Highlights
-
•
Meta-transcriptomics (bulk RNA-Seq) is a powerful new way to characterise viromes.
-
•
Meta-transcriptomic data are changing our understanding of virus evolution.
-
•
Invertebrates harbor an enormous phylogenetic and genomic diversity of RNA viruses.
-
•
Present sampling schemes have only revealed a miniscule fraction of the virosphere.
-
•
The new wealth of virus genomic data presents a major challenge to classification.
Keywords: Evolution, Metagenomics, Meta-transcriptomics, Phylogeny, Classification, Virosphere
Abstract
Metagenomics is transforming the study of virus evolution, allowing the full assemblage of virus genomes within a host sample to be determined rapidly and cheaply. The genomic analysis of complete transcriptomes, so-called meta-transcriptomics, is providing a particularly rich source of data on the global diversity of RNA viruses and their evolutionary history. Herein we review some of the insights that meta-transcriptomics has provided on the fundamental patterns and processes of virus evolution, with a focus on the recent discovery of a multitude of novel invertebrate viruses. In particular, meta-transcriptomics shows that the RNA virus world is more fluid than previously realized, with relatively frequent changes in genome length and structure. As well as having a transformative impact on studies of virus evolution, meta-transcriptomics presents major new challenges for virus classification, with the greater sampling of host taxa now filling many of the gaps on virus phylogenies that were previously used to define taxonomic groups. Given that most viruses in the future will likely be characterized using metagenomics approaches, and that we have evidently only sampled a tiny fraction of the total virosphere, we suggest that proposals for virus classification pay careful attention to the wonders unearthed in this new age of virus discovery.
1. Introduction: virology in the age of metagenomics
Our knowledge of the virosphere is scant. Although viruses are the most abundant source of nucleic acid on earth, with every species of cellular life likely harboring multiple viruses, until recently most studies of virus biodiversity and evolution were of limited scope, with a strong focus on aquatic environments and prokaryotic DNA viruses (Angly et al., 2006, Culley et al., 2006, Desnues et al., 2008, Paez-Espino et al., 2016, Philosof et al., 2017). Far less is known about the diversity of RNA viruses in terrestrial organisms. This has begun to change following advances in bulk genome sequencing that have initiated a new age of virus discovery, in which it is now possible to rapidly document the entire virome of groups of host organisms (Li et al., 2015, Shi et al., 2016a). As well as greatly expanding our knowledge of virus diversity, including the ‘dark matter’ of highly divergent viruses that often elude characterization, these new data will enable us to determine the fundamental evolutionary and ecological processes that shape the virosphere, and better understand the virus-host interactions that lead to disease emergence. It is also clear that the virus diversity generated from these genomic studies will radically shake-up attempts to classify the virus world (Simmonds et al., 2017a).
One genomic technique that is already having a major impact on studies of virus diversity and evolution is RNA-Seq – a whole transcriptome shotgun sequencing approach that enables enormous amounts of RNA sequence to be generated rapidly (Palacios et al., 2008; we describe the technique in more detail below). As the transcriptome data generated by RNA-Seq is able to provide an unbiased and likely comprehensive view of all the viruses present within a host sample – that is, their complete virome – it can also be thought of as ‘meta-transcriptomics’. The data generated by meta-transcriptomics is a rich source of evolutionary and ecological information. As a case in point, meta-transcriptomic studies of invertebrates have unearthed remarkable levels of untapped virus genetic diversity, such that the virosphere is evidently far broader and more complex than previously anticipated (Li et al., 2015, Shi et al., 2016a, Webster et al., 2015). For example, an analysis of 220 species from nine invertebrate phyla identified a remarkable 1445 novel RNA viruses, as well as potentially novel genera and families (or orders) (Shi et al., 2016a). Aside from its evolutionary utility which we will discuss in more detail below, meta-transcriptomics allows the identification of novel microbial pathogens – that is, those associated with overt disease in their hosts – on clinically actionable time-scales (Wilson et al., 2014). Indeed, it is possible that with a continually declining cost meta-transcriptomics may eventually be used for routine microbiological diagnostics. A key advantage of this over other diagnostic techniques is that it has the potential to detect, in an unbiased fashion, any pathogen that produces an RNA molecule (DNA viruses, bacteria, fungi, eukaryotes), as well as the obvious case of RNA viruses. Hence, if appropriate tissues are analyzed meta-transcriptomics may provide a one stop diagnostic shop.
As much as metagenomics is transforming studies of virus evolution, it is also the case that it has shone a bright light on fundamental gaps in our understanding of the virus world. Most obviously, it is evident that we have only just begun to scratch the surface of the true diversity of viruses that make up the virosphere, and the factors that shape this diversity and evolution within ecosystems and over long-term evolutionary scales are largely unknown. Herein, we will review what, in our opinion, meta-transcriptomics has told us about virus diversity, evolution and taxonomy, and provide some suggestions for future work in this area.
2. Overview of meta-transcriptomics
Before the advent of DNA sequencing, new viruses were discovered using a variety of approaches, including filtration, cell culture, electron microscopy, and serology. Many of these techniques remain important in virology (Leland and Ginocchio, 2007). Indeed, the propagation of viruses in cells, accompanied by the visualization of virus particles by electron microscopy and the successful replication of infection in animal models, can still be considered the gold standard for virus discovery. However, the substantial time and effort required for work of this kind means that it is often impossible. In addition, most viruses are not culturable and there are not enough cell lines to meet the diversity of viruses.
More modern approaches of virus discovery involve the determination and comparison of viral nucleic acids. This combination of PCR and sequencing can be used to screen for infectious agents using degenerative primers targeting conserved genomic regions, thereby identifying novel, but related, viruses with great sensitivity. This approach has been very successful in virus discovery, with notable examples including bat influenza A virus (Tong et al., 2012) and rodent hepaciviruses (Drexler et al., 2013). However, the drawback of consensus PCR is that it is heavily dependent on currently available sequences and hence has limited capability to detect more divergent viruses. It can also be tedious to design and run consensus PCR for a large number of different virus families.
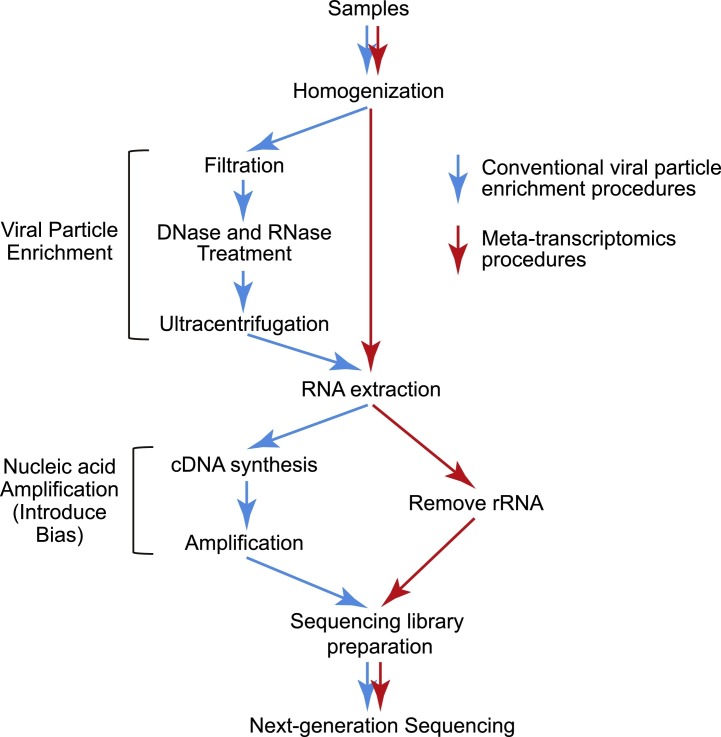
The most robust, although costly, method of virus discovery is through a coupling of metagenomics and high-throughput sequencing technology. Indeed, metagenomics provides an unbiased survey of the genetic material within a sample, and has revolutionized virus discovery in terms of speed, accuracy, sensitivity, and the amount of information generated (Firth and Lipkin, 2013). Among the various metagenomics approaches are available, meta-transcriptomics has recently come to the fore. This approach involves gathering total transcriptome information from a host sample after depletion of ribosomal (r) RNA, as this is the dominant component of the host transcriptome. Compared to metagenomics protocols that involve viral particle enrichment (reviewed in Kumar et al., 2017), this method is far simpler yet still achieves a high level of sensitivity, generality, and efficiency for virus discovery (Fig. 1 ). Previous methodologies were often based on removing as much nucleic acid outside viral particles as possible by filtering, centrifugation, lysis, and nuclease treatment, although this seldom results in a complete depletion of host RNA (Firth and Lipkin, 2013, Mokili et al., 2012). In contrast, in meta-transcriptomics total RNA (i.e. the transcriptome) is directly extracted from untreated homogenates and used for library preparation without filtering and nuclease digestion steps.
Fig. 1.
Comparisons of virus enrichment and meta-transcriptomics approaches for RNA virus discovery. The workflow of a typical virus enrichment approach is marked in blue, whereas that of a meta-transcriptomics approach is marked in red.
Another benefit of meta-transcriptomics is that it provides a ready way to quantify each virus present in a sample. Specifically, the percentage of reads that map to a particular virus genome is a good indication of how abundant any virus is, especially in the context of conserved host genes (Shi et al., 2016a, Shi et al., 2017). In turn, abundance level can provide important pointers to disease associations, whether viruses are segmented (such that genomic components have similar or different expression levels), and help identify those viruses that are in fact derived from other eukaryotic organisms present in the host sampled, such as in undigested food or prey, gut micro flora, and parasites, or simply contamination (and the greater the virus abundance, the more likely that active viral infection has occurred in the host under consideration). In addition, compared to genomic nucleic acid, the transcriptome comprises compact information that is more balanced across domains of life, thereby preventing the over-dominance of genetic information from large cellular organisms.
3. Implications of meta-transcriptomics for virus evolution
3.1. A new view of virus diversity
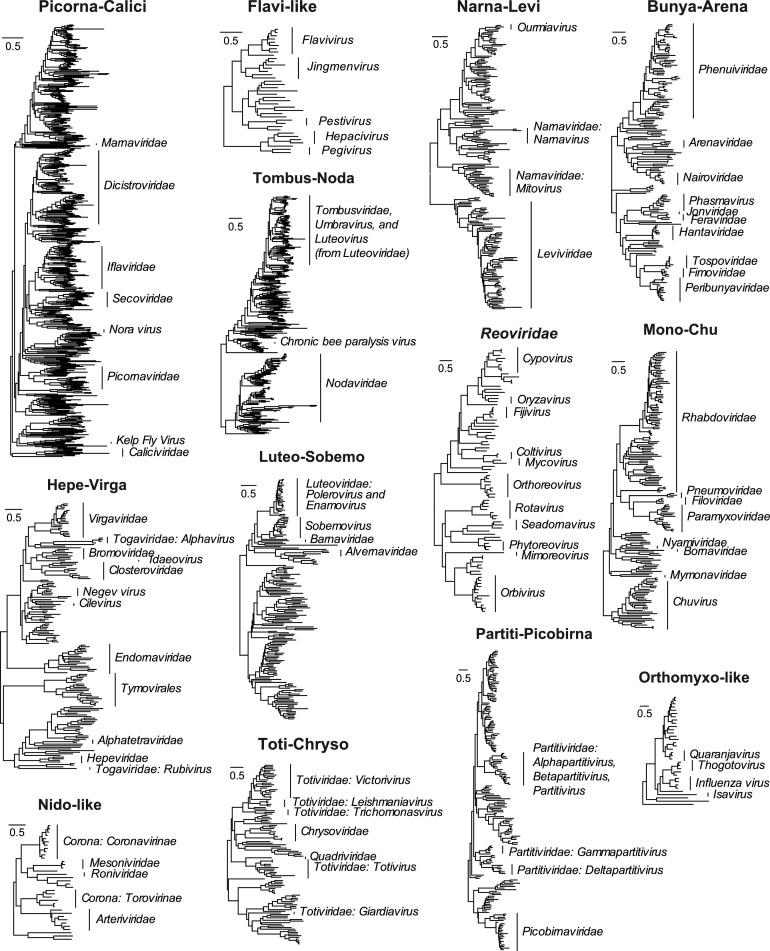
Those meta-transcriptomic studies undertaken to date have transformed our understanding of the extent and nature of viral biodiversity, making it abundantly clear that we have only sampled a tiny fraction of RNA virus biodiversity (as will also be true of DNA viruses). Indeed, it is likely that the diversity of uncharacterized viruses far exceeds that of those that have been classified to date (Fig. 2 ). These studies also highlight the inherent bias toward studying viruses that can be cultured, or associated with overt disease, which in turn reflects a longer-standing historical preference to studying viral infections in humans and economically important plants and animals. As is discussed in more detail below, it is possible that such highly biased sampling has distorted our view of virus evolution.
Fig. 2.
Current taxonomy of RNA viruses in the context of the genetic diversity revealed by meta-transcriptomics. The phylogenies are based on RdRp amino acid sequences from a broader analysis as performed by Shi et al., 2016a, Shi et al., 2016b (and see this paper for a description of branch lengths and rooting schemes). The taxonomic groups (i.e. genus, family, and order) established by ICTV are shown to the left of each phylogeny.
What is perhaps more daunting is that these studies have only been conducted in a small number of sampling locations, often in China. It is therefore simple to predict that we will identify a legion of new viruses in the near future, especially given that only a minuscule fraction of the perhaps eight million eukaryotic species (many of which are marine) have ever been sampled for viruses. Indeed, it was recently estimated that approximately 99.995% of the eukaryotic virosphere remains undiscovered or unclassified (Geoghegan and Holmes, 2017). The reality, therefore, is that our study of virus diversity and evolution, and hence taxonomy, has only just begun.
A powerful example of how meta-transcriptomics is changing our understanding of virus diversity was the discovery of chuviruses in 2015 (Li et al., 2015), that have recently and rapidly been accepted as a new family of negative-sense RNA viruses by the International Committee on Taxonomy of Viruses (ICTV). Although the chuviruses form a monophyletic group in phylogenetic trees of the RNA-dependent RNA polymerase (RdRp), they contain a diverse array of genome structures, including both segmented and unsegmented representatives, as well as a potentially circular form that would be unique among RNA viruses. It is highly likely that similarly diverse new families will be identified in the future.
There are also huge differences between the diversity revealed by previous culturing and PCR-base methods and by metagenomics, again highlighting the biases that detection method may have introduced into our understanding of natural viromes. For example, considerable effort has been directed toward isolating and culturing mosquito viruses that are relevant to humans, such as flaviviruses, alphaviruses and orthobunyaviruses. In reality, however, these disease agents represent a tiny fraction of the mosquito virome (Hall et al., 2017, Junglen and Drosten, 2013, Vasilakis and Tesh, 2015), which in fact comprises representatives from every major virus group, that are more prevalent in the mosquito population, have much higher abundance, and are often transmitted vertically (Cook et al., 2013, Vasilakis and Tesh, 2015, Shi et al., 2017).
The new wealth of diversity revealed by meta-transcriptomics also shows that the virus world is far more connected than we previously thought. New broad-scale RdRp phylogenies have shown that virus families, orders, floating genera, and undefined lineages can often be amalgamated into larger groups, such that they exhibit an evolutionary continuity (Shi et al., 2016a), in turn providing compelling evidence for their common origin (Koonin et al., 2015). It is obvious that the increasing number of newly described viruses from diverse hosts will continue to fill ‘gaps’ in phylogenetic diversity (i.e. the long branches present in inter-virus phylogenies) resulting in a more robust and stable depiction of virus evolutionary history.
3.2. Linking the vertebrate and invertebrate worlds
It is now clear that invertebrates carry a huge diversity of RNA viruses, including the potential ancestors of many those viruses found in vertebrates (Junglen and Drosten, 2013, Li et al., 2015, Marklewitz et al., 2015, Nga et al., 2011, Shi et al., 2016a, Webster et al., 2015). Given their vast diversity, abundance and often huge population sizes, it is no surprise that invertebrates harbor such a high number and diversity of RNA viruses. Although they are the most sampled group, arthropods may be especially important in this evolutionary arena because of their strong ecological relationship with both plants and vertebrates, and a phylogenetic mix between these taxa is becoming increasingly apparent (Li et al., 2015, Shi et al., 2016a). What is far less clear is how frequently this huge array of invertebrate viruses is associated with overt disease in their hosts and, if invertebrates are largely refractory to disease, how this is mediated.
The orthomyxo-like viruses provide an informative example of how the sampling of invertebrate viruses has changed our perspective on virus evolution. Prior to 2015 the orthomyxoviruses comprised a small group of vertebrate (mammal and bird) and tick-associated RNA viruses that were best known through influenza virus and classified into five genera (Allison et al., 2015, Presti et al., 2009). However, subsequent studies have revealed a remarkable diversity of orthomyxo-like viruses in invertebrates, including mosquitoes, cockroaches and earthworms, that fell both basal to, and interleaved among, the previously known genera on phylogenetic trees (Li et al., 2015). Hence, the gaps on the tree have been dramatically filled and the previous genera no long appear as phylogenetically distinct groups. In addition, that all orthomyxo-like viruses currently sampled are segmented shows that this form of genome organization is an ancient innovation in this group.
Despite the recent dramatic expansion in the number of invertebrate viruses, it is striking that some families RNA viruses remain vertebrate-specific and contain no invertebrate viruses, with the Arenaviridae, Paramyxoviridae and Picornaviridae providing important examples. Clearly, the monophyletic nature of vertebrate-specific viruses implies that have had a long-term evolutionary association with vertebrate hosts. Also, although some invertebrate viruses appear basal to vertebrate viruses, the distance between them are often substantial and phylogenetic relationships are not always stable. Therefore, while it is tempting to conclude that most, if not all, families of vertebrate viruses will have their ultimate ancestry with invertebrates, particularly as so very few of the latter have been sampled, it would be wrong to think that this a forgone conclusion.
3.3. Cross-species transmission and emergence
Determining the host range of viruses is essential to understanding the process of cross-species transmission that underpins disease emergence. Meta-transcriptomic data provide a ready means to determine what viruses are present in which hosts and allows a simple measure of virus abundance. Equally important is that the meta-transcriptomic sampling of an increasing number and diverse set of hosts has fundamentally changed the view of the host structure of major virus groups. Before the metagenomics revolution the virus diversity within a specific family was often dominated by particular host groups; so, for example, vertebrate, insect, and plant viruses often fell into distinct taxonomic groups. This has changed dramatically with meta-transcriptomics. For example, the family Totiviridae, previously thought to be largely associated with fungi, are now commonly found in metazoa. Similarly, some previously defined families of plant viruses, such as the Tombusviridae and Luteoviridae, have expanded to include viruses from arthropods, nematodes, molluscs, and protists (Shi et al., 2016a). Given such a complexity of host structure, combined with still very sparse sampling, it is dangerous to construct detailed ancestor-descendant relationships on the currently available data. For example, arthropods were initially proposed to be the ancestral hosts of bunyaviruses (Marklewitz et al., 2015), although more divergent viruses in this group have now been discovered in other invertebrates, fungi, and protists (Akopyants et al., 2016, Shi et al., 2016a).
The combination of meta-transcriptomics and phylogenetics has also told us that virus evolution is a complex interaction between cross-species transmission and virus-host co-divergence, with the evolutionary history of many virus groups reflecting an interweaving of both processes (Geoghegan et al., 2017). However, given their complexity and the often great genetic distances between virus genomes, determining the precise sequence of cross-species transmission and co-divergence events that have shaped the evolutionary history of a particular group will undoubtedly be challenging and require a denser sampling of host taxa. Indeed, the greater diversity of hosts sampled, the more cases of species jumping we are likely to document (Geoghegan et al., 2017). Although the occurrence of virus-host co-divergence has long been suggested, meta-transcriptomic-based studies indicate that this may extend even further back in time than previously suspected. For example, one interpretation of the evolutionary relationships within the Narna–Levi clade of RNA viruses is that there has been virus-host co-divergence since the α-proteobacteria became endosymbionts (Shi et al., 2016a). At the same time, however, it is clear that cross-species transmission has occurred frequently, even among phylogenetically divergent taxa, and is likely the dominant mode of RNA virus evolution.
Finally, although meta-transcriptomics has profound implications for our understanding of virus evolution, it likely undermines biodiversity-based attempts to predict the virus source of the next major disease pandemic (Olival et al., 2017). Although the bulk sequencing of potential animal reservoir species as been proposed as a way to better predict of what types of virus may emerge in human populations in the future, and where this may occur, in reality disease emergence is a nuanced process that entails a complex interaction of ecological and genetic factors (Parrish et al., 2008, Plowright et al., 2017). Meta-transcriptomics tells us that there are so many viruses in nature that trying to establish which will ultimately appear in a new host from diversity sampling alone is almost certainly a futile exercise. This is apparent in the current vogue to study bat viruses. Since the emergence of SARS coronavirus in humans – a pathogen that has its ultimate ancestry in bats – sampling bat viruses as a means to determine which next might emerge in humans has received considerable attention (Smith and Wang, 2013). While these studies have made it clear that bats indeed harbor an enormous number of viruses (Anthony et al., 2017, Luis et al., 2013, Olival et al., 2017), at the same time they clearly show that the vast majority of these viruses have not jumped to humans. The true goal of studies of disease emergence should therefore be to reveal that combination of genetic and ecological factors that underpins successful cross-species transmission and emergence.
3.4. The evolution of genome structures
One of the most important impacts of metagenomic data has been to change our understanding of the structure of virus genomes and the evolutionary processes that have given rise to them. Suffice to say, RNA virus genomes are more diverse, have more complex structures, and a wider range of lengths than previously anticipated. Although the reasons for this diversity and the birth of individual genes are uncertain, one process of undoubted importance is inter-specific recombination, including lateral gene transfer (Krupovic et al., 2012). This evidently occurs more frequently than previously anticipated, and can involve both structural and non-structural genes, with even evidence that cellular genes can be integrated into viral genomes (Shi et al., 2016a). Indeed, an emerging view is that RNA viruses experience as complex processes of genome evolution as in DNA organisms. To better determine the evolutionary processes that shape viral genome structures, and hence how new viruses are created, it is important to use the new wealth of meta-transcriptomic data to carefully determine the frequency, pattern and history of gene duplications and losses, lateral gene transfers, and genomic rearrangements; combined, these will provide a more complete picture of genome-scale evolutionary processes obtained.
Another component of RNA virus genome organization that has proven more fluid than previously envisioned is segmentation. Families of RNA viruses were generally thought to be characterized by a specific segmentation type, such as the presence/absence of segmented genomes or certain number of segments. However, segmentation no longer appears to be a strong taxon defining trait, and a combination of segmented and unsegmented genomes has now been observed within families of RNA viruses. An informative example is presented by the Flaviviridae and their relatives – the so-called ‘flavi-like’ viruses. Traditionally, flaviviruses were considered to be small (∼10 kb) unsegmented positive-sense RNA viruses that infected vertebrates; if invertebrates were involved then it was as vectors of these viruses among vertebrates, particularly mosquitoes and ticks (Simmonds et al., 2017b). Meta-transcriptomic studies have radically changed this view, including the identification of a large number of ‘insect-specific’ flaviviruses (Bolling et al., 2015, May et al., 2013, Qin et al., 2014, Shi et al., 2016b). Indeed, flavi-like viruses now appear to be a group of predominantly invertebrate RNA viruses with the potential to have very large genomes (∼26 kb) and which can be arranged in four or five segments (Ladner et al., 2016, Qin et al., 2014). Even more dramatic is that some of these flavi-like viruses appear to comprise distinct virus particles such that they are multipartite, a form of genome organization that was previously thought to be the exclusive domain of plant RNA viruses (Ladner et al., 2016).
Despite such a data revolution, one key feature of RNA virus genomes that has held firm in the metagenomics revolution is an upper-limit on genome length of <35 kb, with ball python nidovirus exhibiting the largest RNA virus genome reported to date – at 33.5 kb (Stenglein et al., 2014). Although there is still debate as to the cause of this size limit, it is tempting to think that it reflects the high rate of RNA virus evolution and the mutational burden this entails, particularly since single-stranded DNA viruses, that also mutate rapidly, similarly possess small genomes (Holmes, 2009). Of course, it is possible that the length profiles of viruses will radically change with increased sampling, and an RNA virus with the length and complexity of a large double-strand DNA virus stands represents something of a virological holy grail.
4. Implications of meta-transcriptomics for virus taxonomy
The lessons learned from evolutionary studies of meta-transcriptomic data clearly have important implications for RNA virus taxonomy and classification, and we will consider some of these here. Most obviously, that the virosphere is vast and we have only searched a tiny fraction of it leads us to believe that the ‘traditional’ way to perform virus taxonomy is dead. Given the huge number of viruses that exist in nature (Geoghegan and Holmes, 2017), it is both practically impossible and inherently pointless to isolate of all these, determine their structure, and measure their ability of replicate in cells of different types. Indeed, there is now a growing recognition that the primary way in which viruses will be characterized in the future will be through metagenomic surveys (Simmonds et al., 2017a), with complete ‘classical’ virological investigations only being performed on that subset of viruses that may be of special interest or that can be considered as markers of specific groups.
Metagenomics has already revealed the challenges facing current virus classification, with increased sampling challenging the criteria proposed to define many groups (Simmonds et al., 2017a). A key issue is that the genome structures that have been used as criteria for classification, such as segmentation and ORF arrangement, are no longer ‘conservative’ enough over broad evolutionary timescales. An informative example is provided by the Mononegavirales – an order of viruses originally characterized by unsegmented negative-sense RNA genomes and which has recently been the subject of considerable attention from the ICTV. Although use of the taxonomic term ‘Mononegavirales’ is growing in popularity, it now makes little sense in its strict literal definition as RdRp-based phylogenies show that this group contains segmented viruses, so that they no longer fulfil the criterion of possess a single (’mono’) negative-sense RNA molecule (Li et al., 2015), with genome segmentation evolving a number of times independently. Similar stories can be told for the Flaviviridae and the Totiviridae that were originally defined based on single segment but are now found to be closely related to viruses with multiple segments (Li et al., 2015, Qin et al., 2014, Sasaya et al., 2002), and the Partitiviridae and Picobirnaviridae that were thought to be bisegmented yet now include viruses containing one to six segments. These growing number of these ‘exceptions’ have often been classified as separate families or floating genera, in doing so ignoring their evolutionary relationships.
Another important limitation of the current classification system is that equivalent taxonomic groups can vary enormously in their component genetic diversity. Although this is a common problem in classification, and in large part reflects the fact that some families have a much longer evolutionary history than others, it is especially prominent in RNA viruses. The reason for such imbalance again points to the sometimes shaky criteria used for viral classification. For example, the ‘Hepe-Virga’ clade (also known as the alpha-like supergroup) are relatively closely related in RdRp phylogenies yet the ICTV divides them into one order (Tymovirales), eight families (the Virgaviridae, Togaviridae, Bromoviridae, Closteroviridae, Endoranviridae, Alphatetraviridae, Hepeviridae, and Benyviridae), and three floating genera (Negevirus, Idaeovirus, and Cilevirus). Although this clade does possess some divergent genome structures, with differences in segmentation, ORF arrangement, genome length, and even the genome sense, its RdRp diversity is no larger than that of reoviruses that are still classified as a single family thanks to a stable genome plan. In other cases these taxonomic differences appear to be largely arbitrary. For example, in the newly established order Bunyavirales (https://talk.ictvonline.org/taxonomy/), the Jonviridae, Feraviridae and Phasmaviridae are defined as separate families, although they form a single RdRp cluster whose diversity is significantly smaller than those of some individual families, such as the Phenuiviridae and the Peribunyaviridae. Although there have been clear improvements in making virus classifications more compatible with underlying phylogenetic relationships, there are notable exceptions. For example, the Togaviridae comprise two genera, Alphavirus and Rubivirus, that do not share common ancestry in phylogenies of either their replicase or structural proteins. At the very least proposals for individual taxonomic groups should be monophyletic, which is not always the case (Kuhn et al., 2013).
We also contend that it is naive to think that the structure of virus diversity in nature, and the phylogenetic analysis of this data, will necessarily produce a simple and stable classification scheme. First, the boundaries we draw to mark higher virus taxa are inherently arbitrary, rather than reflecting a hard evolutionary ‘rule’, and we should not expect nature to provide neat boundaries for classification. As noted above, the gaps apparent in many phylogenetic trees will likely be filled by newly discovered lineages as our sampling becomes more extensive. Hence, phylogenetic gaps do not necessary reflect a fundamental evolutionary process, but are likely an artefact of sparse and inadequate sampling. Indeed, from a metagenomic perspective virus species will simply be points in phylogenetic space, and viruses ‘species’ differ fundamentally from those in diploid outcrossing animals in which the term has a real biological meaning. At a lower taxonomic level, using genetic distance cut-offs to determine taxonomic differences within virus species, particularly genotypes, is also fraught with difficulties as different schemes are used in different viruses and all such rules of distance may break down if there is extensive rate variation among taxa and if our sampling is biased toward specific geographic locations.
It is also important to recall that virus gene trees are not the same as species trees, such that phylogeny-based classifications will often be only genic in nature. Because of high levels of sequence divergence it is necessarily the case that most deep (particularly inter-family) virus phylogenies are based on the analysis of RdRp alone. However, given the dynamic nature of virus genome organization, particularly the occurrence of lateral gene transfer, it is certain that in many cases the phylogeny of the RdRp will not match that of the virus genome as a whole. For example, the Luteoviridae are currently defined based on the relatedness of the structural proteins, although the replicase sequences of these viruses do not form a monophyletic group. Unfortunately, phylogenetic analyses of other genes, particularly those that encode structural proteins, often present an unsurmountable challenge for sequence-based analytical methods because of the huge sequence distances involved (Holmes, 2009, Zanotto et al., 1996). It is therefore an inconvenient truth that while phylogenies based on the RdRp can sometimes accurately depict the evolutionary history of that gene, they do not necessary reflect that of the virus as a whole. Although there are pros and cons to using either replicase or structural genes to determine phylogenetic relationships, the fact that they often give contrasting views of evolutionary history clearly complicates virus classification.
Most importantly, phylogenetic trees are only ever able to depict the relationship among those viruses that are present in the sample of viruses under study; as our sample is likely negligible, so our classification is necessarily incomplete. A more fundamental question is whether the current classification scheme can withstand the onslaught of metagenomic data? The proliferation of ‘family-like’ viruses revealed from meta-transcriptomic surveys amply highlights the scale of the challenge facing taxonomists.
5. Conclusions and future directions
As emphasized throughout this paper it is clear that we are still only scratching the surface of the virosphere and that we evidently have a great deal to learn about virus diversity and evolution. As well as revealing an abundance of new virus taxa, and determining the evolutionary processes that have shaped this diversity, it is undoubtedly the case that viruses exist in hosts that have not been screened for RNA viruses or that are so divergent in sequence that they cannot readily be detected by standard homology-based methods (such as Blast) or included in phylogenetic analyses. If the nature of this dark matter can be resolved it will surely shed new light on the ultimate origins of viruses as well as their deep phylogenetic relationships. The situation is particularly acute in the case of the archaea in which only a single putative RNA virus has been described to date (Bolduc et al., 2012), and which in large part may reflect our current inability to identify viruses that possess highly divergent genome sequences. It is therefore of critical importance to perform unbiased metagenomics surveys of prokaryotic taxa that have not been examined to date, followed by novel bioinformatics analyses that are able to accurately identify viruses and reveal their phylogenetic relationships. This will entail the characterization of the unknown biodiversity of RNA viruses in prokaryotes and basal eukaryotes and, in parallel, developing and utilizing new computational tools to robustly extract sequence information from highly divergent genome sequences. Similarly, the increasingly frequent detection of recombination and lateral gene transfer also poses a major challenge to current phylogenetic protocols and may require a new computational tool-kit (Iranzo et al., 2016, Koonin and Dolja, 2014).
Our knowledge of the evolutionary processes that have generated the diversity of the virosphere has been strongly skewed by a focus on those viruses that act as agents of disease in economically important animals and plants and those that can be easily cultured. Importantly, recent work has shown that animals harbor enormous uncharacterized viral diversity, only some of which has been associated with disease. However, these viruses still only reflect a tiny proportion of those in nature and therefore provide an incomplete picture of the major processes of virus ecology and evolution. Key questions for future research that can be addressed with the new wealth of meta-transcriptomic data include (i) determining the flow of viruses between host taxa and the processes that shape virus ecosystems; (ii) revealing the mechanisms of long-term virus macroevolution, particularly lineage birth and death, and (iii) revealing the mechanisms and evolutionary processes that structure viral genomes. Rather than simply surveying biodiversity and classifying, the goal for the future should be to perform more ecology-focused studies to reveal fundamental patterns and processes. It is critical that studies of virus diversity evolution shape our attempts to classify these infectious agents, rather than classification schemes guiding how we think that viruses have evolved. Finally, we contend that is perhaps premature to construct inflexible and overly hierarchical classification schemes for RNA viruses when we have clearly sampled so little of what is there in nature. The new age of virus discovery will undoubtedly provide many new challenges for the science of virus classification.
Conflict of interests
The authors declare that no competing interests exist.
Acknowledgements
This study was supported by the Special National Project on Research and Development of Key Biosafety Technologies (2016YFC1201900), the 12th Five-Year Major National Science and Technology Projects of China (2014ZX10004001-005), and National Natural Science Foundation of China (Grants 81290343, 81672057). ECH is funded by an NHMRC Australia Fellowship (GNT1037231).
References
- Akopyants N.S., Lye L.F., Dobson D.E., Lukeš J., Beverley S.M. A novel bunyavirus-like virus of trypanosomatid protist parasites. Genome Announc. 2016;4:e00715–16. doi: 10.1128/genomeA.00715-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison A.B., Ballard J.R., Tesh R.B., Brown J.D., Ruder M.G., Keel M.K., Munk B.A., Mickley R.M., Travassos da Rosa A.P.A., Ellis J.C., Ip H., Shern-Bochsler V.I., Rodgers M.B., Ghedin E., Holmes E.C., Parrish C.R., Dwyer C. Cyclic avian mass mortality in the northeastern United States is associated with a novel orthomyxovirus. J. Virol. 2015;89:1389–1403. doi: 10.1128/JVI.02019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly F.E., Felts B., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Mahaffy J.M., Mueller J.E., Nulton J., Olson R., Parsons R., Rayhawk S., Suttle C.A., Rohwer F. The marine virospheres of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Johnson C.K., Greig D.J., Kramer S., Che X., Wells H., Hicks A.L., Joly D.O., Wolfe N.D., Daszak P., Karesh W., Lipkin W.I., Morse S.S., PREDICT Consortium Mazet J.A.K., Goldstein T. Global patterns in coronavirus diversity. Virus Evol. 2017;3 doi: 10.1093/ve/vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc B., Shaughnessy D.P., Wolf Y.I., Koonin E.V., Roberto F.F., Young M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J. Virol. 2012;86:5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling B.G., Weaver S.C., Tesh R.B., Vasilakis N. Insect-specific virus discovery: significance for the arbovirus community. Viruses. 2015;7:4911–4928. doi: 10.3390/v7092851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S., Chung B.Y., Bass D., Moureau G., Tang S., McAlister E., Culverwell C.L., Glücksman E., Wang H., Brown T.D., Gould E.A., Harbach R.E., de Lamballerie X., Firth A.E. Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts. PLoS One. 2013;18:e80720. doi: 10.1371/journal.pone.0080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A.I., Lang A.S., Suttle C.A. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Desnues C., Rodriguez-Brito B., Rayhawk S., Kelley S., Tran T., Haynes M., Liu H., Furlan M., Wegley L., Chau B., Ruan Y., Hall D., Angly F.E., Edwards R.A., Li L., Thurber R.V., Reid R.P., Siefert J., Souza V., Valentine D.L., Swan B.K., Breitbart M., Rohwer F. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Muller M.A., Lukashev A.N., Gmyl A., Coutard B., Adam A., Ritz D., Leijten L.M., van Riel D., Kallies R., Klose S.M., Gloza-Rausch F., Binger T., Annan A., Adu-Sarkodie Y., Oppong S., Bourgarel M., Rupp D., Hoffmann B., Schlegel M., Kummerer B.M., Kruger D.H., Schmidt-Chanasit J., Setien A.A., Cottontail V.M., Hemachudha T., Wacharapluesadee S., Osterrieder K., Bartenschlager R., Matthee S., Beer M., Kuiken T., Reusken C., Leroy E.M., Ulrich R.G., Drosten C. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013;9:e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth C., Lipkin W.I. The genomics of emerging pathogens. Annu. Rev. Genom. Hum. Genet. 2013;14:281–300. doi: 10.1146/annurev-genom-091212-153446. [DOI] [PubMed] [Google Scholar]
- Geoghegan J.L., Holmes E.C. Predicting virus emergence amidst evolutionary noise. Open Biol. 2017;7 doi: 10.1098/rsob.170189. 170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J.L., Duchêne S., Holmes E.C. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 2017;13:e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Bielefeldt-Ohmann H., McLean B.J., O'Brien C.A., Colmant A.M., Piyasena T.B., Harrison J.J., Newton N.D., Barnard R.T., Prow N.A., Deerain J.M., Mah M.G., Hobson-Peters J. Commensal viruses of mosquitoes: host restriction, transmission, and interaction with arboviral pathogens. Evol. Bioinform. Online. 2017;12:35–44. doi: 10.4137/EBO.S40740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C. In: The Evolution and Emergence of RNA Viruses. Oxford Series in Ecology and Evolution (OSEE) Harvey P.H., May R.M., editors. Oxford University Press; Oxford: 2009. [Google Scholar]
- Iranzo J., Krupovic M., Koonin E.V. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio. 2016;7:e00978–16. doi: 10.1128/mBio.00978-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junglen S., Drosten C. Virus discovery and recent insights into virus diversity in arthropods. Curr. Opin. Microbiol. 2013;16:507–513. doi: 10.1016/j.mib.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. 2014;78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V., Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Dolja V.V., Koonin E.V. Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol. Direct. 2012;10:12. doi: 10.1186/s13062-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Bekal S., Caì Y., Clawson A.N., Domier L.L., Herrel M., Jahrling P.B., Kondo H., Lambert K.N., Mihindukulasuriya K.A., Nowotny N., Radoshitzky S.R., Schneider U., Staeheli P., Suzuki N., Tesh R.B., Wang D., Wang L.F., Dietzgen R.G. Nyamiviridae: proposal for a new family in the order Mononegavirales. Arch. Virol. 2013;158:1621–1629. doi: 10.1007/s00705-013-1674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Murthy S., Kapoor A. Evolution of selective-sequencing approaches for virus discovery and virome analysis. Virus Res. 2017;239:172–179. doi: 10.1016/j.virusres.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Wiley M.R., Beitzel B., Auguste A.J., Dupuis A.P.2nd Lindquist M.E., Sibley S.D., Kota K.P., Fetterer D., Eastwood G., Kimmel D., Prieto K., Guzman H., Aliota M.T., Reyes D., Brueggemann E.E., St John L., Hyeroba D., Lauck M., Friedrich T.C., O'Connor D.H., Gestole M.C., Cazares L.H., Popov V.L., Castro-Llanos F., Kochel T.J., Kenny T., White B., Ward M.D., Loaiza J.R., Goldberg T.L., Weaver S.C., Kramer L.D., Tesh R.B., Palacios G. A multicomponent animal virus isolated from mosquitoes. Cell Host Microbe. 2016;20:357–367. doi: 10.1016/j.chom.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland D.S., Ginocchio C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007;20:49–59. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-X., Shi M., Tian J.-H., Lin X.-D., Kang Y.-J., Qin X.-C., Chen L.-J., Xu J., Holmes E.C., Zhang Y.-Z. Unprecedented RNA virus diversity in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis A.D., Hayman D.T., O'Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R., Mills J.N., Timonin M.E., Willis C.K., Cunningham A.A., Fooks A.R., Rupprecht C.E., Wood J.L., Webb C.T. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. Biol. Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklewitz M., Zirkel F., Kurth A., Drosten C., Junglen S. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7536–7541. doi: 10.1073/pnas.1502036112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May F.J., Clark D.C., Pham K., Diviney S.M., Williams D.T., Field E.J., Kuno G., Chang G.J., Cheah W.Y., Setoh Y.X., Prow N.A., Hobson-Peters J., Hall R.A. Genetic divergence among members of the Kokobera group of flaviviruses supports their separation into distinct species. J. Gen. Virol. 2013;94:1462–1467. doi: 10.1099/vir.0.049940-0. [DOI] [PubMed] [Google Scholar]
- Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., Parquet Mdel C., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N.T., Inoue S., Ito T., Okamoto K., Ichinose A., Snijder E.J., Morita K., Gorbalenya A.E. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7:e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C. Uncovering earth's virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- Palacios G., Druce J., Du L., Tran T., Birch C., Briese T., Conlan S., Quan P.L., Hui J., Marshall J., Simons J.F., Egholm M., Paddock C.D., Shieh W.J., Goldsmith C.S., Zaki S.R., Catton M., Lipkin W.I. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- Parrish C.R., Holmes E.C., Morens D.M., Park E.-C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-species viral transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philosof A., Yutin N., Flores-Uribe J., Sharon I., Koonin E.V., Béjà O. Novel abundant oceanic viruses of uncultured marine group II euryarchaeota. Curr. Biol. 2017;27:1362–1368. doi: 10.1016/j.cub.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright R.K., Parrish C.R., McCallum H., Hudson P.J., Ko A.I., Graham A.L., Lloyd-Smith J.O. Pathways to zoonotic spillover. Nat. Rev. Micro. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presti R.M., Zhao G., Beatty W.L., Mihindukulasuriya K.A., da Rosa A.P., Popov V.L., Tesh R.B., Virgin H.W., Wang D. Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J. Virol. 2009;83:11599–11606. doi: 10.1128/JVI.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.-C., Shi M., Tian J.-H., Lin X.-D., Gao D.-Y., He J.-R., Wang J.-B., Li C.-X., Kang Y.-J., Yu B., Zhou D.-J., Xu J., Plyusnin A., Holmes E.C., Zhang Y.-Z. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6744–6749. doi: 10.1073/pnas.1324194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaya T., Ishikawa K., Koganezawa H. The nucleotide sequence of RNA1 of Lettuce big-vein virus, genus Varicosavirus, reveals its relation to nonsegmented negative-strand RNA viruses. Virology. 2002;297:289–297. doi: 10.1006/viro.2002.1420. [DOI] [PubMed] [Google Scholar]
- Shi M., Lin X.-D., Tian J.-H., Chen L.-J., Chen X., Li C.-X., Qin X.-C., Li J., Cao J.-P., Eden J.-S., Buchmann J.P., Wang W., Xu J., Holmes E.C., Zhang Y.-Z. Redefining the invertebrate virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- Shi M., Lin X.-D., Vasilakis N., Tian J.-H., Li C.-X., Chen L.-J., Eastwood J., Diao X.-N., Chen M.-H., Chen X., Qin X.-X., Widen S.G., Wood T.G., Tesh R.B., Xu J., Holmes E.C., Zhang Y.-Z. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J. Virol. 2016;90:659–669. doi: 10.1128/JVI.02036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Neville P., Nicholson J., Eden J.-S., Imrie A., Holmes E.C. High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J. Virol. 2017;91:e00680–17. doi: 10.1128/JVI.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Adams M.J., Benkő M., Breitbart M., Brister J.R., Carstens E.B., Davison A.J., Delwart E., Gorbalenya A.E., Harrach B., Hull R., King A.M., Koonin E.V., Krupovic M., Kuhn J.H., Lefkowitz E.J., Nibert M.L., Orton R., Roossinck M.J., Sabanadzovic S., Sullivan M.B., Suttle C.A., Tesh R.B., van der Vlugt R.A., Varsani A., Zerbini F.M. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Becher P., Bukh J., Gould E.A., Meyers G., Monath T., Muerhoff S., Pletnev A., Rico-Hesse R., Smith D.B., Stapleton J.T., ICTV Report Consortium ICTV virus taxonomy profile: Flaviviridae. J. Gen. Virol. 2017;98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I., Wang L.F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013;3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein M.D., Jacobson E.R., Wozniak E.J., Wellehan J.F., Kincaid A., Gordon M., Porter B.F., Baumgartner W., Stahl S., Kelley K., Towner J.S., DeRisi J.L. Ball python nidovirus: a candidate etiologic agent for severe respiratory disease in Python regius. mBio. 2014;5:e01484–14. doi: 10.1128/mBio.01484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Li Y., Rivailler P., Conrardy C., Castillo D.A., Chen L.M., Recuenco S., Ellison J.A., Davis C.T., York I.A., Turmelle A.S., Moran D., Rogers S., Shi M., Tao Y., Weil M.R., Tang K., Rowe L.A., Sammons S., Xu X., Frace M., Lindblade K.A., Cox N.J., Anderson L.J., Rupprecht C.E., Donis R.O. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Tesh R.B. Insect-specific viruses and their potential impact on arbovirus transmission. Curr. Opin. Virol. 2015;15:69–74. doi: 10.1016/j.coviro.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C.L., Waldron F.M., Robertson S., Crowson D., Ferrari G., Quintana J.F., Brouqui J.M., Bayne E.H., Longdon B., Buck A.H., Lazzaro B.P., Akorli J., Haddrill P.R., Obbard D.J. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 2015;13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.R., Naccache S.N., Samayoa E., Biagtan M., Bashir H., Yu G., Salamat S.M., Somasekar S., Federman S., Miller S., Sokolic R., Garabedian E., Candotti F., Buckley R.H., Reed K.D., Meyer T.L., Seroogy C.M., Galloway R., Henderson S.L., Gern J.E., DeRisi J.L., Chiu C.Y. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotto P.M. de A., Gibbs M.J., Gould E.A., Holmes E.C. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 1996;70:6083–6096. doi: 10.1128/jvi.70.9.6083-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]