w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan G., Kang P.D., Pei F.X. Glucocorticoids affect the metabolism of bone marrow stromal cells and lead to osteonecrosis of the femoral head: a review. Chin. Med. J. 2012;125:134–139. [PubMed] [Google Scholar]
- 3.Tang P., Xiong Q., Ge W., Zhang L. The role of microRNAs in osteoclasts and osteoporosis. RNA Biol. 2014;11:1355–1363. doi: 10.1080/15476286.2014.996462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devaux Y., Stammet P., Friberg H., Hassager C., Kuiper M.A., Wise M.P. MicroRNAs: new biomarkers and therapeutic targets after cardiac arrest. Crit. Care. 2015;19:54. doi: 10.1186/s13054-015-0767-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L.Q., Kwong Y.L., Wong K.F., Kho C.S., Jin D.Y., Tse E. Epigenetic inactivation of mir-34b/c in addition to mir-34a and DAPK1 in chronic lymphocytic leukemia. J. Transl. Med. 2014;12:52. doi: 10.1186/1479-5876-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kafchinski L.A., Jones K.B. MicroRNAs in osteosarcomagenesis. Adv. Exp. Med. Biol. 2014;804:119–127. doi: 10.1007/978-3-319-04843-7_6. [DOI] [PubMed] [Google Scholar]
- 7.Sugatani T., Hruska K.A. MicroRNA-223 is a key factor in osteoclast differentiation. J. Cell. Biochem. 2007;101:996–999. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 8.Sugatani T., Vacher J., Hruska K.A. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117:3648–3657. doi: 10.1182/blood-2010-10-311415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugatani T., Hruska K.A. Down-regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J. Cell. Biochem. 2013;114:1217–1222. doi: 10.1002/jcb.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Xie R.L., Croce C.M., Stein J.L., Lian J.B., van Wijnen A.J. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamasaki K., Nakasa T., Miyaki S., Yamasaki T., Yasunaga Y., Ochi M. Angiogenic microRNA-210 is present in cells surrounding osteonecrosis. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2012;30:1263–1270. doi: 10.1002/jor.22079. [DOI] [PubMed] [Google Scholar]
- 12.Jia J., Feng X., Xu W., Yang S., Zhang Q., Liu X. MiR-17-5p modulates osteoblastic differentiation and cell proliferation by targeting SMAD7 in non-traumatic osteonecrosis. Exp. Mol. Med. 2014;46:e107. doi: 10.1038/emm.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuai B., Shen L., Yang Y.P., Xie J., Shou Z.X., Wei B. Low plasma adiponectin as a potential biomarker for osteonecrosis of the femoral head. J. Rheumatol. 2010;37:2151–2155. doi: 10.3899/jrheum.100342. [DOI] [PubMed] [Google Scholar]
- 14.Prats-Puig A., Ortega F.J., Mercader J.M., Moreno-Navarrete J.M., Moreno M., Bonet N. Changes in circulating microRNAs are associated with childhood obesity. J. Clin. Endocrinol. Metab. 2013;98:E1655–E1660. doi: 10.1210/jc.2013-1496. [DOI] [PubMed] [Google Scholar]
- 15.Yuan H.F., Von Roemeling C., Gao H.D., Zhang J., Guo C.A., Yan Z.Q. Analysis of altered microRNA expression profile in the reparative interface of the femoral head with osteonecrosis. Exp. Mol. Pathol. 2015;98:158–163. doi: 10.1016/j.yexmp.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Martin T.J. Bone biology and anabolic therapies for bone: current status and future prospects. J. Bone Metab. 2014;21:8–20. doi: 10.11005/jbm.2014.21.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hupkes M., Sotoca A.M., Hendriks J.M., van Zoelen E.J., Dechering K.J. MicroRNA miR-378 promotes BMP2-induced osteogenic differentiation of mesenchymal progenitor cells. BMC Mol. Biol. 2014;15:1. doi: 10.1186/1471-2199-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong J., Cui X., Jiang Z., Sun J. MicroRNA-23a modulates tumor necrosis factor-alpha-induced osteoblasts apoptosis by directly targeting Fas. J. Cell. Biochem. 2013;114:2738–2745. doi: 10.1002/jcb.24622. [DOI] [PubMed] [Google Scholar]
- 19.Huang J., Zhao L., Xing L., Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeliger C., Karpinski K., Haug A.T., Vester H., Schmitt A., Bauer J.S. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2014;29:1718–1728. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C., Yao C., Li H., Wang G., He X. Combined elevation of microRNA-196a and microRNA-196b in sera predicts unfavorable prognosis in patients with osteosarcomas. Int. J. Mol. Sci. 2014;15:6544–6555. doi: 10.3390/ijms15046544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F.S., Chuang P.C., Lin C.L., Chen M.W., Ke H.J., Chang Y.H. MicroRNA-29a protects against glucocorticoid-induced bone loss and fragility in rats by orrating bone acquisition and resorption. Arthritis Rheum. 2013;65:1530–1540. doi: 10.1002/art.37948. [DOI] [PubMed] [Google Scholar]
- 23.Berner H.S., Lyngstadaas S.P., Spahr A., Monjo M., Thommesen L., Drevon C.A. Adiponectin and its receptors are expressed in bone-forming cells. Bone. 2004;35:842–849. doi: 10.1016/j.bone.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Glueck C.J., Fontaine R.N., Gruppo R., Stroop D., Sieve-Smith L., Tracy T. The plasminogen activator inhibitor-1 gene, hypofibrinolysis, and osteonecrosis. Clin. Orthop. Related Res. 1999:133–146. doi: 10.1097/00003086-199909000-00017. [DOI] [PubMed] [Google Scholar]
- 25.Lenchik L., Register T.C., Hsu F.C., Lohman K., Nicklas B.J., Freedman B.I. Adiponectin as a novel determinant of bone mineral density and visceral fat. Bone. 2003;33:646–651. doi: 10.1016/s8756-3282(03)00237-0. [DOI] [PubMed] [Google Scholar]
- 26.Oshima K., Nampei A., Matsuda M., Iwaki M., Fukuhara A., Hashimoto J. Adiponectin increases bone mass by suppressing osteoclast and activating osteoblast. Biochem. Biophys. Res. Commun. 2005;331:520–526. doi: 10.1016/j.bbrc.2005.03.210. [DOI] [PubMed] [Google Scholar]
- 27.Shinoda Y., Yamaguchi M., Ogata N., Akune T., Kubota N., Yamauchi T. Regulation of bone formation by adiponectin through autocrine/paracrine and endocrine pathways. J. Cell. Biochem. 2006;99:196–208. doi: 10.1002/jcb.20890. [DOI] [PubMed] [Google Scholar]