Abstract
Human rhinovirus (HRV) infections are common but poorly characterized in university students. Thus, we characterized asymptomatic and symptomatic HRV infections by incidence, species diversity, and viral load of 502 university students during September and October of 2010 and 2011 from nasal swabs and electronically submitted symptom questionnaires. We tested all symptomatic students and randomly sampled participants who remained asymptomatic (n = 25/week, over 8 weeks each study year) on a weekly basis by real-time PCR and sequenced HRV positives. HRV was identified in 33/400 (8.3%) and 85/92 (92.4%) of the asymptomatic and symptomatic students, respectively. We identified a higher than previously reported rate of HRV-B in both groups, although the distribution of HRV species was similar (P = 0.37). Asymptomatic viral load averaged 1.2 log10 copies/mL lower than symptomatic HRV (P < 0.001). In conclusion, asymptomatic HRV activity preceded peak symptomatic activity in September and October and was associated with lower viral load.
Keywords: Rhinovirus, Viral load, Asymptomatic, Quantitative PCR, Species diversity
Highlights
-
•
We characterized asymptomatic and symptomatic HRVs in an adult population.
-
•
We examine HRV incidence, species diversity, and viral load.
-
•
Asymptomatic HRV was associated with lower viral load than symptomatic HRV.
-
•
Asymptomatic HRV activity preceded peak symptomatic activity.
-
•
HRV-A and HRV-B are common in both populations.
1. Background
Human rhinoviruses (HRVs) account for the majority of upper respiratory tract infections (URTI) in all age groups (Mackay, 2008). Although recent advances in molecular diagnostic tools have improved our understanding of the impact of HRV infections, they have also revealed confounding factors such as the detection of multiple pathogens and the significance of detecting virus in asymptomatic individuals. As a result, the burden of HRV infections may be overestimated in certain populations (Kieninger et al., 2013).
Few studies have investigated asymptomatic HRV infections prior to the implementation of PCR and other molecular diagnostic techniques. Surveillance of 15 children ages 1–9 years over a 12-month period identified 20% of children with asymptomatic HRV infections over 1 year (Peltola et al., 2008, Brownlee and Turner, 2008, Greenberg, 2011). In children 3 months to 15 years old, incidence of symptomatic HRV was 6 infections per year (Winther et al., 2006). The incidence of asymptomatic HRV has not been reported in adult populations; however, the incidence of symptomatic HRV is estimated to be 2–3 infections per person per year (Madigan et al., 2003). Rates of asymptomatic HRV have varied in the literature, and direct comparisons are difficult because of differences in the study population (most populations observed are children), in the definition of an asymptomatic episode, and in the detection method utilized (Jartti et al., 2004, Johnston et al., 1993, Kusel et al., 2006, Mackay, 2008, Nokoso-Koivisto et al., 2002, Peltola et al., 2008, van Benten et al., 2003, Winther et al., 2006). We sought to detect and characterize asymptomatic and symptomatic HRVs in a cohort of university students enrolled in a clinical trial of vitamin D.
2. Study design
2.1. Study population
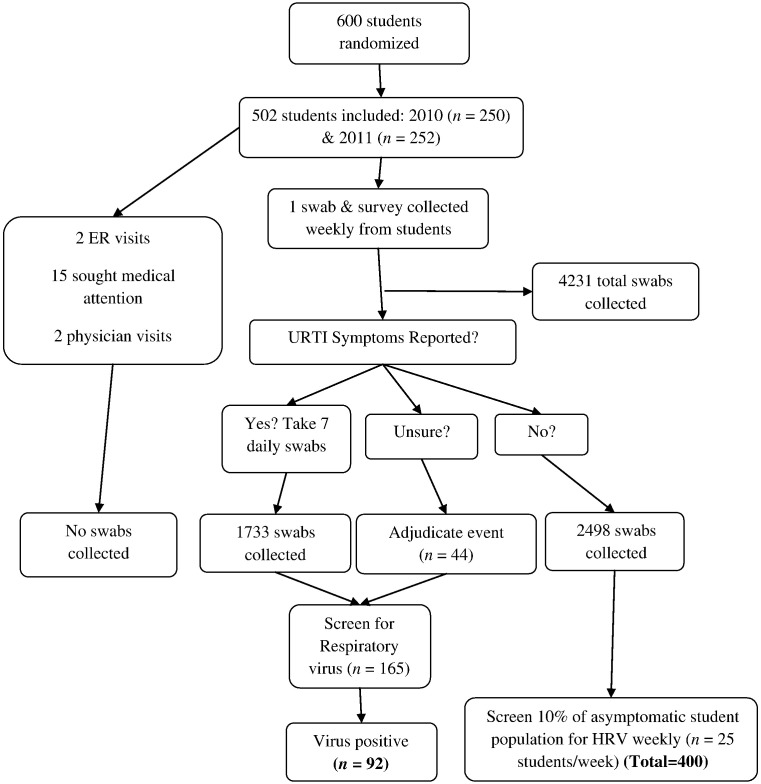
Of 600 McMaster University undergraduate students participating in a randomized controlled trial of vitamin D and gargling (McFlu2 COLD3), 471 (78.5%) completed all weekly surveys discussing URTI symptoms, 86 (14.3%) completed at least 1 but not all, and 43 (7.2%) completed none (Goodall et al., 2014). The median age of participants was 19 years (interquartile range 18–20 years); 64% were female (Goodall et al., 2014). Participants who submitted at least 1 self-collected nasal swab were included in the current laboratory-based study (n = 502). Participants were followed from weeks 36–43 of 2010 (n = 250) or weeks 36–43 of 2011 (n = 252). Study participants self-collected a mid-turbinate flocked nasal swab (FLOQSwabs; Copan Italia, Brescia, Italy) in CyMol™ transport media (Copan Italia) once weekly – regardless of symptomatic status – for a total of 8 weeks (Luinstra et al., 2011, Smieja et al., 2010) (Fig. 1 ). This study was approved by the Hamilton Health Sciences/Faculty of Health Sciences Research Ethics Board, McMaster University, Hamilton, ON, Canada, and all subjects gave written informed consent.
Fig. 1.
Flow charts of study design during the McFlu2 COLD3 Prevention Trial, Hamilton, ON, Canada; weeks 36–43 2010 and weeks 36–43 2011.
2.2. Sample collection
At the time of randomization, study staff oriented each participant with the components of the nasal swab kit and instructed them on the appropriate procedure to self-administer the nasal swab. Participants also received a step-by-step guide describing the procedure as well as a link to an online instructional video developed for the study for a reference at home. There were also weekly visits between the study staff and participants, which provided further opportunities to answer outstanding questions. The FLOQSwab design (Copan Italia) only permits mid-turbinate sample collection; nasopharyngeal specimens were therefore not collected.
2.3. Outcome measures
An episode of symptomatic URTI was determined based on 2 criteria: the participant's report of a “cold” together with 2 or more URTI symptoms (i.e., runny/stuffy nose, congestion, cough, sneeze, sore throat, muscle aches, or fever) (Barret et al., 2009, Li-Ng et al., 2009, Lizogub et al., 2007, Predy et al., 2005, Schulten et al., 2001). Uncertain cases where symptoms were reported and no additional information was provided that attributed the symptoms to another cause were adjudicated by 2 infectious disease clinicians (Goodall et al., 2014). Symptom severity and duration were measured using the 21-item Wisconsin Upper Respiratory Symptom Survey (WURSS-21) (Barret et al., 2009). Fig. 1 identifies the population size and specimens tested for each population. All day 1 nasal swabs from each unique symptomatic event were assessed for viral etiology.
To screen for asymptomatic HRV, we randomly sampled 10% of the nasal swabs collected from participants who never reported a URTI during the entire 8-week study period. Samples were stratified by week (25/week) for a total of 400 swabs (n = 200 in each of 2010 and 2011). A 10% sampling was selected to ensure adequate precision of estimated infection incidence rates. Samples selected for asymptomatic screens were also adjudicated to confirm asymptomatic status.
2.4. Detection of HRV infection and viral load
FLOQ swabs (Copan Italia) placed in 2 mL of CyMol™ transport media (Copan Italia) were extracted using the easyMAG automated extractor according to the manufacturer's instruction (bioMeriéux, Montreal, QC, Canada). Twenty microliters of MS2 bacteriophage was added to each sample prior to extraction as an internal extraction control. Clinical specimens were tested for HRV RNA by reverse transcriptase polymerase chain reaction (RT-PCR) amplification of a 400-bp region of the HRV 5′ untranslated region with an HRV-specific assay (Granados et al., 2012, Kiang et al., 2008); samples were also screened for other respiratory viruses with the xTAG™ RVP assay (Luminex Molecular Diagnostics, Toronto, ON, Canada). Viral load was determined by a previously described assay (Granados et al., 2012). Samples were tested in triplicate; the average of each sample is used in further analysis.
2.5. Identification of HRV species
Presumptive identification of HRV species was conducted by sequencing a portion of the VP1 gene (Lu and Erdman, 2007). Five microliters of amplified nucleic acid was sequenced on the ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) (Mobix, Hamilton, ON, Canada). Genotypic designations were assigned according to threshold divergences of 13%, 12%, and 13% for HRV-A, HRV-B, and HRV-C, respectively (McIntyre et al., 2013, Picornavirus Study Group, 2013).
2.6. Statistical analysis
Group comparisons were performed using the χ2 test for categorical variables and the Student's t test for continuous variables. Multivariable linear regression was used to determine if an association exists between viral load and HRV species/symptomatic status. Results were considered statistically significant with a P < 0.05. All statistical analyses were conducted using SPSS version 20 (IBM, Chicago, IL, USA).
3. Results
3.1. Incidence of HRV in asymptomatic and symptomatic students
Throughout the study, we tested self-collected nasal swabs from 25 randomly selected asymptomatic students per week, equaling a total of 400 asymptomatic students screened for HRV by RT-PCR. HRV was identified in 33/400 (8.3%) of swabs, enterovirus D68 in 4/400 (1.0%), and coxsackievirus A6 in 1/400 (0.25%). By comparison, there were 165 reported symptomatic events (121 self-reported, 44 adjudicated), for which a viral pathogen was identified in 92 (55.8%) events. HRV was the predominant pathogen identified (85/92; 92.4%), followed by enterovirus D68 (7/92; 7.6%), and coronavirus NL63 (1/92; 1.1%).
We compared the distribution of HRV infections in the participants. Peak asymptomatic infections occurred in weeks 37 and 38 in 2010 and 2011, respectively (Fig. 2 ). Peak symptomatic infections occurred a week after. The incidence rates of asymptomatic and symptomatic presentation were calculated as 8.3%/week and 1.9%/week, respectively. These rates were determined by inferring that the same proportion of events occurred in the entire asymptomatic population. The start of an asymptomatic infection was determined by testing the sample given the week before.
Fig. 2.
Weekly positivity rate of asymptomatic and symptomatic HRV in weeks 36–43 in 2010 and 2011. The percent positivity of HRV infections by week in 2010 (a) and 2011 (b) in university students. The percentage of positive asymptomatic HRV each week is determined by the number of HRVs identified in the 10% weekly random sampling (n = 25/week in 2010 and n = 25/week in 2011). The percentage of positive symptomatic HRV was determined by the number of positive HRV identified in the student population each week (n = 250/week in 2010 and n = 252/week in 2011).
3.2. Comparison of HRV species in asymptomatic and symptomatic students
A portion of the VP1 gene of HRV positives was sequenced to determine species. Of the 118 HRV-positive specimens (33 asymptomatic and 85 symptomatic), 113 (95.7%) were sequenced, and 5 had unresolved sequences (4.2%). HRV-A and HRV-B were the most common species among the participants in both the asymptomatic and symptomatic students (Table 1 ). There was no significant difference in HRV-C distribution (P = 0.37).
Table 1.
Distribution of HRV-A, HRV-B, and HRV-C in asymptomatic and symptomatic students.a
| A (%) | B (%)b | C (%)c | Total | |
|---|---|---|---|---|
| Asymptomatic | 15 (48.4) | 14 (45.1) | 2 (6.5) | 31 |
| Symptomatic | 39 (47.6) | 34 (41.4) | 9 (12.3) | 82 |
HRV species determined by partial sequencing of the HRV VP1 gene.
Comparison of HRV-B frequency to HRV-A and HRV-C, χ2 = 0.064, P = 0.80.
Comparison of HRV-C frequency to HRV-A and HRV-B, χ2 = 0.79, P = 0.37.
3.3. Comparison of HRV viral loads in asymptomatic and symptomatic students
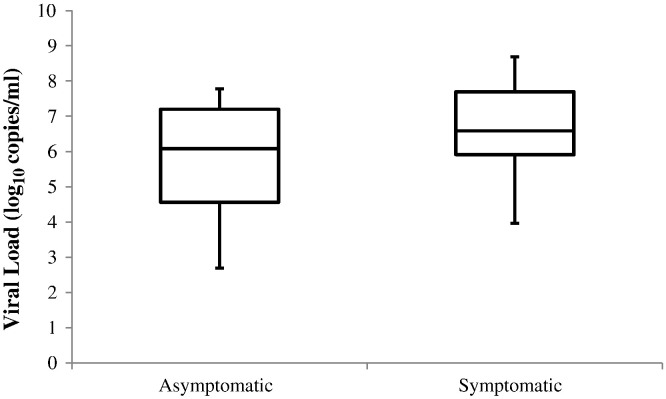
We sought to determine whether a difference in viral load could account for the presence or absence of symptoms. The mean viral load (±SD) of HRV in 33 asymptomatic students was 5.3 (±1.5) log10 copies/mL versus 6.4 (±1.3) log10 copies/mL in 85 symptomatic students. The mean difference was 1.2 log10 copies/mL (95% confidence interval [CI] 0.59–1.72; P < 0.001) (Fig. 3 ). Viral load was higher in the presence of symptoms (β = 0.92; P = 0.004), adjusting for HRV species. However, HRV species was not associated with increased viral load (β = −0.39; P = 0.37).
Fig. 3.
Box and whiskers plot of HRV viral loads (log10 copies/mL) in asymptomatic (n = 33) and symptomatic (n = 85) university students. The solid line represents the median; the bottom and top of the box represent 25th and 75th percentiles, and the whiskers represent the minimum and maximum data points, respectively. Mean difference in viral load was 1.2 log10 copies/mL (95% CI 0.59–1.72; P < 0.001).
3.4. Duration of infection in asymptomatic and symptomatic students
We investigated the duration of asymptomatic HRV episodes by testing swabs provided by HRV-positive asymptomatic students a week before and a week after the PCR positive was first identified. Of the 33 asymptomatic individuals, 10 (30.3%) were positive 1 week later, which indicates a period of at least 8 days of infection. We sequenced a portion of the VP1 gene of HRV in those individuals on days 1 and 8 to determine if the same genotype or a novel genotype was present on day 8. We identified 4/10 (40.0%) with the same genotype, and 6/10 (60.0%) were infected with a new genotype. An electronic symptom diary was sent to the 106 students who reported a symptomatic URTI, of whom 69 (65.1%) completed the diary in full. From the students who completed the diary, the mean duration of symptoms (±SD) was 6.1 (±1.1) days.
4. Discussion
In this study, we characterized asymptomatic and symptomatic HRV infections in 2 cohorts of university students. HRV incidence rates were calculated to be 8.3%/week and 1.9%/week for asymptomatic and symptomatic students, respectively. To our knowledge, our study is the first to determine the incidence of asymptomatic HRV infection in an adult population. In cases of symptomatic HRV, incidence has been documented at 1.6 colds/9 months per student (Brownlee and Turner, 2008, Picornavirus Study Group, 2013). The weekly distributions of HRV illustrated that asymptomatic HRV peaked just prior to the symptomatic infections. This suggests that there is a relationship between the incidence of asymptomatic infections and the spread of symptomatic illness in the community.
Partial sequencing of the VP1 gene determined that the distribution of HRV species was similar in both populations, indicating that species may not account for the presence or absence of symptoms. A larger study would be necessary to confirm this finding as we only identified 11 episodes of HRV-C (2 asymptomatic and 9 symptomatic) in our student cohorts. Interestingly, when we investigated a cohort of hospitalized children with respiratory illness during the same time frame, we identified a higher proportion of HRV-C (Granados, 2012). The lower number of HRV-Cs identified in our student cohort may indicate that HRV-C infections are more common in children than in healthy adults, as previously suggested by Bochkov and Gern (2012). In our study, HRV-B was identified at a higher frequency than has been previously reported (Watanabe et al., 2010, Xiang et al., 2010). In 271 adults presenting with URTI, 25% were infected with HRV-B and HRV-C and had a lower percentage of URTI symptoms compared to HRV-A (χ2 = 7.18; P < 0.05) (Xiang et al., 2010). This indicates that HRV-B may be associated with mild or asymptomatic infection in adults.
We found that the viral load in asymptomatic students was an average of 1.2 log10 copies/mL (P < 0.001) lower than in symptomatic students as has been documented by Jansen et al. (2011) in asymptomatic children. Viral loads of 4.5–5.0 log10 copies/mL or greater have been previously observed in symptomatic children ages 1–6 infected with HRV (Gerna et al., 2009, Jansen et al., 2011). In our study, we could not conclusively distinguish whether a person was asymptomatic or symptomatic by viral load alone. There are likely differences in the level of acquired immunity in different age groups, and research involving HRV viral loads in previously exposed individuals is required (Peltola et al., 2008).
Our study design allowed us to crudely estimate the duration of virus shedding in asymptomatic students by examining weekly swabs; however, we were unable to accurately identify the true start of an infection. In 4 individuals, the same HRV episode was identified 8 days later, consistent with reports stating that HRV infections do not persist beyond day 11 (Greenberg, 2011, Johnston et al., 1993). This also agrees with our previous study of symptomatic university students, who self-sampled 7 times during the first 14 days of their illness. In that study, we found that HRV RNA persisted in a symptomatic individual for 7–12 days with no change in genotype (Granados et al., 2012). Our current observations indicate that asymptomatic HRV RNA was present for a similar length of time.
There are limitations to our study design that need to be taken into account. In 73 symptomatic events, a viral etiology could not be identified; this could be accounted for by symptoms confused for allergic events or a viral pathogen that could not be detected by the xTAG™ RVP assay. Our study period only looked at September and October when HRV is most frequently detected in North America. To accurately estimate the annual rate of asymptomatic HRV incidence, surveillance throughout the year would need to be conducted. We utilized an absolute quantitation method to determine the amount of virus. Differences in methodology and imprecision among any quantitative polymerase chain reaction (qPCR) assay make direct comparisons between reports difficult to conduct unless digital qPCR is conducted. While previous studies have shown that self-sampling with flocked nasal swabs has consistent quality, our viral load results should not be directly compared with previous studies (Smieja et al., 2010). The identification of symptoms is subjective, and there is a risk that some students had such minimal symptoms that they did not perceive and report any, whereas others may have low tolerance and would report symptoms. In order to minimize reporting bias, all participants were required to submit self-collected nasal swabs weekly regardless of symptom status as it became a weekly activity. Additionally, during the weekly visits, study staff could confirm that any symptoms were properly recorded.
In summary, we have shown that HRV infections are common in university students, with a similar distribution of species regardless of symptomatic status. HRV-B was commonly identified, and viral load, regardless of species, was lower in asymptomatic students than in those with symptoms. This stresses the importance of taking preventative measures such as handwashing in order to limit the transmission of HRV from person to person and highlights the need for studies investigating the factors, which lead from asymptomatic to symptomatic infections.
References
- Barret B., Brown R., Mundt M., Thomas G., Barlow S., Highstrom A. Validation of a short Wisconsin Upper Respiratory Symptom Survey (WURSS-21) Health Qual Life Outcomes. 2009;7:76. doi: 10.1186/1477-7525-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov Y., Gern J. Clinical and molecular features of human rhinovirus C. Microbes Infect. 2012;14:485–494. doi: 10.1016/j.micinf.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee J., Turner R. New developments in the epidemiology and clinical spectrum of rhinovirus infections. Curr Opin Pediatr. 2008;20:67–71. doi: 10.1097/MOP.0b013e3282f41cb6. [DOI] [PubMed] [Google Scholar]
- Gerna G., Piralla A., Rovida F., Rognoni V., Marchi A., Locatelli F. Correlation of rhinovirus load in the respiratory tract and clinical symptoms in hospitalized clinical symptoms in hospitalized immunocompetent and immunocompromised patients. J Med Virol. 2009;81:1498–1507. doi: 10.1002/jmv.21548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall E., Granados A., Luinstra K., Pullenayegum E., Coleman B., Loeb M. Vitamin D3 and gargling for the prevention of upper respiratory tract infections: a randomized controlled trial. BMC Infect Dis. 2014;14:273. doi: 10.1186/1471-2334-14-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados A. 2012. Aspects of human rhinovirus infection in hospitalized and non-hospitalized individuals. (Master of Science Thesis) [Retrieved from Open Access Dissertations and Theses 2012a; http://hdl.handle.net/11375/12400] [Google Scholar]
- Granados A., Luinstra K., Chong S., Goodall E., Banh L., Mubareka S. Use of an improved quantitative polymerase chain reaction assay to determine differences in human rhinovirus viral loads in different populations. Diagn Microbiol Infect Dis. 2012;74:384–387. doi: 10.1016/j.diagmicrobio.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S. Update on rhinovirus and coronavirus infections. Semin Respir Crit Care Med. 2011;32:433–446. doi: 10.1055/s-0031-1283283. [DOI] [PubMed] [Google Scholar]
- Jansen R., Wieringa J., Koekkoek S., Visser C., Pajkrt D., Molenkamp R. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49:2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti T., Lehtinen P., Vuorinen T., Koskenvuo M., Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72:695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- Johnston S., Sanderson G., Pattermore P., Smith S., Bardin P., Bruce C. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J Clin Microbiol. 1993;31:111–117. doi: 10.1128/jcm.31.1.111-117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiang D., Kalra I., Yagi S., Louie J., Boushey H., Boothby J. Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J Clin Microbiol. 2008;46:3736–3745. doi: 10.1128/JCM.00674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieninger E., Fuchs O., Latzin P., Frey U., Regamey N. Rhinovirus infections in infancy and early childhood. Eur Respir J. 2013;41:443–452. doi: 10.1183/09031936.00203511. [DOI] [PubMed] [Google Scholar]
- Kusel M., de Klerk N., Holt P., Kebdaze T., Johnston S., Sly P. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr Infect Dis J. 2006;25:680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- Li-Ng J., Pollack S., Cunha B., Mikhail M., Yeh J., Berbari N. A randomized controlled trial of vitamin D3 supplementation for the prevention of symptomatic upper respiratory tract infections. Epidemiol Infect. 2009;137:1396–1404. doi: 10.1017/S0950268809002404. [DOI] [PubMed] [Google Scholar]
- Lizogub V., Riley D., Heger M. Efficacy of a pelargonium sidoides preparation in patients with the common cold: a randomized, double blind, placebo-controlled clinical trial. Explore (NY) 2007;3:573–584. doi: 10.1016/j.explore.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Lu X., Erdman D. Poster International Conference of the American Thoracic Society May 2007; Toronto, ON. 2007. Comparison of three typing methods for human rhinoviruses based on sequence analysis of partial 5′NCR, VP2/4, and VP1 regions. [Google Scholar]
- Luinstra K., Petrich A., Castriciano S., Ackerman M., Chong S., Carruthers S. Evaluation and clinical validation of an alcohol-based transport medium for preservation and inactivation of respiratory viruses. J Clin Microbiol. 2011;49:2138–2142. doi: 10.1128/JCM.00327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I. Human rhinoviruses: the cold wars resume. J Clin Virol. 2008;42:297–320. doi: 10.1016/j.jcv.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan M., Martinko J., Parker J. Brock Biology of Microorganisms. 10th ed. Pearson Education; Upper Saddle River, NJ: 2003. Person-to-person microbial disease: colds and influenza; pp. 889–892. [Google Scholar]
- McIntyre C., Knowles N., Simmonds P. Proposals for the classification of human rhinovirus species A, B, and C into genotypically assigned types. JGV. 2013;94:1791–1806. doi: 10.1099/vir.0.053686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokoso-Koivisto J., Kinnari T., Lindahl P., Hovi T., Pitkaranta A. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J Med Virol. 2002;66:417–420. doi: 10.1002/jmv.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola V., Waris M., Osterback R., Susi P., Ruuskanen O., Hyypia T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- Picornavirus Study Group Enterovirus. 2013. http://www.picornaviridae.com/enterovirus/enterovirus.htm Retrieved from.
- Predy G., Goel V., Lovlin R., Donner A., Stitt L., Basu T. Efficacy of an extract of North American ginseng containing poly-furanosyl-pyranosyl-saccharides for preventing upper respiratory tract infections: a randomized controlled trial. CMAJ. 2005;173:1043–1048. doi: 10.1503/cmaj.1041470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten B., Bulitta M., Ballering-Bruhl B., Koster U., Shaffer M. Efficacy of Echinacea purpurea in patients with a common cold. A placebo-controlled, randomized, double-blind clinical trial. Arzneimittelforschung. 2001;51:563–568. doi: 10.1055/s-0031-1300080. [DOI] [PubMed] [Google Scholar]
- Smieja M., Castriciano S., Carruthers S., So G., Chong S., Luinstra K. Development and evaluation of a flocked nasal mid-turbinate swab for self-collection in respiratory virus infection diagnostic testing. J Clin Microbiol. 2010;48:3340–3342. doi: 10.1128/JCM.02235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z., Gonzalez R., Wan Z., Xiao Y., Chen L., Li T. Human rhinoviruses in Chinese adults with acute respiratory tract infection. J Infect. 2010;61:289–298. doi: 10.1016/j.jinf.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Benten I., Koopman L., Niesters B., Hop W., van Middlekoop B., de Waal L. Predominance of rhinovirus in the nose of symptomatic and asymptomatic infants. Pediatr Allergy Immunol. 2003;14:363–370. doi: 10.1034/j.1399-3038.2003.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., Carraro E., Kamikawa J., Leal E., Granato C., Bellei N. Rhinovirus species and their clinical presentation among different risk groups of non-hospitalized patients. J Med Virol. 2010;82:2110–2115. doi: 10.1002/jmv.21914. [DOI] [PubMed] [Google Scholar]
- Winther B., Hayden F., Hendley J. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol. 2006;78:644–650. doi: 10.1002/jmv.20588. [DOI] [PubMed] [Google Scholar]