Abstract
Bovine respiratory disease (BRD) often occurs when active respiratory virus infections (BHV-1, etc.) impair resistance to Mannheimia haemolytica infection in the lower respiratory tract. The interactions that occur when the respiratory epithelium encounters these viral and bacterial pathogens are poorly understood. We used Agilent bovine gene microarray chips containing 44,000 transcripts to elucidate bovine bronchial epithelial cell (BBEC) responses following in vitro exposure to BHV-1 alone, M. haemolytica alone, or both BHV-1 and M. haemolytica. Microarray analysis revealed differential regulation (>2-fold) of 978 transcripts by BHV-1 alone, 2040 transcripts by M. haemolytica alone, and 2189 genes by BHV-1 and M. haemolytica in combination. M. haemolytica treatment produced significantly greater inductions (>10-fold) of several inflammation associated genes, such as CXCL2, IL-6, IL-1α, e-selectin, and IL-8, than to BHV-1 alone. Functional analysis of the microarray data revealed a significant upregulation of genes involved in important biological processes such as inflammation (TNF-α, IL-8, Tlr-2, IL-1, CXCL2, CSF2), vascular functions (VEGF, EDN2) and leukocyte migration (ICAM1, IL-16) during a co-infection with BHV-1 and M. haemolytica compared to either pathogen alone. This study provides evidence to support that lung epithelial cells are a source of mediators that may promote inflammatory changes observed during bovine respiratory disease.
Keywords: Microarray, Bovine herpesvirus 1, Mannheimia haemolytica, Bronchial epithelial cells
1. Introduction
Bovine respiratory disease (BRD) is a multi-factorial disease complex that involves interactions among stressors, management factors, and viral and bacterial pathogens (Car et al., 1991, Hodgson et al., 2005, Ohmann and Babiuk, 1985). The main bacterial pathogen of BRD is Mannheimia haemolytica, which produces a potent leukotoxin that is its principal virulence factor (Fedorova and Highlander, 1997, Highlander et al., 2000). In its most severe manifestation, infection with M. haemolytica can cause fibrinous pleuropneumonia (Car et al., 1991, Hodgson et al., 2005, Loneragan et al., 2001, Ohmann and Babiuk, 1985, Yates, 1982). It is clear that in cattle, as in humans and other mammalian species, active viral infection dramatically increases susceptibility to bacterial pneumonia. This has been demonstrated experimentally in cattle infected with any one of several bovine respiratory viruses such as bovine herpesvirus 1 (BHV-1) and bovine respiratory syncitial virus (BRSV), which renders them highly susceptible to challenge with M. haemolytica (Hodgson et al., 2005, Yates, 1982). In marked contrast, far greater numbers of M. haemolytica cells are required to cause pneumonia in the absence of viral infection, even when the bacterial cells are inoculated into a bronchus (Ohmann and Babiuk, 1985, Yates, 1982). These observations indicate that viral infection impairs host defense mechanisms against M. haemolytica, or amplifies undesirable aspects of the host response to this bacterial pathogen.
We have a limited understanding of how respiratory virus infection increases the susceptibility of cattle to bacterial pneumonia with M. haemolytica. Earlier in vivo and in vitro studies identified functional defects in bovine leukocytes exposed to BHV-1 or other viruses (Forman et al., 1982, Hinkley et al., 1998, Leite et al., 2002, McGuire and Babiuk, 1984, Noel et al., 1988). However, these relatively modest alterations do not sufficiently explain the increased susceptibility to bacterial pneumonia that is observed in the field.
BHV-1 does not result in a productive infection in bovine leukocytes. However, it infects other cell types including bovine epithelial cells (Babiuk et al., 1996, Thaker et al., 1994). Thus, one might infer that the effects of BHV-1 on resistance to bacterial pneumonia are indirect and may result in part from epithelial cells releasing chemical mediators during viral infection (Babiuk et al., 1996, Brown and Shin, 1990, Leite et al., 2004a, Leite et al., 2004b, Raz et al., 1993). These mediators in turn can alter the activity of bovine neutrophils and mononuclear phagocytes. For example, we demonstrated previously that exposure of leukocytes to BHV-1 virus -induced cytokines, alters leukocyte expression (or activation) of β-integrins in ways that increase their susceptibility to M. haemolytica leukotoxin (LKT) and their adhesion to epithelial cells in vitro (Rivera et al., 2009, Leite et al., 2004a, Leite et al., 2004b).
Previous studies have focused on the direct interactions of bovine respiratory pathogens (both viral and bacterial) with bovine leukocytes (Forman et al., 1982, Leite et al., 2004a, Leite et al., 2004b). Less attention has been paid to the interplay among viral and bacterial pathogens and respiratory epithelial cells. Recent reports from our laboratory and others (Gershwin et al., 2005, Hodgson et al., 2005, Rivera et al., 2009, Leite et al., 2004a, Leite et al., 2004b, Raz et al., 1993, Wilson et al., 2005) provide evidence for viral and bacterial pathogen interactions with bovine respiratory epithelial cells. These findings suggest that BHV-1 infection of respiratory epithelial cells results in the release of mediators that attract leukocytes, and with subsequent exposure to M. haemolytica LKT, intensify the inflammatory process that characterizes BRD.
In this study we investigated the gene expression response of bovine bronchial epithelial cells to the bovine respiratory pathogens BHV-1 and M. haemolytica. We use a bovine gene microarray platform to assess expression of more than 44,000 gene targets by primary bovine bronchial epithelial (BBE) cells exposed in vitro to BHV-1, M. haemolytica, or the two agents in combination (Fig. 1 ). The results of this analysis demonstrate significant changes in gene expression as a result of epithelial cell encounter with BHV-1 and M. haemolytica. These observations will inform subsequent efforts to assess how products of these genes alter the response of bovine leukocytes to respiratory pathogens.
Fig. 1.
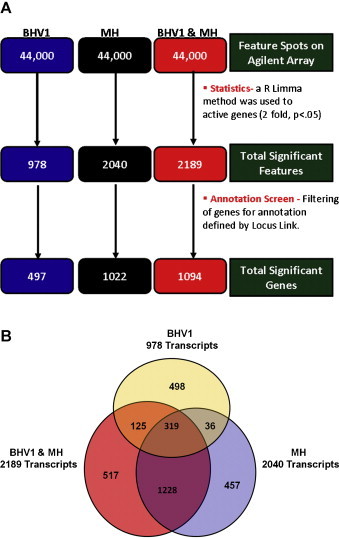
Overview of differential gene expression by bronchial epithelial cells exposed to BHV-1, M. haemolytica (MH) or BHV-1 and M. haemolytica in combination (BHV1 + MH). (A) Identification of differentially expressed features at each treatment by annotation screening (p < 0.05 and fold change ≥ 2.0). (B) There were 978, 2040, and 2189 differentially expressed transcripts identified for BHV-1, MH, and BHV-1 + MH, respectively. Three hundred and nineteen transcripts were differentially regulated by all treatments (p < 0.05, and fold change ≥ 2.0).
2. Methods
2.1. Bovine bronchial epithelial cell culture
Primary bovine bronchial epithelial cells (BBEs), generously provided by Dr. Allen-Gipson (University of Nebraska Medical Center), were maintained in Dulbecco's Modified Eagle's Medium/F12 (DMEM) (Cellgro; Mediatech, Inc., Herndon, VA) containing 10% Fetal Bovine Serum (FBS) (Atlanta Biologicals, Lawrenceville, GA), 2 mM l-glutamine (l-glu) (Mediatech), 200 ng/l epithelial growth factor (EGF) (Sigma, St. Louis, MO), 1% penicillin and streptomycin (Cellgro) at 37 °C and 5% CO2.
2.2. M. haemolytica
M. haemolytica A1 isolate D153 (a plasmid-negative strain isolated from the lung of a steer that died of pneumonia) was a gift from R. Briggs (Ames, IA). M. haemolytica was grown in brain heart infusion (BHI) broth with shaking at 37 °C for 10 h. Bacterial cells were pelleted at 3750 × g, washed three times in phosphate-buffered saline (PBS). The number of CFU in each broth culture was extrapolated from growth curves performed in our laboratory, and confirmed by dilution plating on tryptic soy agar with 5% sheep red blood cells (Becton Dickinson, Franklin Lakes, NJ) to enumerate CFU.
2.3. Bovine herpesvirus 1 propagation
Cooper strain of bovine herpesvirus 1 (BHV-1), was propagated on Simian Virus 40 Large T (SV-40 LT) antigen transformed BBE cells. When cytopathic effects were evident, conditioned media was collected and centrifuged at 250 × g for 5 min. The virus titer was determined by standard plaque assay on transformed BBE cells and expressed as plaque-forming units (PFU)/ml. Alternatively the Reed and Muench method was used to determine the Tissue Culture Infectious Dose 50 (TCID50) per ml of stock virus. Aliquots (200 μl) of viral stocks were stored at −80 °C. A fresh aliquot of stock was thawed and used for each experiment. Cell culture media and conditioned media were free of endotoxin or mycoplasma contamination as determined by the Limulus Amoebocyte Lysate kit (Cambrex, East Rutherford, NJ) and by culture on Mycoplasma agar, respectively.
2.4. BBE cell responses to BHV-1 or M. haemolytica
BBE cell gene responses to BHV-1, M. haemolytica, or both, were obtained via gene microarray analyses. BBE cells were either untreated, or exposed to BHV-1 (1 h viral absorption with 1 × 106 PFU, 6 h total), M. haemolytica (1 × 107 CFU for 3 h), or both BHV-1 and M. haemolytica. Total RNA was isolated from treated and untreated BBE cells (1 × 106 cells) via RNeasy mini kit according to the recommended protocols (QIAGEN, Valencia, CA). RNA was quantified spectrophotometrically (A 260) to estimate concentration. RNA purity was determined by Agilent Bioanalyzer (University of Wisconsin McArdle Labs Macromolecular Analysis Facility).
2.5. Microarray analysis
All microarray experiments were performed with 3 biological replicates of treated and untreated cells. A reference design labeling of each sample was employed with the use of Agilent bovine genome cDNA arrays containing 44,000 features. For reference design, all RNA samples (treated and untreated) from BBE cells were labeled with cy3 dye. In addition, a reference control pool of 3 untreated BBE cell RNA samples was labeled with cy5 and used as a control sample. Detailed protocols for microarray preparation, cy3/cy5 labeling of the cDNA probe, sample hybridization, washing, and scanning of Agilent arrays can be found at Bradfield Laboratory (http://mcardle.oncology.wisc.edu/bradfield/default.html, University of Wisconsin Madison McArdle Cancer Center Labs). All arrays were normalized, quality checked and are publicly available through NIH funded EDGE Bioinformatics software housed internally in the Bradfield Laboratory (http://mcardle.oncology.wisc.edu/bradfield/default.html; University of Wisconsin Madison McArdle Cancer Center Labs). For differential gene expression analysis, log base 2 ratios of cy3/cy5 signal intensity were used to estimate fold changes and significance (p values) by an empirical Bayes method using R Bioconductor Limma package (http://bioinf.wehi.edu.au/limma/) and p-values adjusted for false discovery rate (FDR) using Benjamini-Hochberg FDR approach for multiple comparisons. Genes with a fold change greater than or equal to 2 (log2 = 1) and p-value of less than 0.05 were used to filter differentially expressed genes.
2.6. Microarray gene functional analysis
Implications of the gene expression changes induced by BHV-1, M. haemolytica, and BHV1 + M. haemolytica for biological processes were examined by performing a group wise Gene ontology term analysis using DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/) and Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA; http://www.ingenuity.com/). For DAVID analysis, the GenBank accession numbers of genes which were up or down-regulated by when comparing treated versus untreated BBE cells were analyzed using DAVID default settings. In addition to the enrichment scores for each annotation cluster produced by DAVID, we also determined the cluster size (i.e. the number of individual annotations which satisfied an FDR adjusted p-value <0.01).
Functional and pathway enrichment analyses were further performed with Interactive Pathway Analysis (IPA) software (Ingenuity Systems, Redwood, CA, USA), after obtaining GenBank accession identifiers for the Agilent Bovine array genes. For all functional and pathway enrichments, we required the Benjamini-Hochberg corrected p-value to be <0.05. For cell-type specific functional enrichment analysis, significant function annotations were separated into their function and respective cell types. Functions that were related were grouped together; one cell-type may be significant more than once in each category.
2.7. Quantitative real-time PCR
Quantitative real-time PCR verification of microarray responses was performed for BBE cells (106 cells) that were untreated or exposed to BHV-1, M. haemolytica, or both BHV-1 and M. haemolytica as described above. Total RNA was isolated from treated and untreated BBECs using an RNeasy mini kit according to the recommended protocols (QIAGEN, Valencia, CA). Total RNA (1.5 μg) was heated at 70 °C for 10 min, and then transcribed to cDNA with a Reverse Transcription System kit (Promega, Madison, WI). Relative expression of IL-6, IL-8, COX-2 and BDNF were measured by real-time PCR using the 7300 real-time PCR System (Applied Biosystems, Foster City, CA). The primer sequences used for real-time PCR were constructed by the University of Wisconsin Biotechnology Center (Madison, WI).
3. Results
3.1. BBE cell gene microarray analysis
Microarray analysis revealed differential BBE cell gene regulation (>2-fold, p < 0.05) of 978 transcripts by BHV-1 alone, 2040 transcripts by M. haemolytica alone, and 2189 genes by co-exposure to BHV-1 and M. haemolytica, as compared to samples from uninfected BBE cell (Fig. 1A). Comparison of the differentially expressed gene lists identified 319 genes regulated by all 3 treatments (Fig. 1B). Differential gene expression elicited by M. haemolytica was comparable to that elicited by co-exposure to BHV-1 and M. haemolytica. There was significantly less overlap in gene expression between BHV-1 alone- and M. haemolytica alone-responsive genes. Pathway analysis to determine the top 5 canonical pathways, and top 5 up stream regulators are presented in Table 1A, Table 1B .
Table 1A.
Top canonical pathways as demonstrated by interactive pathway analysis (Ingenuity System).
| M. haemolytica + BHV-1 | BHV-1 | M. haemolytica |
|---|---|---|
| 1. Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis 2.51E−11*; 38/332 (0.114)a |
1. Role of Cytokines in Mediating Communication between Immune Cells 6.17E−06; 8/55 (0.145) |
1. Hepatic Fibrosis/Hepatic Stellate Cell Activation 2.41E−11; 24/146 (0.164) |
| 2. Hepatic Fibrosis/Hepatic Stellate Cell Activation 2.13E−10; 24/146 (0.164) |
2. IL-6 Signaling 8.4E−06; 11/124 (0.089) |
2. Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis 1.39E−09; 33/332 (0.099) |
| 3. Triggering receptor expressed on myeloid cells 1 (TREM1) Signaling 2.13E−10; 16/71 (0.225) |
3. Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis 1.02E−05; 18/332 (0.054) |
3. Differential Regulation of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F 1.57E−09; 9/18 (0.5) |
| 4. Tumor Necrosis Factor Receptor 2 (TNFR2) Signaling 2.86E−08; 10/33 (0.303) |
4. Atherosclerosis Signaling 1.16E−05; 11/136 (0.081) |
4. Death Receptor Signaling 3.92E−09; 14/64 (0.219) |
| 5. Altered T Cell and B Cell Signaling in Rheumatoid Arthritis 7.5E−08; 16/92 (0.174) |
5. Role of Hypercytokinemia or hyperchemokinemia in the Pathogenesis of Influenza 1.29E−05; 7/44 (0.159) |
5.TREM1 Signaling 4.99E−09; 14/71 (0.197) |
Target/total pathway ratio.
p-Value.
Table 1B.
Top upstream regulators as demonstrated by interactive pathway analysis (Ingenuity System).
| M. haemolytica + BHV-1 | BHV-1 | M. haemolytica |
|---|---|---|
| IL-1β – 1.47E−52* (Activated)a | U0126 – 1.03E−26 (Inhibited) | IL-1β – 2.86E−57 (Activated) |
| TNF – 5.21E−51 (Activated) | EGF – 1.31E−25 (Activated) | TNF – 2.71E−54 (Activated) |
| LPS – 4.74E−47 (Activated) | PDGF – 1.38E−24 (Activated) | LPS – 5.94E−47 (Activated) |
| NFkβ – 4.23E−40 (Activated) | CREB1 – 6.59E−22 (Activated) | NFkβ – 2.89E−45 (Activated) |
| poly rI: – 8.11E−37 (Activated) | ERK – 1.32E−20 (Activated) | IFNγ – 4.53E−34 (Activated) |
| rC-RNA |
Predicted state.
p-Value.
3.2. Analysis of BBE cell gene responses to BHV-1
The top 70 of the 978 BHV-1 responsive BBE cell genes (Fig. 1) are indicated in Supplemental Table 1. BHV-1 alone induced greater than 10-fold (p < 0.05) changes in expression of EGR1 (>20-fold), CCRN4L, IL6, AVPR1B, and PRKCG; and more than 5-fold changes in FOS, PRM1, TNF, CSF2, FUT2, OXT, CYR61, CSF3, IL1A, and CLDN3 (Supplemental Table 1). Functional analysis of these top 70 genes using the DAVID bioinformatics tool revealed 52 annotated bovine IDs. Functional annotation of the 52 annotated DAVID IDs show significant enrichment of genes involved in disulfide bonds (15/52), glycoproteins (14/52), secreted proteins (11/52), cell proliferation (9/52), cytokine activity (7/52), lipoproteins (6/52), developmental factors (4/52), and growth regulation (2/52).
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetimm.2013.06.012.
3.3. Analysis of BBE cell gene responses to M. haemolytica
The top 70 of the 2040 M. haemolytica responsive BBE cell genes identified (Fig. 1) are indicated in Supplemental Table 2. M. haemolytica alone induced substantial changes in expression of NOS2A (>100-fold), IL6 (>100-fold), IL1A (>90-fold), and CXCL2 (>40-fold); more than 30-fold increases for IL-8, CSF3, and CCL5; and more than 20-fold for CXCL1, PTGS2, SERPINB2, SELE, and TNF. The induction of TNF (>20-fold), IL1A (>90-fold), and IL6 (>100-fold) were significantly greater than those seen following exposure to BHV-1 alone. Functional analysis of the top 70 genes in the DAVID bioinformatics tool revealed 50 annotated bovine IDs. Gene profiling of the 50 annotated DAVID IDs shows significant enrichment of genes involved in cytokine activity (10/50), glycoproteins (21/50), secreted proteins (17/50), disulfide bonds (22/50), signal transduction (19/50), and inflammatory response (6/50).
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetimm.2013.06.012.
3.4. Analysis of BBE cell gene responses resulting from co-exposure to BHV-1 and M. haemolytica
The top 70 of the 2189 BBE cell responses to co-exposure with BHV-1 and M. haemolytica (Fig. 1) are indicated in Supplemental Table 3. Co-exposure to BHV-1 and M. haemolytica induced substantial changes (p < 0.05) in expression of NOS2A (120-fold), IL6 (>100-fold), IL1A (>90-fold), and CXCL2 (>40-fold); 30-fold or greater increases in CSF3, IL8, CCL5, and SERPINB2; and >20-fold increases in CXCL1, PTGS2, and SELE. Functional analysis of these top 70 genes in the DAVID bioinformatics tool revealed 53 annotated bovine IDs. Gene profiling of the 53 annotated DAVID IDs shows significant enrichment of genes involved in cytokine activity (10/53), glycoproteins (21/53), secreted proteins (18/53), disulfide bonds (22/53), signal transduction (20/53), and inflammatory response (6/53).
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetimm.2013.06.012.
Analysis of gene expression profiles (by multivariate pairwise correlation analysis) indicated a strong correlation between genes expressed in BBE cells treated with M. haemolytica alone and cells co-infected with M. haemolytica and BHV-1. A weak correlation was shown between BBE cells treated with BHV-1 alone and those co-infected with BHV-1 and M. haemolytica (Fig. 2 ).
Fig. 2.
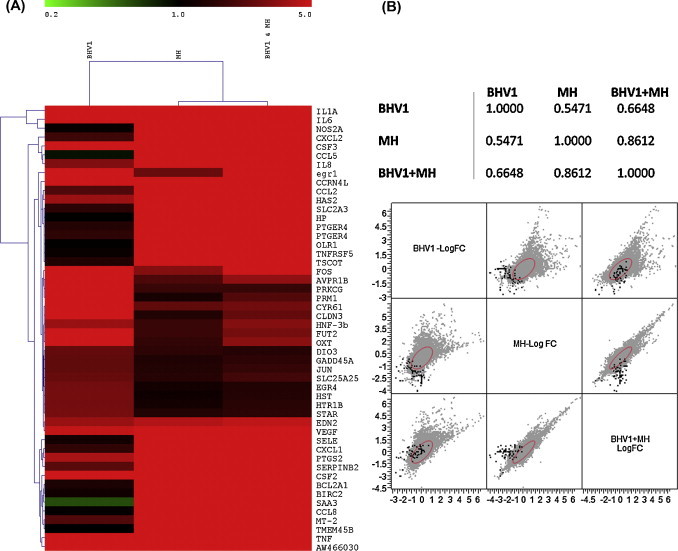
Hierarchical clustering of the top 50 differentially expressed genes differentially regulated by BHV-1, MH and BHV-1 + MH. (A) This heat map depicts a qualitative assessment of the similarities of the gene expression of the top 50 of the 319 differentially expressed genes in response to the 3 treatments. Red indicates up-regulation while green indicates down- regulation of selected genes. (B) Multivariate pairwise correlation analysis of the BBE cell gene expression patterns elicited by BHV-1, MH, and BHV-1 + MH. This analysis provides a more quantitative examination of similarities in expression patterns of the differentially expressed genes by determining the correlation between gene expression (as fold change) and significance (p < 0.05) profiles of the genes differentially regulated by all treatments. This analysis indicates that the responses elicited by MH are highly correlated with gene expression patterns for BHV-1 + MH.
Overall analysis of genes indicated significantly increased gene expression in the BHV-1 and M. haemolytica co-infected group, suggesting some synergism between BHV-1 and M. haemolytica (Fig. 3 ). Scatter plot matrix analysis of a selected group of highly expressed genes, however, failed to demonstrate a clear synergism for BBE cells co-infected with BHV-1 and M. haemolytica versus either pathogen alone (Fig. 4 ).
Fig. 3.
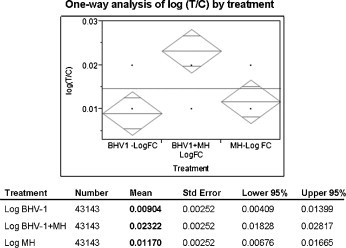
One-way analysis of BBE cell gene responses elicited by BHV-1, MH or BHV-1 + MH. Total significant bovine genes responding to BHV-1, MH or BHV-1 + MH were analyzed and presented as a mean log ratio of test/control. The mean log ratio for (BHV-1 + MH), 0.023227 is greater than the sum of individual log mean ratios for MH and BHV-1, (0.009041 + 0.011703). As a result these data suggest a synergistic effect of BHV-1 and MH.
Fig. 4.
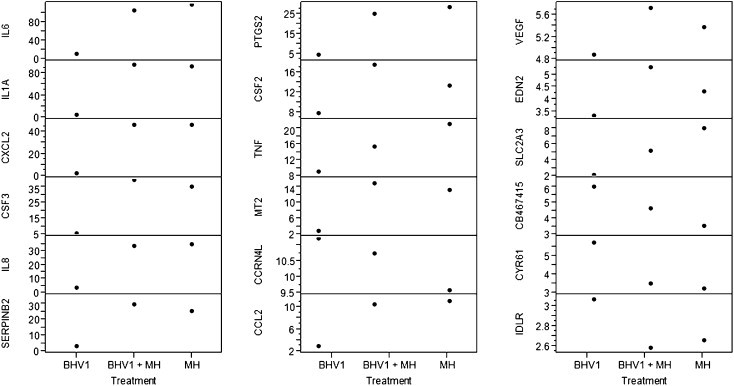
Scatter plot matrix expression profiles of select differentially regulated genes. This analysis illustrates mean fold increase in expression, as compared to unstimualted cells, for selected genes of interest by BBE cell incubated with BHV-1, MH or BHV-1 + MH.
3.5. Quantitative real-time PCR analysis of BBE cells exposed to BHV-1 or M. haemolytica co-infections
RT-PCR analysis of five selected inflammatory cytokine genes revealed a relative increase in gene expression during M. haemolytica infection compared to BHV-1 infection. Co-infection with both M. haemolytica and BHV-1 did not increase expression to a greater extent than did exposure to BHV-1 or M. haemolytica alone (Fig. 5 ). These results confirm the general responses observed in the gene microarray analysis.
Fig. 5.
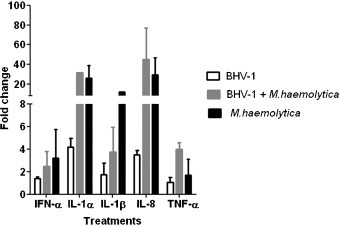
Real-time PCR analysis of cytokine mRNA production by BBE cells in response to BHV-1, MH, or BHV-1 + MH. Quantitative real time PCR was used to confirm BBE cell cytokine gene expression in response to BHV-1, MH, or both BHV-1 + MH. The figure illustrates the relative expression levels (fold increase) of IFN-α, IL-1β, IL-1α, IL-8 and TNF-α (early BHV-1 transcript) by BBE cells compared to unstimulated control cells.
4. Discussion
BRD involves complex interactions among viral and bacterial pathogens that can lead to intense pulmonary inflammation and ultimately fibrinous pleuropneumonia (Yates, 1982). Identifying underlying mechanisms responsible for the increased susceptibility of BHV-1 infected cattle to M. haemolytica has been difficult (Hodgson et al., 2005). A previous study used bovine microarrays and real-time PCR analysis to identify the genetic changes with enteric infections (by rotavirus and corona viruses) and respiratory infections by BHV-1 from intestinal loop tissue and monocytes from cattle (Wilson et al., 2005). We have reported that inflammatory cytokines released from BHV-1-infected BBE cells can alter the adherence and migration of bovine neutrophils and mononuclear phagocytes (Rivera et al., 2009, Noel et al., 1988). In the present study, we employed a gene microarray strategy to investigate whether BBE cells respond differently to BHV-1and M. haemolytica, than to co-infection with either pathogen.
Cytokines and other mediators released by primary BBE cells can contribute to the pathogenesis of BRD through leukocyte recruitment, attachment and activation (Cudd et al., 2001, Leite et al., 2004a, Leite et al., 2004b). Activated leukocytes in turn secrete mediators that increase vascular permeability and tissue injury (Bielefeldt-Ohmann et al., 1991). The BBE cell represents both a primary site of viral infection (Forman et al., 1982), and a physical barrier against microbial pathogens (Galdiero et al., 2002, Malazdrewich et al., 2001, Lee et al., 2000). Adherence of the bacterial cells to respiratory epithelium is a crucial and early event in the pathogenesis of BRD (Auger et al., 2009, Kisiela and Czuprynski, 2009). We previously reported that pro-inflammatory cytokines produced by BBE cells in response to BHV-1 infection can activate bovine neutrophils, increase their adherence to respiratory epithelium, and exacerbate their response (e.g. apoptosis) to the leukotoxin of M. haemolytica (Leite et al., 2004a, Leite et al., 2004b, Rivera et al., 2009).
In this study we demonstrate that gene expression by primary BBE cells infected with M. haemolytica is further amplified by a co-infection with BHV-1 and M. haemolytica (Fig. 3). For example, the gene expression responses for the cytokines IL-1, IL-6, and IL-8 were 20-fold, 10-fold, and 9-fold greater, respectively following M. haemolytica and BHV-1 co-infection than with BHV-1 infection. These data indicate that co-infection contributes to pro-inflammatory cytokine production that in turn could exacerbate the pathogenesis of BRD. We also noted that gene expression following BHV-1 and M. haemolytica co-infection correlates more strongly with M. haemolytica infection than with exposure to BHV-1 (Fig. 2).
This study also shows that there are several genes that are less up regulated by a co-infection with M. haemolytica and BHV-1 than BHV-1 or M. haemolytica infection alone. One such gene is CYR61, a pro-angiogenic protein that is responsible for wound healing, cell migration and IL-6 induction (Perbal, 2004). Similarly, TNF and SLC2A3 (GLUT3) were also up regulated less by co-infection than by infection with M. haemolytica alone. TNF is a potent pro-inflammatory cytokine produced by and having a variety of effects on leukocytes, endothelial cells and fibroblasts. In regards to bronchial epithelial cells, TNF is a double edged sword in the pathogenesis of acute lung injury. On one hand, it has a positive effect by promoting clearance of alveolar sac liquid and restoring the integrity of the endothelial barrier (Yang et al., 2010). Conversely it can interfere with tight junction integrity leading to increased permeability of alveolar epithelial cells and the capillary endothelium. A balance between these two functions influences the outcome of pulmonary infection (Yang et al., 2010).
Our study reveals that VEGF, EDN2 and CSF2 are three genes that are highly up-regulated by co-infection with BHV-1 and M. haemolytica. VEGF, which promotes vasculogenesis and increases endothelial cell permeability is induced by hypoxia and a variety of different cytokines (Neufeld and Kessler, 2006). EDN2 is a secreted vasoconstrictive peptide that is responsible for hypertension. CSF2 is a cytokine which stimulates growth and differentiation of granulocytes, macrophages, eosinophils and erythrocytes (Yap et al., 2000, Egea et al., 2010). Thus, it is noteworthy that during co-infection with BHV and M. haemolytica, these upregulated factors could contribute to enhanced acute lung injury by several pathways and complex mechanisms.
Interestingly, we also demonstrate by overall gene analysis a synergistic effect on gene expression following co-exposure to M. haemolytica and BHV-1 infections (Fig. 3). This is compatible with the clinical outcome of respiratory tract infections in which bacterial pneumonia is often augmented by viral infection of the respiratory tract in both animals and humans.
We also observed that some genes are differentially regulated during co-infection with both M. haemolytica and BHV-1. CYR61 is up regulated to a lesser extent than IL-6, and the latter is highly upregulated, by co-infection with BHV-1 and M. haemolytica as compared to BHV-1 or M. haemolytica alone. CYR61 is a matrix associated signaling protein that induces IL-6 production (Perbal, 2004). These gene expression patterns indicate the complex nature of responses elicited by viral–bacterial co-infection. Such infections may be associated with gene expression profiles that promote pathogenesis of disease while the host counters by enhancing expression of genes that mediate protection mechanisms. The balance between these two responses governs the clinical outcome of the infection. Understanding the complex nature of factors contributing to tissue injury requires investigating multiple factors at a given point of time. Gene expression profiling is a useful method to achieve this goal. Because BBE cells resemble the respiratory epithelium in cattle we infer that they provide a model system to assess relevant responses to infection with BHV-1 and M. haemolytica.
We also analyzed these gene expression data using interactive pathway analysis, to reveal signaling pathways and their complex interactions following exposure to BHV-1, M. haemolytica or both BHV-1 and M. haemolytica. The most interesting relationships were noted in the top 5 canonical pathways and the top 5 upstream regulators for the three treatment groups. Consistent with the data in Fig. 2, these pathways confirmed an overall similarity in pro-inflammatory cytokine (IL-1β, TNF, LPS induction and NFkβ) up regulation during exposure to M. haemolytica, or co-infection with M. haemolytica and BHV-1 (Table 1A, Table 1B). Interestingly, pathway analysis of BBE cells exposed to BHV-1 alone revealed increased generalized immunological activity due to increased cytokine mediated communication and hyperchemokinemia (Table 1A). These data, however, revealed generalized immunological activity involving systems other than the respiratory tract (e.g. hepato-billiary and musculo-skeletal).
In summary, our study identifies mediators and their regulatory pathway interactions in BBE cells exposed to the important bovine respiratory pathogens BHV-1 and M. haemolytica. Insights gained in this study may lead to novel therapeutic and preventive strategies to reduce the losses associated with bovine respiratory diseases.
Acknowledgements
We thank Dr. Christopher A. Bradfield and his research group at the University of Wisconsin – Madison for their assistance with collecting the gene microarray data. This work was funded by USDA NIFA Grant# 2011-67015-30171.
References
- Auger E., Deslandes V., Ramjeet M., Contreras I., Nash J.H., Harel J., Gottschalk M., Olivier M., Jacques M. Host-pathogen interactions of Actinobacillus pleuropneumoniae with porcine lung and tracheal epithelial cells. Infect. Immun. 2009;77:1426–1441. doi: 10.1128/IAI.00297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiuk L.A., van Drunen Little-van den Hurk S., Tikoo S.K. Immunology of bovine herpesvirus 1 infection. Vet. Microbiol. 1996;53:31–42. doi: 10.1016/s0378-1135(96)01232-1. [DOI] [PubMed] [Google Scholar]
- Bielefeldt-Ohmann H., Babiuk L.A., Harland R. Cytokine synergy with viral cytopathic effects and bacterial products during the pathogenesis of respiratory tract infection. Clin. Immunol. Immunopathol. 1991;60:153–170. doi: 10.1016/0090-1229(91)90060-n. [DOI] [PubMed] [Google Scholar]
- Brown T.T., Jr., Shin K. Effect of bovine herpesvirus-1 or parainfluenza-3 virus on immune receptor-mediated functions of bovine alveolar macrophages in the presence or absence of virus-specific serum or pulmonary lavage fluids collected after virus infection. Am. J. Vet. Res. 1990;51:1616–1622. [PubMed] [Google Scholar]
- Car B.D., Suyemoto M., Neilsen N.R., Slauson D.O. The role of leukocytes in the pathogenesis of fibrin deposition in bovine acute lung injury. Am. J. Pathol. 1991;138:1191–1197. [PMC free article] [PubMed] [Google Scholar]
- Cudd L.A., Ownby C.L., Clarke C.R., Sun Y., Clinkenbeard K.D. Effects of Mannheimia haemolytica leukotoxin on apoptosis and oncosis of bovine neutrophils. Am. J. Vet. Res. 2001;62:136–141. doi: 10.2460/ajvr.2001.62.136. [DOI] [PubMed] [Google Scholar]
- Egea L., Hirata Y., Kagnoff M.F. GM-CSF: a role in immune and inflammatory reactions in the intestine. Expert. Rev. Gastroenterol. Hepatol. 2010;4:723–731. doi: 10.1586/egh.10.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova N.D., Highlander S.K. Generation of targeted nonpolar gene insertions and operon fusions in Pasteurella haemolytica and creation of a strain that produces and secretes inactive leukotoxin. Infect. Immun. 1997;65:2593–2598. doi: 10.1128/iai.65.7.2593-2598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman A.J., Babiuk L.A., Misra V., Baldwin F. Susceptibility of bovine macrophages to infectious bovine rhinotracheitis virus infection. Infect. Immun. 1982;35:1048–1057. doi: 10.1128/iai.35.3.1048-1057.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero M., Pisciotta M.G., Marinelli A., Petrillo G., Galdiero E. Coinfection with BHV-1 modulates cell adhesion and invasion by P. multocida and Mannheimia (Pasteurella) haemolytica. New Microbiol. 2002;25:427–436. [PubMed] [Google Scholar]
- Gershwin L.J., Berghaus L.J., Arnold K., Anderson M.L., Corbeil L.B. Immune mechanisms of pathogenetic synergy in concurrent bovine pulmonary infection with Haemophilus somnus and bovine respiratory syncytial virus. Vet. Immunol. Immunopathol. 2005;15:119–130. doi: 10.1016/j.vetimm.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Highlander S.K., Fedorova N.D., Dusek D.M., Panciera R., Alvarez L.E., Rinehart C. Inactivation of Pasteurella (Mannheimia) haemolytica leukotoxin causes partial attenuation of virulence in a calf challenge model. Infect. Immun. 2000;68:3916–3922. doi: 10.1128/iai.68.7.3916-3922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkley S., Hill A.B., Srikumaran S. Bovine herpesvirus-1 infection affects the peptide transport activity in bovine cells. Virus Res. 1998;53:91–96. doi: 10.1016/s0168-1702(97)00128-7. [DOI] [PubMed] [Google Scholar]
- Hodgson P.D., Aich P., Manuja A., Hokamp K., Roche F.M., Brinkman F.S., Potter A., Babiuk L.A., Griebel P.J. Effect of stress on viral–bacterial synergy in bovine respiratory disease: novel mechanisms to regulate inflammation. Comp. Funct. Genomics. 2005;6:244–250. doi: 10.1002/cfg.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiela D., Czuprynski C.J. Outer membrane proteins of Mannheimia haemolytica bind to bovine bronchial epithelial cells. Infect. Immun. 2009;77:446–455. doi: 10.1128/IAI.00312-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.Y., Kehrli M.E., Jr., Brogden K.A., Gallup J.M., Ackermann M.R. Influence of β2-integrin adhesion molecule expression and pulmonary infection with Pasteurella haemolytica on cytokine gene expression in cattle. Infect. Immun. 2000;68:4274–4281. doi: 10.1128/iai.68.7.4274-4281.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite F., O’Brien S., Sylte M.J., Page T., Atapattu D., Czuprynski C.J. Inflammatory cytokines enhance the interaction of Mannheimia haemolytica leukotoxin with bovine peripheral blood neutrophils in vitro. Infect. Immun. 2002;70:4336–4343. doi: 10.1128/IAI.70.8.4336-4343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite F., Kuckleburg C., Atapattu D., Schultz R.D., Czuprynski C.J. BHV-1 infection and inflammatory cytokines amplify the interaction of Mannheimia haemolytica leukotoxin with bovine leukocytes in vitro. Vet. Immunol. Immunopathol. 2004;99:193–202. doi: 10.1016/j.vetimm.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Leite F., Atapattu D., Kuckleburg C., Schultz R., Czuprynski C.J. Incubation of bovine PMNs with conditioned medium from BHV-1 infected peripheral blood mononuclear cells increases their susceptibility to Mannheimia haemolytica leukotoxin. Vet. Immunol. Immunopathol. 2004;103:187–193. doi: 10.1016/j.vetimm.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Loneragan G.H., Dargatz D.A., Morley P.S., Smith M.A. Trends in mortality ratios among cattle in US feedlots. J. Am. Vet. Med. Assoc. 2001;219:1122–1127. doi: 10.2460/javma.2001.219.1122. [DOI] [PubMed] [Google Scholar]
- Malazdrewich C., Ames T.R., Abrahamsen M.S., Maheswaran S.K. Pulmonary expression of tumor necrosis factor alpha, interleukin-1 beta, and interleukin-8 in the acute phase of bovine pneumonic pasteurellosis. Vet. Pathol. 2001;38:297–310. doi: 10.1354/vp.38-3-297. [DOI] [PubMed] [Google Scholar]
- McGuire R.L., Babiuk L.A. Evidence for evidence for defective neutrophil function in lungs of calves exposed to infectious bovine rhinotracheitis virus. Vet. Immunol. Immunopathol. 1984;5:259–271. doi: 10.1016/0165-2427(84)90039-4. [DOI] [PubMed] [Google Scholar]
- Neufeld G., Kessler O. Pro-angiogenic cytokines and their role in tumor angiogenesis. Cancer Metastasis Rev. 2006;25:373–385. doi: 10.1007/s10555-006-9011-5. [DOI] [PubMed] [Google Scholar]
- Noel E.J., Israel B.A., Letchworth G.J., III, Czuprynski C.J. Preincubation of bovine blood neutrophils with bovine herpesvirus-1 does not impair neutrophil interaction with Pasteurella haemolytica A1 in vitro. Vet. Immunol. Immunopathol. 1988;19:273–284. doi: 10.1016/0165-2427(88)90114-6. [DOI] [PubMed] [Google Scholar]
- Ohmann H.B., Babiuk L.A. Viral–bacterial pneumonia in calves: effect of bovine herpesvirus-1 on immunologic functions. J. Infect. Dis. 1985;151:937–947. doi: 10.1093/infdis/151.5.937. [DOI] [PubMed] [Google Scholar]
- Perbal B. CCN proteins: multifunctional signaling regulators. Lancet. 2004;363:62–64. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- Raz M., Robbins R.A., Kelling C.L., Stine L.C., Leikauf G.D., Rennard S.I., Spurzem J.R. Viral infection of bovine bronchial epithelial cells induces increased neutrophil chemotactic activity and neutrophil adhesion. Clin. Sci. (Lond.) 1993;85:753–760. doi: 10.1042/cs0850753. [DOI] [PubMed] [Google Scholar]
- Rivera J., Kisiela D., Czuprynski C.J. Bovine herpesvirus type 1 infection of bovine bronchial epithelial cells increases neutrophil adhesion and activation. Vet. Immunol. Immunopathol. 2009;131:167–176. doi: 10.1016/j.vetimm.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Thaker S.R., Stine D.L., Zamb T.J., Srikumaran S. Identification of a putative cellular receptor for bovine herpesvirus 1. J. Gen. Virol. 1994;75:2303–2309. doi: 10.1099/0022-1317-75-9-2303. [DOI] [PubMed] [Google Scholar]
- Wilson H.L., Aich P., Roche F.M., Jala l.S., Hodgson P.D., Brinkman F.S., Potter A., Babiuk L.A., Griebel P.J. Molecular analyses of disease pathogenesis: application of bovine microarrays. Vet. Immunol. Immunopathol. 2005;105:277–287. doi: 10.1016/j.vetimm.2005.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Hamacher J., Gorshkov B., White R., Sridhar S., Verin A., Chakraborty T., Lucas R. The dual role of TNF in pulmonary edema. Cardiovasc. Dis. Res. 2010;1:29–36. doi: 10.4103/0975-3583.59983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap E.Y.S., Battistini B., McKay K.O. Contraction to big endothelin-1, big endothelin-2 and big endothelin-3, and endothelin-converting enzyme inhibition in human isolated bronchi. Br. J. Pharmacol. 2000;1291:170–176. doi: 10.1038/sj.bjp.0703006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates W.D.G. A review of infectious bovine rhinotracheitis, shipping fever pneumonia and viral–bacterial synergism in respiratory disease of cattle. Can. J. Comp. Med. 1982;46:225–263. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


