Abstract
Bioactive paper includes a range of potential paper-based materials that can perform analytical functions normally reserved for multi-well plates in the laboratory or for portable electronic devices. Pathogen detection is the most compelling application. Simple paper-based detection, not requiring hardware, has the potential to have impacts in society, ranging from the kitchen to disasters in the developing world. Bioactive-paper research is an emerging field with significant efforts in Canada, USA (Harvard), Finland and Australia.
Following a brief introduction to the material and surface properties of paper, I review the literature. Some of the early work exploits the porosity of paper to generate paper-based microfluidics (“paperfluidics”) devices. I exclude from this review printed electronic devices and plastics-supported devices.
Keywords: Antibody, Antimicrobial paper, Bacteriophage, Bioactive paper, Biosensor, DNA aptamer, Lateral flow, Paperfluidics, Paper-supported assay, Pathogen detection
1. Introduction
This article reviews initial developments in a new field called “bioactive paper”. At the interface between enormous global biomedical-biotechnology research activities and the small, esoteric world of paper science, bioactive-paper research targets exciting new paper products, aiming to improve the quality of life world-wide. VTT (a Finnish research organization) defined bioactive paper as “paper-like products, cardboard, fabrics and their combinations, etc., with active recognition and/or functional material capabilities” [1].
The most exciting potential implementations of bioactive paper involve leading-edge concepts in genetic engineering, biochemistry, and microbiology. There are major bioactive-paper research initiatives in a research consortium in Canada [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31] and Whiteside’s group at Harvard [32], [33], [34], [35], [36], [37], with smaller efforts currently in Scandinavia [38], Australia [39], [40] and Japan [41]. I summarize below initial results from these groups, with emphasis on the links between paper properties and bioactivity.
Issues involving tainted food and water, resistant bacteria in hospitals, the global spread of disease and the threat of bioterrorism receive almost daily coverage in the Canadian media. Although Canada is a rich country with a good health-care system and large fresh water reserves, poor water quality (Walkerton, Ontario, May 2000) and tainted food (Ontario, October 2008) recently have killed Canadians. The Severe Acute Respiratory Syndrome (SARS) epidemic of a few years ago highlighted our vulnerabilities. Hospital workers following existing protocols died from SARS exposure. Inexpensive bioactive-paper assays could help control such outbreaks.
The need for bioactive paper is obvious. How close are we? Existing products include tissues that claim to kill viruses, and non-woven fabrics with antibacterial photochemical elements. However, apart from dipstick test kits and similar devices, I am not aware of any commercial paper products that indicate the presence of pathogens.
Over the decades, plastics materials have replaced paper products for some food packaging, grocery bags and general packaging applications. This begs the question – why bioactive paper instead of bioactive plastic? Paper offers unique advantages over plastic including:
-
1)
paper is very inexpensive and is manufactured locally in nearly every part of the world from renewable and recyclable resources;
-
2)
paper has a long and successful history performing as filter media and barrier media, and can even function as sterile packaging;
-
3)
paper is easily printed, coated and impregnated;
-
4)
cellulose is particularly protein and biomolecule friendly;
-
5)
paper is a good filter;
-
6)
paper is biodegradable or easily burned; and,
-
7)
the porous structure facilitates lateral-flow assays, chromatographic separations and inexpensive microfluidics devices.
The focus of this review is research leading to large-scale implementation of inexpensive bioactive-paper assays that can function without laboratories or significant instrumentation. I view the “paper” part of bioactive paper in the broadest context to include non-woven fabrics, and coated and uncoated paperboard. I do not consider examples involving small cellulosic adsorption pads and nitrocellulose films, which are widely used in “dip-stick” lateral-flow devices (e.g., over-the-counter (OTC) pregnancy test kits, printed electronic devices or instrumentation [42]) and “intelligent” or “active” packaging involving plastic films.
2. Paper properties – a primer
2.1. Introduction
Paper filters have a long history in the biochemistry laboratory, both for filtration and as chromatography supports – these are simple materials often based on pure cellulose. By contrast, everyday packaging, writing, tissues, boxboard and other paper products are complex materials that have a wide range of porosity and surface chemistries. To understand the differences, we need to discuss the fibers and other building blocks, as well as how they are assembled on a paper machine.
Paper is made by filtering a dilute (∼1 wt%) aqueous suspension of fibers, colloidal filler particles and soluble polymers. Modern paper machines produce a 10-m wide continuous sheet of paper at expressway speeds (∼100 km/h). The dried paper sheet may be coated either directly on the paper machine or in a separate operation. From the perspective of bioactive paper, critical properties include surface chemistry, porosity and optical properties. In subsequent sections, I show that surface chemistry influences biomolecule immobilization, non-specific binding and color expression in enzyme-catalyzed reporting assays. Porosity, together with surface chemistry, influences wet properties, which are important for bioactive-paper fabrication by printing or coating. Finally, the optical properties may influence color-based or fluorescence-based reporting schemes. For example, many commercial paper types are treated with fluorescent molecules, called optical brightening agents, which are added to make paper appear white. Such surfaces would give a very high background in fluorescence-detection assays.
2.2. Composition
Cellulose fibers are the major component of many paper types, with most of the fibers coming from wood. Wood-fiber composition depends upon the type of wood and the pulping process. In mechanical pulping, the wood is simply ground to a pulp, so the final paper contains cellulose, lignin and pitch (resin and fatty acids). Newsprint is a typical example of paper made from mechanical pulp.
In chemical pulping, high pH (kraft pulping) or low pH (sulfite pulping) is used to decompose the lignin in wood selectively, releasing fibers from wood chips. Kraft pulp contains some residual lignin, and is brown. Paper grocery bags and corrugated boxes are examples of papers made with unbleached or semi-bleached kraft pulp. Finally, fully bleached pulp is white and is used to make filter paper and copy paper. Only trace quantities of lignin are present in fully bleached pulp.
Printing and office papers usually contain up to 30 wt% mineral fillers. Traditionally, kaolin was the filler of choice, whereas calcium carbonate is the dominant filler in modern papers, so any paper-supported analytical assay employing filled paper must function in the carbonate buffer that forms when the paper is wetted.
Finally, several polymeric additives are used, either to facilitate the paper-making process or to influence the final paper properties. From the perspective of bioactive paper, these additives are important, since they influence the surface chemistry.
Cationic starch is widely used to strengthen printing papers, which have been weakened by the presence of fillers, and in packaging paper and boxboard. Low concentrations of quaternary-ammonium groups promote starch adsorption onto pulp fibers before the papermaking step. Starch strengthens dry paper by promoting interfiber adhesion; it is not effective at strengthening wet paper.
Bioactive-paper products are likely to require wet strength, since most biology is wet. The two most common wet-strength enhancing polymers are glyoxal-modified polyacrylamide (GPAM) and polyamide–epichlorohydrin (PAE) (for chemical structures, see Fig. 1 ) [43]. GPAM gives temporary wet strength and is used in facial tissues and packaging applications where moisture exposures are short. PAE is used in applications requiring longer-term wet strength (e.g., kitchen towels and coffee filters). In a later section, I describe some recent results on the influence of these polymers on antibody and DNA-aptamer immobilization on paper.
Figure 1.

The chemical structures of polyamide-epichlorohydrin (PAE) and glyoxal-modified polyacrylamide (GPAM) wet-strength resins. PAE gives permanent wet strength whereas GPAM cross-links hydrolyze with exposure time to water.
Cellulose fibers are porous, hydrophilic materials that take up more than their own mass of water. Printing, writing and some packaging papers are routinely treated with “size” that lowers surface energy, as evidenced by increased water-contact angle and lower rates of water penetration [44]. Wood rosin-aluminum sulfate combinations were the original sizing materials, whereas most modern papers are sized with either alkyl succinic anhydride or alkyl ketene dimer (AKD), which provides cellulose-reactive treatments. Fig. 2 shows the structure and the cellulose-coupling chemistry of AKD. We will see that sizing influences the performance of printed enzymes.
Figure 2.

Alkyl ketene dimer (AKD) and alkyl succinic anhydride “size” react with fibers to lower surface energy and water-penetration rates.
Glossy-magazine covers, packaging for expensive products and magazine paper are examples of coated papers. Most coatings are mixtures of kaolin embedded in styrene butadiene rubber latex. The surface properties of such papers are closer to those of plastics films than uncoated paper.
2.3. Structure
Cellulose fibers, the building blocks of paper, are hollow tubes ∼1.5 mm long, 20-μm wide, with a wall thickness of ∼2 μm. Since paper is formed in a filtration process, the fibers are approximately layered in the x,y plane [45], [46].
Paper is described by two macroscopic properties – the thickness or caliper, τ (m), and the basis weight, bw (g/m2), which is the mass of dry paper per square meter. Obviously, these parameters are correlated. An estimate of the total pore volume in paper can be estimated from τ and bw. The density of the solid component of wood fiber is ρfiber ∼1540 kg/m3, whereas the corresponding bulk density of paper is given by the caliper and basis weight, ρpaper = bw/τ. For example, Whatman No. 1 filter paper has τ 180 μm and bw 87 g/m2, which translates into a density of ρpaper = 483 kg/m3. The corresponding pore-volume fraction of Whatman No. 1 is approximately 0.69:
| (1) |
The porosity of paper arises from spaces between the fibers, un-collapsed fiber lumens and the intrinsic porosity of the fiber walls. Kraft pulping removes the lignin from the fiber walls, leaving a porous structure. There are many reports of the fiber-wall pore-size distribution in the literature. The results depend upon tree species, pulping type and whether or not the fibers have been dried after pulping. Drying causes some of the pores to collapse in an irreversible process called hornification [47]. Thus, dry, wet, and never-dried samples of the same paper could have very different pore-volume distributions. Alince gives compelling arguments that, in wet-pulp fibers, the average pore size is around 100 nm and, controversially, that the pore-size distribution is rather narrow [48].
Finally, it is important to recognize paper as a very anisotropic material. The mass distribution in a paper sheet is usually not constant in the z (thickness) dimension. The maximum density is in the center, usually decreasing near the surfaces. Fiber orientation is another artifact of the papermaking process. Usually, fibers have a slightly preferred orientation along the direction that the paper was made on the paper machine. Thus, fluid transport along a strip of paper in a lateral-flow device may depend upon the angle at which the paper strip was cut. Conventional 8.5x11 copy paper is cut so that the longest dimension is parallel to the papermaking direction.
Paper structure influences the maximum quantity of biosensors that can be attached to cellulose. Specifically, the maximum biosensor content is Γ.σ where Γ is the maximum density of the immobilized sensor and σ is the specific surface area of the paper structure accessible to the biosensor. Most polymers and proteins have Γ values in the range 0.1–1 mg/m2 [49]. Paper structures can have a wide range of σ. For non-porous paper, such as glassine, only the macroscopic external surface is accessible. In this case, σ = 2/bw. Thus for a glassine with a bw of 50 g/m2, the corresponding accessible σ = 0.04 m2/g. By contrast, Hong et al. recently measured σ of various forms of cellulose accessible to proteins using a probe protein, which was a fusion of cellulose-binding domain and green fluorescent protein [50]. They found that the accessible σ of Whatman No. 1 filter papers was 9.5 m2/g. Of course, smaller proteins will access smaller pores, giving a higher σ and vice versa.
Consider two extreme cases – a biosensor with a low saturation coverage, Γ of 0.1 mg/m2 coated on glassine, described above, gives a maximum biosensor coating of 0.004 mg of sensor per g of paper. A more compact biosensor, giving a higher Γ of 1 mg/m2, can be taken up by the Whatman No. 1 filter paper to give 9.5 mg of biosensor per g of paper, so we see that the capacity of conventional paper substrates to take up biosensors can range over four orders of magnitude.
2.4. Surface chemistry
For bio-analytical applications, the surface chemistry of paper must facilitate biosensor immobilization, minimize non-specific sorption and be compatible with reporting strategies. The following paragraphs summarize the relevant surface-chemical properties of wet paper. We shall start with the simplest case, bleached cellulose fibers with no additives, typically found in laboratory filter paper.
The degree of crystallinity of cellulosic fibers is ∼50% [51]. The crystalline domains do not swell with water. By contrast, amorphous cellulose swells in water and is more susceptible to chemical reaction. In addition, most fibers are coated with hemicellulose that is also water swollen. All pulp fibers have low concentrations of carboxyl groups. Unbleached kraft pulps typically have 0.1–0.2 milli-equivalent per gram (meq/g), whereas fully bleached fibers have about an order of magnitude fewer carboxyl groups [52], [53].
There are many publications addressing the surface energy of cellulose. Printing [54] and adhesion in cellulose-plastic composites [55] have driven much of this work. Cunha et al. pointed out that carbohydrates (e.g., cellulose) have among the highest surface energies of any macromolecules – approximately 30 mJ/m2 for the dispersive component and a high of 30 mJ/m2 for the polar component, giving a total surface energy of ∼60 mJ/m2 [56]. Surface modification of cellulose with hydrophobic sizing agents (see Fig. 2) lowers paper-surface energy by decreasing the polar component to near 0 mJ/m2 [57].
The water-wetting properties reflect the surface energetics of paper. Pure cellulose fibers have advancing water-contact angles of ∼55°, whereas the advancing water-contact angle is ∼43° for lignin-containing fibers [58]. The corresponding receding water-contact angles are 0° [58]. Sized papers have a higher advancing water-contact angle, whereas the receding water-contact angle of sized papers is also 0° [59].
For many bioactive-paper applications, paper surfaces are saturated with water or immersed in water. In this situation, these paper surfaces comprise a slightly anionic water swollen hydrogel of amorphous cellulose and hemicellulose. Colloidal particles, negatively charged water-soluble polymers, non-ionic water-soluble polymers [60], and DNA [8] have little tendency to adsorb on wet lignocellulose surfaces. Only cationic molecules and particles [60] have a strong tendency to adsorb onto unaltered wet cellulose fibers.
3. Potential formats of bioactive paper
One can imagine a number of formats in which a pathogen-detecting bioactive paper might function. In direct-contact format, the bioactive paper would report the presence of a pathogen coming into contact with its surface. Applications could include protective clothing that could warn the user of contamination.
Filters are another important format for bioactive paper. Bacteria are easily trapped on filters. This is a form of amplification (actually concentration). A filter with built-in pathogen detection could demonstrate the presence of dangerous bacteria in water.
Finally, lateral-flow, bioactive-paper devices are also likely to be important. The idea is that an aqueous sample is introduced onto one end of a dry sample of bioactive paper. Capillary forces pull the liquid along the paper strip. Many bioanalytical assays employ lateral-flow devices (e.g., OTC pregnancy test kits). Lateral flow offers the following advantages that are unique to paper and similar porous substrates.
-
(1)
Liquid will move, whereas larger particles will be trapped in the paper matrix, a form of sample filtration. Chromatographic separation of soluble components on the paper surface is also possible.
-
(2)
The paper can be treated to give hydrophilic channels that can split the sample into two or more parts transported to different locations on the paper surface. Whiteside’s group have recently illustrated this concept [32].
-
(3)
An important application of lateral flow is the ability to expose a sample consecutively to a series of binding sites along the eluted surface.
-
(4)
Finally, lateral flow can be used to remove unbound components from a region of paper with surface-capture groups.
4. Paper-supported assays
4.1. Introduction
The detection of pathogens in our food, water and air is receiving a lot of attention because of the enormous public-health implications [42], [61]. The challenge is to achieve sensitive, selective detection using rapid inexpensive assays. Similarly, point-of-care biomedical diagnostics require the same characteristics – sensitivity, selectivity and speed – while being inexpensive. It is in these applications that bioactive paper offers the greatest promise. Diagnostic food packaging, disposable medical protective coverings and consumer products would have a big positive impact in the developed world. Most exciting are the potential impacts of simple, inexpensive point-of-care diagnostics for the developing world.
Litmus paper is a spectacular example of a sensing paper – it is inexpensive, requires no amplification or equipment and little training, has a long shelf life and is very sensitive. A litmus paper indicating pH 11 is reporting a hydrogen-ion concentration of 10−11 mol/L, which is picomolar sensitivity. Pathogen-detecting paper with these characteristics does not yet exist.
A pathogen-detecting paper must perform two important functions (i.e. biorecognition and reporting). Biorecognition refers to the capture of the target pathogen, or a chemical marker indicating the presence of a dangerous bacterium or virus. To be useful, the capture must be specific – there are many benign microbes, which are ignored by a useful biosensor. Note, there exist dyes that report the presence of bacteria on paper, but these are non-specific, giving no information about the nature of the microbe [62]. Recognition is a critical component of pathogen-sensing bioactive paper.
Biorecognition agents, herein called biosensors, likely to be important for bioactive paper are antibodies, enzymes, bacteriophages and DNA aptamers. I describe examples of each below. The mainstream analytical literature describes other potential capture or recognition agents, including molecular imprinted polymers [63] and whole cells [64], which have not yet been applied to paper. In addition, this review does not consider paper-supported electronic devices.
Reporting is the second step in pathogen detection. When a target is captured by an antibody or one of the other types of capture or recognition agents, the paper must signal (report) the occurrence of the capture event to the human user. The development of robust, sensitive reporting is the greatest challenge in the development of bioactive paper.
4.2. Biosensor immobilization on paper
4.2.1. Introduction to immobilization
In this section, I describe various approaches to attachment of the biosensing agents to paper. The goal is to control:
-
a)
the location of the biosensors in or on the paper structure;
-
b)
the density of biosensors; and,
-
c)
the tertiary structure and the orientation of the immobilized sensor molecules.
The immobilization process can be considered as two steps – transport and attachment. Transport is the process by which a buffer solution of sensor molecules is brought to the surface. Because of the importance in printing and coating, the transport of aqueous solutions into paper structures is much studied and well understood [44], [54], [65], [66]. Capillary-force-driven liquid flow is the major process. Paper-surface chemistry, controlling contact angle and pore-structure distribution in the paper influence both the rate and the extent of penetration of water into paper.
Attachment is not a strict requirement for incorporation of biosensors or any other water-soluble component into filter paper. A dry filter paper will sorb more than its dry mass of aqueous solution when immersed in a bath. Indeed, the cellulose fiber walls typically adsorb 0.5–2.5 g of water/g dry fiber [67]. Removal of the paper and drying will leave all of the non-volatile components of the bath solution in the paper structure. However, impregnation without attachment is not recommended, because it is difficult to control the distribution of the biosensor molecules in the paper structure. Furthermore, subsequent exposure to water is likely to leach the biosensors.
The location of the biosensing elements in the paper structure is important – biosensing molecules are expensive and must be used efficiently (e.g., an antibody hidden in a cellulose fiber pore will never be able to contact the surface of a 1-μm diameter bacterium, so, although filter paper may have the capacity to adsorb a large quantity of biosensor, much of it could be inaccessible to the target).
In summary, we face wide ranges of potential paper-substrate types, biosensor types, and immobilization strategies. Table 1 attempts to simplify the immobilization landscape by defining four categories, as described in the following four sub-sections:
-
(1)
physical immobilization, where the biosensor adheres to the paper surface because of van der Waals and electrostatic forces;
-
(2)
chemical immobilization, where covalent bonds fix the biosensor to the paper surface;
-
(3)
biochemical coupling, where cellulose binding modules (CBMs) or other biochemical binding agents are employed; and,
-
(4)
bioactive pigments, where biosensors are coated on colloidal particles that are then printed or coated onto the paper.
Table 1.
Four approaches to immobilizing biosensors onto dry or wet cellulose
| Biosensor | Physical | Chemical | Biochemical | Carrier particles⁎ |
|---|---|---|---|---|
| Antibodies | [77] | Film [105] | [77], [85], [86], [88], [106] | Microgel [17] |
| Silica [30] | ||||
| Enzyme | [5], [27], [37], [41], [71] | Film [105], [108], [109] | [86] | Silica [9] |
| Extruded with regenerated cellulose film [107] | Paper [110], [111] | |||
| Layer-by-layer [81] | ||||
| Phage | [19], [28] | Phage [19] | ||
| DNA Aptamer | [8], [23] | Paper [8], [112] | [90] | Microgel [17] |
| Nitrocellulose film [113] | ||||
| Cells | [114] | [87] | ||
| Biotin, streptavidin | [115] |
There are many examples of biomacromolecule immobilization on particles – these references are restricted to cases where the particles were subsequently put onto paper.
4.2.2. Physical immobilization – direct application to wet or dry paper
Printing and coating technologies allow application of almost any fluid onto dry paper. Aqueous solutions are particularly easy because capillary forces and the hydrophilic nature of cellulose promote rapid sorption. Antibodies, enzymes, aptamers and phages can be spotted or printed onto dry filter paper without denaturation. However, in most cases, the biosensors are not firmly anchored. The following paragraphs briefly review the adsorption behaviors of synthetic polymers, proteins, DNA aptamers and phages onto pure cellulose. Adsorption experiments indicate whether physical forces are sufficient to fix the biosensor to cellulose.
There have been many studies of the adsorption of synthetic polymers onto both pure cellulose [68] and papermaking fibers [60]. Clean cellulose is a hydrophilic, slightly anionic surface with a low, negative, surface-charge density [69]. Cationic polymers readily adsorb onto cellulose from aqueous solution, whereas anionic and non-ionic water-soluble polymers tend not to. Electrostatic interactions between cationic patches on proteins and anionic cellulose are also an important driving force for protein adsorption onto paper [70] and regenerated cellulose [71]. There is some evidence of attractive interactions between tyrosine groups and cellulose [72], [73], which may also contribute to binding.
Halder et al. reported adsorption isotherms for a number of proteins on cellulose powder, and found that ranking of proteins in terms of moles of adsorbed protein per mass of cellulose was gelatin > β-lactoglobulin > lysozyme > bovine serum albumin (BSA) under one set of conditions [74]. Note that the properties of both proteins and paper substrates are sensitive to pH, ionic strength and specific ion effects, so the details are important.
Many researchers have investigated blood-plasma-protein adsorption onto regenerated cellulose, a potential membrane material for hemodialysis (e.g., Brash showed that while fibrinogen did adsorb onto cellulose, the rate and extent of adsorption were low compared to hydrophobic surfaces, such as silicone, PVC and polyethylene [75]). In summary, proteins are not strongly adsorbed onto pure cellulose, so protein-based sensors are likely to require a more aggressive immobilization strategy. Other types of biosensors do not adsorb proteins strongly either.
Halder et al. showed that high-molecular-weight DNA did not adsorb onto cellulose at pH 6 and pH 8, whereas adsorption was observed at pH 4 [74].
Su et al reported adsorption isotherms for low-molecular-weight DNA aptamers onto microcrystalline cellulose [8]. However, the utility of direct DNA-aptamer application is limited because the aptamers were easily washed off. The low affinity of these low-molecular-weight oligonucleotides is consistent with the synthetic polymer adsorption literature, which shows that anionic polymers tend not to adsorb onto cellulose [76].
Tolba et al.’s recent publication is currently the only report of the direct application of phages to cellulosic surfaces [19]. They showed that wild T4 phage does bind to cellulose, but the subsequent activity of the bound wild T4 phage is lower than genetically-engineered T4, which binds via its head. They speculated that the wild T4 phage interacted with cellulose via the binding sites on the phage’s long tail fibers, which are the bacterial binding sites (see Fig. 3 ).
Figure 3.
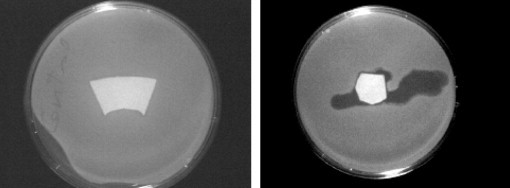
Killing bacteria with filter paper impregnated with wild T4 phage (left), and filter paper with genetically-engineered T4 phage with cellulose-binding module (CBM) (right) giving spontaneous adhesion of the phage head to cellulose. The CBM-immobilized phage infects and kills the bacteria giving transparent regions in the media [19].
Most laboratory filter papers are pure cellulose that is slightly anionic and very hydrophilic. By contrast, paper products that are expected to function while wet are usually treated with wet-strength resins that are reactive polymers added to maintain paper strength in water [43]. Kitchen towels and coffee filters are everyday examples of paper with high contents of wet-strength resin. Fig. 1 shows the chemical structure of PAE, the most widely used wet-strength treatment. Paper treated with PAE has a net positive surface charge [43] and there may be residual chemically reactive groups that can couple to proteins. Polyvinylamine is another polymer used in papermaking, which renders cellulose cationic and reactive due to the large number of primary amine groups.
PAE wet-strength resin can influence both biosensor immobilization and activity. Cationic PAE promotes the adsorption of DNA aptamers, but the bound sensors are denatured and thus inactive [17]. PAE also promotes antibody adsorption onto cellulose, but it does not seem to lower antibody activity significantly [77].
Paper surfaces are also often covered with a grafted layer of a hydrophobic chemical, called “size” in the paper industry [44]. Recently, Yan’s group reported the influence of sizing on ink-jet-printed horseradish peroxidase (HRP) [5], [27]. They showed that moderate sizing increased the color intensity from HRP-catalyzed reactions, whereas excessive sizing lowered enzyme efficiency. Presumably, excessively hydrophobic surfaces denatured the adsorbed antibody.
Finally, a variation of physical immobilization is Decher’s [78] layer-by-layer (LBL) assembly, where surfaces are consecutively exposed to oppositely charged polymer solutions, followed by washing to give multilayer adsorbed structures. The driving force for sorption is usually electrostatic, but hydrogen bonding and other interactions can also drive assembly [79]. LBL assembly can be used to embed particles, viruses [80] or cells onto surfaces. Lvov’s group has demonstrated that LBL assembly can be used to fix enzymes onto cellulose surfaces while maintaining enzyme activity [81]. LBL assembly has also been used to deposit large quantities of antimicrobial polymers on surfaces [38].
In summary, proteins, phages and DNA aptamers are weakly bound on pure cellulose paper. It seems that paper treated with wet-strength resin may be a good substrate for the direct immobilization of biosensors. There are two caveats. The cationic surfaces will adsorb most biomacromolecules, so it may be necessary to use some kind of blocking to prevent non-specific adsorption [82]. Common blocking chemicals include Tween 20 (a non-ionic surfactant), BSA, casein or fat-free milk [83]. Second, there is no control of biosensor orientation and we might expect very cationic and reactive surfaces to denature protein-based and DNA-based sensors. The general sense from the literature is therefore that direct application is not a robust strategy because every biosensor/paper combination would have to be optimized before use.
4.2.3. Chemical immobilization – covalent coupling
Bioconjugation is a large, mature field that has been summarized in an excellent text [84]. Ideally, chemical-coupling reactions should achieve very high yields in water under mild conditions with few side reactions and little denaturation of biomacromolecules. I summarize some examples relevant to bioactive-paper fabrication.
Pure cellulose offers few functional groups for direct bioconjugation. The backbone hydroxyl groups are too unreactive for specific reactions in water at low temperature. Low concentrations of carboxyl groups from inadvertent oxidation of the C6 hydroxyls and the oxidizing end of cellulose chains are the only available functional groups on pure cellulose. Of course, practical paper surfaces may also have hemicellulose, lignin and other extractives, offering a wider range of potentially reactive centers.
The lack of reactivity means that most cellulose substrates need to be activated by reaction with a small molecule or polymer to give surface functional groups suitable for a subsequent bioconjugation reaction. For example, my colleagues and I oxidized regenerated cellulose to give aldehyde groups, which reacted with amine groups on a DNA aptamer to form a Schiff base, which was reduced to give a stable covalent bond (Fig. 4 ) [8].
Figure 4.
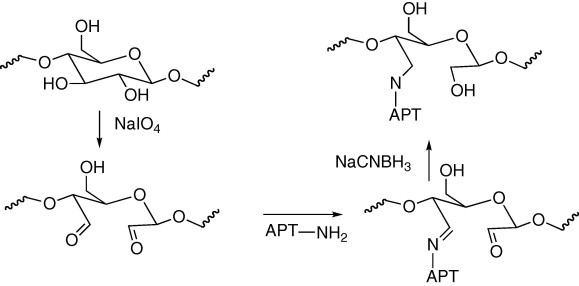
Coupling a DNA aptamer to oxidized cellulose.
Table 2 gives examples of the coupling of biomacromolecules to activated cellulose surfaces. The literature contains many more examples, particularly for coupling to nitrocellulose. However, I believe that these methods contribute little to practical production of bioactive paper. Since most of these approaches involve multiple chemical steps, they are not very attractive as a route to commodity paper products with inexpensive pathogen detection. However, chemical coupling could be effective in the preparation of bioactive pigments for bioactive inks – see below.
Table 2.
Examples of conjugation to activated cellulose surfaces
| Surface activation | Biosensor | Surface | Ref. |
|---|---|---|---|
| Epichlorohydrin reaction to give epoxy groups | DNA for antibody removal from blood | Regenerated cellulose | [112] |
| Periodate oxidation to give aldehyde groups | DNA aptamers | Regenerated cellulose and microcrystalline cellulose | [8] |
| 1-fluoro-2-nitro-4-azidobenzene photo-activated linker | Antibodies | Regenerated cellulose | [105] |
| Epichlorohydrin followed by pentaethylenehexamine | Invertase | Cellulose regenerated from diacetylcellulose | [110] |
| 1,4-diaminobenzene reaction with C6-tos | Glucose oxidase, HRP, and lactate oxidase | Regenerated cellulose | [109] |
| 1,4-butanediol diglycidyl ether | Glucoamylase | Bacterial cellulose | [116] |
4.2.4. Biochemical immobilization
Genetic engineering approaches have been used to couple CBMs to antibodies [85], [86], enzymes and bacteriophages (Fig. 3) [19] or cells [87], which adhere spontaneously to cellulose and/or hemicellulose. For example, Cao described a fusion protein comprising CBM bound to protein A [85]. The protein A end or the bifunctional protein specifically binds to a wide variety of antibody fragments, whereas the CBM spontaneously binds to cellulose. In another example, Lewis and co-workers described an elegant approach in which CBMs were engineered onto llama antibodies to give a construct which spontaneously bonded to cotton, a form of nearly pure cellulose [88]. In their experiments, the llama-antibody chain was bound to antigen-coated coacervate spheres, which then spontaneously deposited onto cellulose. This is an excellent paper, which gives much information regarding the strength of CBM-cellulose adhesion.
Wang et al. recently reported the influence of PAE treatment on the activity of paper-supported antibodies [77]. They evaluated two types of antibodies:
-
•
a construct consisting of 5 llama-antibody chains fused to 5 cellulose-binding domains; and,
-
•
a conventional anti-mouse antibody with no specific binding sites for cellulose.
They reported the influence of wet-strength resin on the efficiency of the antibody immobilization and function. Using enzyme-linked immunosorbent assay (ELISA) (see Section 4 below for an explanation), they showed that antibody activity was only slightly decreased by high loadings of PAE (Fig. 5 ). Indeed, low PAE loadings typical of commercial papers improved the antibody activities. This is a surprising result; one would anticipate catastrophic denaturation of the antibody on the cationic polymer.
Figure 5.
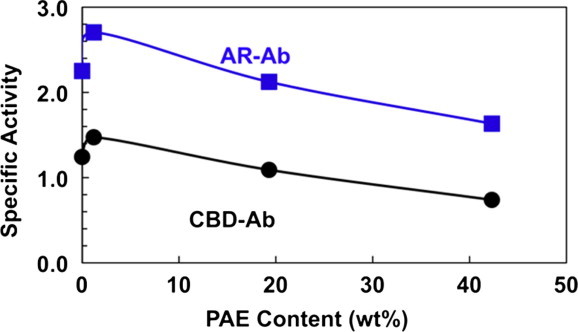
The influence of PAE wet-strength resin on paper-supported ELISA. AR-Ab is a conventional anti-mouse antibody (Ab) immobilized by non-specific adsorption. CBD-Ab is complex structure with 5 Abs fused to 5 cellulose-binding modules (CBMs) (Adapted from [77]).
A recent publication from Ye and co-workers demonstrates the simultaneous application of three types of biological immobilization in a process used to attach TiO2 nanoparticles to cellulose [30].
Fig. 6 summarizes their approach. In the first step, cellulose was coated with a bifunctional fusion protein based on CBM and strep tag. The CBM ensured irreversible binding. The surface was exposed to streptavidin in a second step. In a final step, exposure to biotinylated TiO2 [89] gave specific attachment of the nanoparticles to cellulose.
Figure 6.
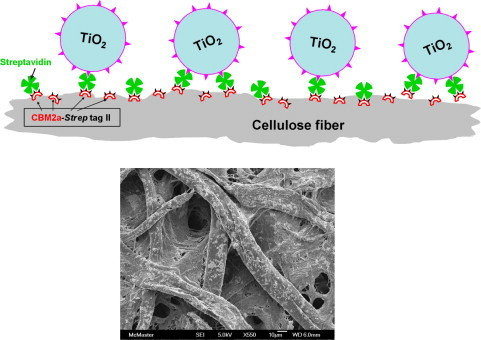
Cellulose-binding-module protein used to attach TiO2 to cellulose (Adapted from [30]).
Finally, there are reports of DNA aptamers designed to give specific binding to cellulose [90]. Presumably, complex aptamers with binding and detection functions could be designed.
4.2.5. Carrier particles – bioactive pigments for bioactive inks and coating colors
A very good approach to biosensor immobilization is to couple covalently (conjugate) the biosensor to colloidal particles that can then be printed, coated or even added during the papermaking process. This approach has the following advantages.
-
(1)
Coupling processes involving difficult, expensive and sensitive reagents can be performed in suitable bioprocessing facilities far from papermaking, printing, coating or converting operations.
-
(2)
Compared with small, water-soluble biomacromolecules, it is easier to concentrate colloidal particles onto exterior surfaces of porous papers.
-
(3)
The microenvironment around the biosensor is determined by the support-particle chemistry, not the paper surface. Thus, supported biosensors should be less sensitive to variations in paper-substrate properties compared to small, water-soluble biosensors. Blocking and reporting functions can be built into the carrier particles.
Attaching biosensors to particles is a mature subject (e.g., there have been commercial latex-agglutination assays for decades). When dilute suspensions of latex particles, coated with antibodies, are exposed to antigen, antibody-antigen binding causes the latex dispersion to aggregate giving a visible response [91]. The early work involved polystyrene latexes, available as monodispersed particles, which had clean surfaces and could be magnetic. Streptavidin adsorbs spontaneously and irreversibly, giving particles that will bind biotinylated biosensors [92], [93]. Pichot’s group reported extensively on the preparation and the characterization of a wide variety of polymer colloids as potential support particles for biosensors [94]. They concluded that colloidal microgels based on poly(N-isopropylacrylamide) (PNIPAM) [95] were superior because PNIPAM gives little non-specific protein binding [96]. Following from Pichot’s work, my colleagues and I covalently coupled DNA aptamers and antibodies to carboxylated PNIPAM microgels [17]. Fig. 7 shows the conjugation chemistry. We were surprised to observe that simply air-drying the microgels after spotting them on filter paper immobilized the gels. They did not come off or move when the dried paper was subsequently immersed in buffer or eluted with buffer.
Figure 7.

Coupling antibodies or DNA aptamers to carboxylated microgels (Adapted from [17]).
Fig. 8 shows examples of elution experiments. The microgels did not move. Examples of signals given by the microgel-supported sensors are given in the Section 4 below.
Figure 8.
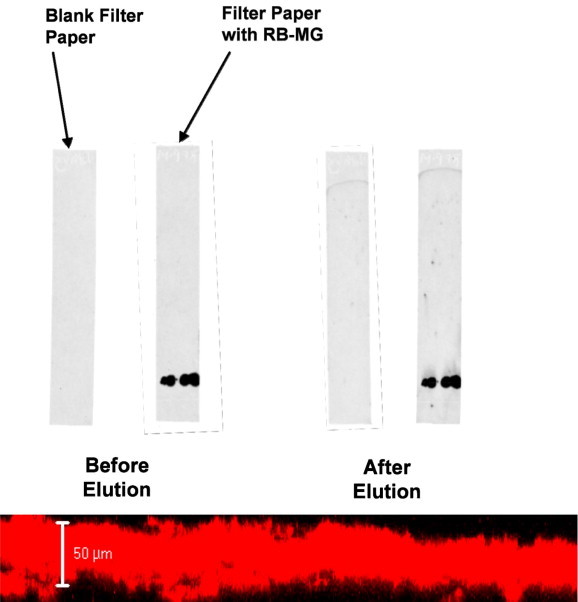
Chromatographic elution of filter paper spotted with fluorescently-labeled microgel (Adapted from [17]).
Silica is also a convenient surface for biosensor immobilization and silica is available as either solid or porous nanoparticles. Voss et al. described the preparation of porous-silica particles with active HRP in the pores [9], so one can imagine support particles with the biosensor functions on the exterior surface and the reporting chemistry embedded in interior pores.
Bang et al. described porous sol-gel particles, prepared in an aerosol process. [97] They printed arrays of 16 particle types each with a different dye. The arrays could distinguish amines. Such a paper-supported assay may be useful for detecting food spoilage.
Finally, a recent report describes the inkjet printing of reacting sol-gel impregnated with enzymes and giving good detection limits (paraoxon ∼100 nM; aflatoxin B1 ∼1 M) and rapid response times (<5 min) [31]. Spots of strongly cationic polyvinylamine on the paper served to fix and thus concentrated the colored 5-thio-2-nitrobenzoate from the Ellman assay. Without PVAm, the colored Ellman product was present as large, faint, diffuse stains.
Microcapsules have a long history of use in paper products (e.g., carbonless paper). A recent paper describes laccase encapsulated in cross-linked polyethyleneimine and deposited on paper [98].
5. Reporting
It is relatively straightforward to attach a biosensor (e.g., an antibody) to a paper surface and then capture a specific target. The challenge is to report the capture event to the human observer. Bodenhamer’s recent patent describes an interesting displacement assay for transparent plastic packaging, which has many of the desired attributes for bioactive paper [99], [100]. Fig. 9 illustrates their approach. A heat killed or facsimile antigen is immobilized in a printed pattern such as a “√”. The surface is then saturated with an antibody bearing a pigment giving a √ when the excess is washed away. Upon exposure to pathogenic antigens in contacting liquor, the dyed antibodies are released from the surface in favor of forming stronger complexes with the pathogenic target. The disappearance of the √ is the reporting event.
Figure 9.

Bodenhamer’s displacement assay for pathogen detection on transparent films. Initially a dye-conjugated antibody is bound to an immobilized facsimile antigen, giving a colored √. Antigen exposure strips the antibody-dye conjugate from the surface, giving a color change [99], [100].
ELISA is one of the most important detection/reporting combinations. Fig. 10 illustrates the main features of ELISA. The biosensor (capture agent) is usually an antibody or antibody fragment that is immobilized on a support surface. Exposure of the test solution leads to antigen capture. After washing, a second antibody (secondary antibody) bearing an enzyme is introduced. Finally, the test surface is exposed to the substrate solution for the immobilized enzyme that catalyzes the production of a colored or fluorescent product. With care and some form of instrumentation for accessing color or fluorescence, ELISA can be quantitative. To avoid high backgrounds, it is often necessary to block the support surface by adsorbing a polymer, a biomacromolecule and/or a surfactant to prevent the non-specific binding of the antigen or the secondary antibody. Although it will be shown below that ELISA detection of paper works well, the multiple steps doom ELISA to “offline” applications, where the exposed bioactive paper must be processed to generate a signal – not the best solution.
Figure 10.

Enzyme-linked immunosorbent assay (ELISA) scheme for detection and reporting.
The effectiveness of ELISA was shown to be very sensitive to nature of the paper substrate. Wang et al. compared 50 commercial papers, from a single supplier, in a filtration capture ELISA [77]. The signal intensity varied by more than an order of magnitude amongst the filter papers, emphasizing the importance of paper properties.
The use of enzymes to generate a color is a common approach to reporting. Whiteside’s group have published a series of papers demonstrating the use of paper to segregate a sample into different chambers, where a different target is probed in each chamber using a variety of assays, some involving enzyme reporting [32], [33], [37]. They describe hydrophobic paper with a hydrophilic channel feeding three chambers. The channels and chambers were created by printing hydrophobic patterns onto filter paper. Color-developing enzymes were spotted and dried onto one of the chambers for glucose detection. Capillary forces were used to carry the sample solution into the three chambers. Such devices could be very inexpensive and suitable for both point-of-care and developing-world applications. Whiteside’s work and that of others [41] emphasize an important feature of paper – the ability to generate complex “macro” fluidic devices useful for sample conditioning, separation and transport prior to the pathogen-detection step.
Fluorescence-based reporting is the workhorse of the modern bio-analytical laboratory. When used with suitable instrumentation, fluorescence is very sensitive. From the perspective of bioactive paper, fluorescence reporting is a challenge for two reasons. First, instrumentation is required. Second, many commercial papers fluoresce, giving a high background. It is common practice to include fluorescent agents in papermaking to increase the appearance of whiteness.
Li’s group have developed DNA aptamers with built-in fluorescent reporters [101] (e.g. Fig. 11 ). Initially, the DNA aptamer is present as a duplex with a short DNA molecule labeled with a quencher (Q). The aptamer is also made to form a second duplex with another short DNA molecule labeled with a fluorescent group, so the fluorescence of the aptamer in the unbound state is quenched. When the target, ATP in this case, is introduced, the duplex dissociates in favor of forming an aptamer-ATP complex which fluoresces. Su et al. have shown that Li’s aptamers can be immobilized on microgels that can be inkjet printed onto paper, giving an aptamer-based sensor-reporter combination [17].
Figure 11.
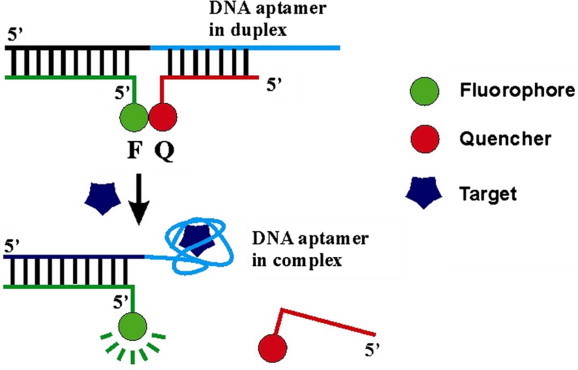
DNA aptamer with built-in fluorescent reporting. In the initial duplex, the fluorescent aptamer is quenched. Upon exposure to the target, the duplex dissociates, giving a fluorescent signal (Adapted from [101]).
Nanoparticle reporters show some promise. Gold nanoparticles in size range 10–50 nm have an intense red color when the particles are separated by a distance equivalent to a few particle diameters. The color changes to purple when aggregation brings the particles closer together. In a typical sensing application, the particles are coated antibodies or aptamers [102] and a color change is observed when the target antigen induces aggregation of the gold particles. This is an attractive approach because the colors are very intense, typically nanogold-extinction coefficients are more than 1000 times greater than those of organic dyes [10]. In addition, the ability of thiols to chemisorb onto gold provides a simple route to immobilization. Fig. 12 shows a schematic representation of a DNA-aptamer biosensor employing a nanogold sensor [10].
Figure 12.

Gold-nanoparticle reporting with a DNA-aptamer biosensor. Initially the aptamers prevent the nanoparticles from aggregating (Adapted from [10]). Exposure to target strips the aptamers from the particles causing aggregation and color change.
Most publications describe nanogold sensors that are dispersions in buffers. For gold to be useful on bioactive paper, the nanoparticles must function after drying and subsequent wetting of the sensor. Zhao et al. recently describe a paper-supported gold biosensor capable of detecting the presence of DNase I, an endonuclease, and adenosine, a small biomolecule [23]. For both targets, the biosensor functions by causing the dissociation of a gold-nanoparticle aggregate to give an intense red color (see Fig. 13 ). The DNase I sensor functions by degrading the DNA chains bridging the nanoparticles, whereas, with the adenosine sensor, the gold particles are weakly aggregated with an adenosine aptamer. The presence of adenosine strips the aptamer from the gold, causing the particles to disperse. This is an important publication because it demonstrates that a sensitive biosensor can be dried on paper, heated and stored while retaining activity upon subsequent exposure to the target solution. From a paper-science perspective, Zhao’s work is important because it shows that the details of the paper substrate are important. Papers coated with hydrophilic or hydrophobic polymers were suitable for the nanoparticle assay, whereas untreated filter paper was not because capillary forces caused the spotted nanoparticles to bleed over too great an area.
Figure 13.

A paper-supported sensor measuring the presence of DNase. Decomposition of the stabilizing chains on the nanoparticles causes them to aggregate, giving a color change (Adapted from [23]).
Quantum dots are semiconductor nanoparticles and are intensely fluorescent, offering many advantages for biosensing [103]. Fig. 14 shows one example, taken from a patent application [104]. The biosensor consists of quantum dots with immobilized antibodies on the surface, which, in turn, are weakly bound to a surrogate facsimile antigen bearing a fluorescent quenching molecule. The quencher prevents fluorescent emissions from the quantum dot. Reporting occurs when the target antigen displaces the surrogate, separating the quencher from the quantum dot, which is then free to fluoresce.
Figure 14.

Antigen displaces facsimile antibody-quencher from quantum dot, producing fluorescence [104].
6. Conclusion
There are promising approaches that could lead to bioactive paper that can detect pathogens. Although it is early days, it seems that fabrication and biomolecule stability of bioactive paper are not major problems. Immediate, sensitive detection and reporting without instrumentation or a laboratory environment remain significant challenges.
Acknowledgements
I thank the Sentinel Bioactive Paper NSERC Network for funding much of the research summarized herein and our industrial partners, including Ahlstrom, Buckman Laboratories, Cascades, Domtar, FPInnovations, Fujifilm Dimatix, Stora Enso, and Sun Chemical. Finally, I thank Professors Yingfu Li, Carlos Filipe and Mansel Griffiths for comments.
References
- 1.S. Aikio, S. Grönqvist, L. Hakola, E. Hurme, S. Jussila, O. Kaukoniemi, H. Kopola, M. Känsäkoski, M. Leinonen, S. Lippo, R. Mahlberg, S. Peltonen, P. Qvintus-Leino, T. Rajamäki, A. Ritschkoff, M. Smolander, J. Vartiainen, L. Viikari, M. Vilkman, Bioactive paper and fibre products patent and literary survey, VTT Working Paper 51, VTT Technical Research Centre of Finland, Oulu, Finland, 2006.
- 2.Shen Y., Chiuman W., Brennan J.D., Li Y. Chembiochem. 2006;7:1343. doi: 10.1002/cbic.200600195. [DOI] [PubMed] [Google Scholar]
- 3.Ali M.M., Kandadai S.A., Li Y. Can. J. Chem. 2007;85:261. [Google Scholar]
- 4.Ali M.M., Su S., Filipe C.D.M., Pelton R., Li Y. Chem. Commun. 2007:4459. doi: 10.1039/b709817k. [DOI] [PubMed] [Google Scholar]
- 5.Di Risio S., Yan N. Macromol. Rapid Commun. 2007;28:1934. [Google Scholar]
- 6.Kavoosi M., Creagh A.L., Kilburn D.G., Haynes C.A. Biotechnol. Bioeng. 2007;98:599. doi: 10.1002/bit.21396. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y.T., Mackey G., Rupcich N., Gloster D., Chiuman W., Li Y., Brennan J.D. Anal. Chem. 2007;79:3494. doi: 10.1021/ac070235u. [DOI] [PubMed] [Google Scholar]
- 8.Su S., Nutiu R., Filipe C.D.M., Li Y., Pelton R. Langmuir. 2007;23:1300. doi: 10.1021/la060961c. [DOI] [PubMed] [Google Scholar]
- 9.Voss R., Brook M.A., Thompson J., Chen Y., Pelton R.H., Brennan J.D. J. Mater. Chem. 2007;17:4854. [Google Scholar]
- 10.Zhao W.A., Chiuman W., Brook M.A., Li Y. Chembiochem. 2007;8:727. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- 11.Zhao W.A., Chiuman W., Lam J.C.F., Brook M.A., Li Y. Chem. Commun. 2007:3729. doi: 10.1039/b705335e. [DOI] [PubMed] [Google Scholar]
- 12.Zhao W.A., Gonzaga F., Li Y., Brook M.A. Adv. Mater. 2007;19:1766. [Google Scholar]
- 13.Jabrane T., Jeaidi J., Dube M., Mangin P.J. Adv. Printing Media Technol. 2008;35:279. [Google Scholar]
- 14.Mao J., Grgic B., Finlay W.A., Kerekes R.J. Nordic Pulp Paper Res. J. 2008;23:420. [Google Scholar]
- 15.G. Pianet, D. Vidal, B. Mallet, F. Bertrand, Modeling the compression of particle packings using the discrete element method, TAPPI, 10th Advanced Coating Fundamentals Symp., Montreal, Canada, 2008.
- 16.Qian L.Y., Guan Y., He B.H., Xiao H.N. Polymer. 2008;49:2471. [Google Scholar]
- 17.Su S., Ali M.M., Filipe C.D.M., Li Y., Pelton R. Biomacromolecules. 2008;9:935. doi: 10.1021/bm7013608. [DOI] [PubMed] [Google Scholar]
- 18.Tan H., Xiao H. Tetrahedron Lett. 2008;49:1759. [Google Scholar]
- 19.M. Tolba, L.Y. Brovko, O. Minikh, M.W. Griffiths, NSTI, Nanotech 2008, Boston, USA, 2008, pp. 449-452.
- 20.Vyhnalkova R., Eisenberg A., Van De Ven T.G.M. J. Phys. Chem. B. 2008;112:8477. doi: 10.1021/jp8009707. [DOI] [PubMed] [Google Scholar]
- 21.Zhao W., Brook M.A., Li Y.F. Chembiochem. 2008;9:2363. doi: 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W., Chiuman W., Lam J.C.F., Mcmanus S.A., Chen W., Cui Y., Pelton R., Brook M.A., Li Y. J. Am. Chem. Soc. 2008;130:3610. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- 23.Zhao W.A., Ali M.M., Aguirre S.D., Brook M.A., Li Y.F. Anal. Chem. 2008;80:8431. doi: 10.1021/ac801008q. [DOI] [PubMed] [Google Scholar]
- 24.Zhao W.A., Ali M.M., Brook M.A., Li Y.F. Angew. Chem., Int. Ed. Engl. 2008;47:6330. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 25.Brovko L.Y., Meyer A., Tiwana A.S., Chen W., Liu H., Filipe C.D.M., Griffiths M. J. Food Prot. 2009;72:1020. doi: 10.4315/0362-028x-72.5.1020. [DOI] [PubMed] [Google Scholar]
- 26.Cademartiri R., Brook M.A., Pelton R., Brennan J.D. J. Mater. Chem. 2009;19:1583. [Google Scholar]
- 27.S. Di Risio, N. Yan, J. Pulp Paper Sci. (2009), in press.
- 28.T. Jabrane, M. Dubé, P.J. Mangin, Bacteriophage immobilization on paper surface: Effect of cationic pre-coat layer, PAPTAC 95th Annual Meeting, Montreal, Canada, 2009, pp. 311–315.
- 29.Vidal D., Ridgway C., Pianet G., Schoelkopf J., Roy R., Bertrand F. Comput. Chem. Eng. 2009;33:256. [Google Scholar]
- 30.Ye L., Filipe C.D.M., Kavoosi M., Haynes C.A., Pelton R., Brook M. J. Mater. Chem. 2009;19:2189. [Google Scholar]
- 31.Hossain S.M.Z., Luckham R.E., Smith A.M., Lebert J.M., Davies L.M., Pelton R., Filipe C.D.M., Brennan J.D. Anal. Chem. 2009;81:5474. doi: 10.1021/ac900660p. [DOI] [PubMed] [Google Scholar]
- 32.Bruzewicz D.A., Reches M., Whitesides G.M. Anal. Chem. 2008;80:3387. doi: 10.1021/ac702605a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez A.W., Phillips S.T., Carrilho E., Thomas S.W., Sindi H., Whitesides G.M. Anal. Chem. 2008;80:3699. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez A.W., Phillips S.T., Whitesides G.M. Proc. Natl. Acad. Sci. USA. 2008;105:19606. doi: 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez A.W., Phillips S.T., Wiley B.J., Gupta M., Whitesides G.M. Lab Chip. 2008;8:2146. doi: 10.1039/b811135a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracher P.J., Gupta M., Whitesides G.M. Adv. Mater. 2009;21:445. doi: 10.1002/adma.200801186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez A.W., Phillips S.T., Butte M.J., Whitesides G.M. Angew. Chem., Int. Ed. Engl. 2007;46:1318. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westman E., Ek M., Wagberg L. Holzforschung. 2009;63:33. [Google Scholar]
- 39.X. Li, J. Tian, W. Shen, Paper as a low-cost base material for diagnostic and environmental sensing applications, 63rd Appita Annual Conference, Melbourne, Australia, 2009, pp. 267–271.
- 40.M.S. Khan, W. Shen, G. Garnier, 63rd Appita Annual Conference, Melbourne, Australia, 2009, pp. 273–280.
- 41.Abe K., Suzuki K., Citterio D. Anal. Chem. 2008;80:6928. doi: 10.1021/ac800604v. [DOI] [PubMed] [Google Scholar]
- 42.Leonard P., Hearty S., Brennan J., Dunne L., Quinn J., Chakraborty T., O’Kennedy R. Enzyme Microb. Technol. 2003;32:3. [Google Scholar]
- 43.Espy H.H. Tappi J. 1995;78:90. [Google Scholar]
- 44.Hubbe M.A. BioResources. 2007;2:106. [Google Scholar]
- 45.Niskanen K. Fapet Oy; Helsinki, Finland: 1998. Paper Physics. [Google Scholar]
- 46.Sampson W. Springer; Berlin, Germany: 2008. Modelling Stochastic Fibrous Materials with Mathematics. [Google Scholar]
- 47.Weise U. Paper Timber. 1998;80:110. [Google Scholar]
- 48.Alince B. Nordic Pulp Paper Res. J. 2002;17:71. [Google Scholar]
- 49.Fleer G.J., Cohen Stuart M.A., Scheutjens J.M.H.M., Cosgrove T., Vincent B. Chapman & Hall; London, UK: 1993. Polymers at Interfaces. [Google Scholar]
- 50.Hong J., Ye X.H., Zhang Y.H.P. Langmuir. 2007;23:12535. doi: 10.1021/la7025686. [DOI] [PubMed] [Google Scholar]
- 51.Klemm D., Heublein B., Fink H.P., Bohn A. Angew. Chem., Int. Ed. Engl. 2005;44:3358. doi: 10.1002/anie.200460587. [DOI] [PubMed] [Google Scholar]
- 52.Towers M., Scallan A.M. J. Pulp Paper Sci. 1996;22:J332. [Google Scholar]
- 53.Katz S., Scallan A.M. Svensk Papperstidning. 1984;87:48. [Google Scholar]
- 54.Lyne M.B., Aspler J.S. ACS Symp. Ser. 1982;200:385. [Google Scholar]
- 55.Heng J.Y.Y., Pearse D.F., Thielmann F., Lampke T., Bismarck A. Compos. Interfaces. 2007;14:581. [Google Scholar]
- 56.Cunha A.G., Fernandes S.C.M., Freire C.S.R., Silvestre A.J.D., Pascoal C., Gandini A. Biomacromolecules. 2008;9:610. doi: 10.1021/bm701199g. [DOI] [PubMed] [Google Scholar]
- 57.Seppanen R., Von Bahr M., Tiberg F., Zhmud B. J. Pulp Paper Sci. 2004;30:70. [Google Scholar]
- 58.Berg J. Nordic Pulp Paper Res. J. 1993;8:75. [Google Scholar]
- 59.Deng Y.L., Abazeri M. Wood Fiber Sci. 1998;30:155. [Google Scholar]
- 60.Wagberg L., Odberg L. Nordic Pulp Paper Res. J. 1989;4:135. [Google Scholar]
- 61.Lopez-Rubio A., Gavara R., Lagaron J.A. Trends Food Sci. Technol. 2006;17:567. [Google Scholar]
- 62.J. Lye, J.G. Macdonald, N. Wei, Solvatochromatic bacterial detection, US Patent, 7, 282, 349 B2 (2007).
- 63.Bossi A., Bonini F., Turner A.P.F., Piletsky S.A. Biosens. Bioelectron. 2007;22:1131. doi: 10.1016/j.bios.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 64.S. Nicklin, M. Cooper, N. D’Souza, WO Patent Application 083137 Al (2007).
- 65.Senden T.J., Knackstedt M.A., Lyne M.B. Nordic Pulp Paper Res. J. 2000;15:554. [Google Scholar]
- 66.Roberts R.J., Senden T.J., Knackstedt M.A., Lyne M.B. J. Pulp Paper Sci. 2003;29:123. [Google Scholar]
- 67.Scallan A.M., Carles J.E. Svensk Papperstidning. 1972;17:699. [Google Scholar]
- 68.Geffroy C., Labeau M.P., Wong K., Cabane B., Cohen Stuart M.A.C. Colloids Surf., A. 2000;172:47. [Google Scholar]
- 69.Pelton R. Nordic Pulp Paper Res. J. 1993;11:113. [Google Scholar]
- 70.Jones K.L., O’Melia C.R. J. Membr. Sci. 2000;165:31. [Google Scholar]
- 71.Karra-Chaabouni M., Bouaziz I., Boufi S., Botelho Do Rego A.M., Gargouri Y. Colloids Surf., B. 2008;66:168. doi: 10.1016/j.colsurfb.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 72.Lehtio J., Sugiyama J., Gustavsson M., Fransson L., Linder M., Teeri T.T. Proc. Natl. Acad. Sci. USA. 2003;100:484. doi: 10.1073/pnas.212651999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mattinen M.-L., Linder M., Teleman A., Annila A. FEBS Lett. 1997;407:291. doi: 10.1016/s0014-5793(97)00356-6. [DOI] [PubMed] [Google Scholar]
- 74.Halder E., Chattoraj D.K., Das K.P. Biopolymers. 2005;77:286. doi: 10.1002/bip.20232. [DOI] [PubMed] [Google Scholar]
- 75.Brash J.L., Tenhove P. J. Biomater. Sci. Polym. Ed. 1993;4:591. doi: 10.1163/156856293x00230. [DOI] [PubMed] [Google Scholar]
- 76.Hendrickson E.R., Neuman R.D. J. Colloid Interface Sci. 1986;110:243. [Google Scholar]
- 77.J. Wang, R. Pelton, L. Veldhuis, C.R. Mackenzie, J.C. Hall, C.D.M. Filipe, Appita J., submitted for publication.
- 78.Decher G., Schlenoff J.B. Wiley-VCH; Weinheim, Germany: 2002. Multilayer Thin Films: Sequential Assembly of Nanocomposite Materials. [Google Scholar]
- 79.Zhang D., Tanaka H., Pelton R. Langmuir. 2007;23:8806. doi: 10.1021/la700711a. [DOI] [PubMed] [Google Scholar]
- 80.Yoo P.J., Nam K.T., Qi J.F., Lee S.K., Park J., Belcher A.M., Hammond P.T. Nat. Mater. 2006;5:234. doi: 10.1038/nmat1596. [DOI] [PubMed] [Google Scholar]
- 81.Xing Q., Eadula S.R., Lvov Y.M. Biomacromolecules. 2007;8:1987. doi: 10.1021/bm070125x. [DOI] [PubMed] [Google Scholar]
- 82.Reimhult K., Petersson K., Krozer A. Langmuir. 2008;24:8695. doi: 10.1021/la800224s. [DOI] [PubMed] [Google Scholar]
- 83.Vikholm-Lundin I. Langmuir. 2005;21:6473. doi: 10.1021/la046992u. [DOI] [PubMed] [Google Scholar]
- 84.Hermanson G.T. Academic Press; San Diego, California, USA: 1996. Bioconjugate Techniques. [Google Scholar]
- 85.Cao Y., Zhang Q., Wang C., Zhu Y.Y., Bai G. J. Chromatogr., A. 2007;1149:228. doi: 10.1016/j.chroma.2007.03.032. [DOI] [PubMed] [Google Scholar]
- 86.Calixto-Romo M.D.A., Santiago-Hernandez J.A., Vallejo-Becerra V., Amaya-Delgado L., Montes-Horcasitas M.D., Hidalgo-Lara M.E. J. Ind. Microbiol. Biotechnol. 2008;35:1455. doi: 10.1007/s10295-008-0447-1. [DOI] [PubMed] [Google Scholar]
- 87.Craig S.J., Shu A., Xu Y., Foong F.C., Nordon R. Protein Eng. Design Selection. 2007;20:235. doi: 10.1093/protein/gzm016. [DOI] [PubMed] [Google Scholar]
- 88.Lewis W., Keshavarz-Moore E., Windust J., Bushell D., Parry N. Biotechnol. Bioeng. 2006;94:625. doi: 10.1002/bit.20849. [DOI] [PubMed] [Google Scholar]
- 89.Ye L., Pelton R., Brook M.A. Langmuir. 2007;23:5630. doi: 10.1021/la0626656. [DOI] [PubMed] [Google Scholar]
- 90.Boese B.J., Corbino K., Breaker R.R. Nucleosides Nucleotides Nucleic Acids. 2008;27:949. doi: 10.1080/15257770802257903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ouali L., Stoll S., Pefferkorn E., Elaissari A., Lanet V., Pichot C., Mandrand B. Polym. Adv. Technol. 1995;6:541. [Google Scholar]
- 92.Huang S.C., Swerdlow H., Caldwell K.D. Anal. Biochem. 1994;222:441. doi: 10.1006/abio.1994.1514. [DOI] [PubMed] [Google Scholar]
- 93.Huang S.C., Stump M.D., Weiss R., Caldwell K.D. Anal. Biochem. 1996;237:115. doi: 10.1006/abio.1996.0208. [DOI] [PubMed] [Google Scholar]
- 94.Pichot C., Taniguchi T., Delair T., Elaissari A. J. Dispersion Sci. Technol. 2003;24:423. [Google Scholar]
- 95.Pelton R. Adv. Colloid Interface Sci. 2000;85:1. doi: 10.1016/s0001-8686(99)00023-8. [DOI] [PubMed] [Google Scholar]
- 96.Kawaguchi H., Fujimoto K., Mizuhara Y. Colloid Polym. Sci. 1992;270:53. [Google Scholar]
- 97.Bang J.H., Lim S.H., Park E., Suslick K.S. Langmuir. 2008;24:13168. doi: 10.1021/la802029m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kouisni L., Rochefort D. J. Appl. Polym. Sci. 2009;111:1. [Google Scholar]
- 99.W. Bodenhamer, US Patent 7, 226, 753 B2 (2007).
- 100.W. Bodenhamer, G. Jackowski, E. Davies, US Patent 6, 051, 388 (2004).
- 101.Nutiu R., Li Y.F. J. Am. Chem. Soc. 2003;125:4771. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 102.Elghanian R., Storhoff J.J., Mucic R.C., Letsinger R.L., Mirkin C.A. Science (Washington, DC) 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 103.Jovin T.M. Nat. Biotechnol. 2003;21:32. doi: 10.1038/nbt0103-32. [DOI] [PubMed] [Google Scholar]
- 104.R. Mogul, Luminescent nanosensors composed of quantum dots and fluorophores, WO Application, 083269 A2 (2006).
- 105.Bora U., Sharma P., Kannan K., Nahar P. J. Biotechnol. 2006;126:220. doi: 10.1016/j.jbiotec.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 106.Lappalainen A., Tenkanen M., Pere J. ACS Symp. Ser. 2003;864:140. [Google Scholar]
- 107.Turner M.B., Spear S.K., Holbrey J.D., Rogers R.D. Biomacromolecules. 2004;5:1379. doi: 10.1021/bm049748q. [DOI] [PubMed] [Google Scholar]
- 108.Dikmans A., Beutling U., Schmeisser E., Thiele S., Frank R. QSAR Combinatorial Sci. 2006;25:1069. [Google Scholar]
- 109.Tiller J.C., Rieseler R., Berlin P., Klemm D. Biomacromolecules. 2002;3:1021. doi: 10.1021/bm020041i. [DOI] [PubMed] [Google Scholar]
- 110.Bryjak J., Liesiene J., Stefuca V. Cellulose. 2008;15:631. [Google Scholar]
- 111.Tyagi C., Tomar L.K., Singh H. J. Appl. Polym. Sci. 2009;111:1381. [Google Scholar]
- 112.Kong D.L., Schuett W., Dai J., Kunkel S., Holtz M., Yamada R., Yu Y.T., Klinkmann H. Artificial Organs. 2002;26:200. doi: 10.1046/j.1525-1594.2002.06721.x. [DOI] [PubMed] [Google Scholar]
- 113.Reinhartz A., Alajem S., Samson A., Herzberg M. Gene. 1993;136:221. doi: 10.1016/0378-1119(93)90468-i. [DOI] [PubMed] [Google Scholar]
- 114.De Taillac L.B., Porte-Durrieu M.C., Labrugere C., Bareille R., Amedee J., Baquey C. Compos. Sci. Technol. 2004;64:827. [Google Scholar]
- 115.Goldstein L., Niv A., Yankofsky S.A. J. Chromatogr. 1990;510:23. [Google Scholar]
- 116.Wu S.C., Lia Y.K. J. Mol. Catal. B: Enzymatic. 2008;54:103. [Google Scholar]


