Abstract
The dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) is a type II C-type lectin whose expression is restricted to the most potent antigen-presenting cells (APCs), the dendritic cells (DCs). In recent years, DC-SIGN has gained an exponential increase in attention because of its involvement in multiple aspects of immune function. Besides being an adhesion molecule, particularly in binding ICAM-2 and ICAM-3, it is also crucial in recognizing several endogenous and exogenous antigens. Additionally, the intracellular domain of DC-SIGN includes molecular motifs, which enable the activation of signal transduction pathways involving Raf-1 and subsequent modulation of DC-maturation status, through direct modification of nuclear factor Nf-κB in DCs. Upon DC-SIGN engagement by mannose- or fucose-containing oligosaccharides, the latter leads to a tailored Toll-like receptor signalling, resulting in an altered DC-cytokine profile and skewing of Th1/Th2 responses. In this article, we will discuss recent advances on a broad perspective concerning DC-SIGN structure, signalling and immune function.
Abbreviations: (APCs), antigen-presenting cells; (CRD), carbohydrate recognition domain; C(CLRs), C-type lectin receptors; (CBP), CREB-binding protein; (CRD), cysteine-rich domain; (DC-SIGN), dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin; (HATs), histone acetyltransferases; (HDACs), histone deacetylases; (ICAM), intracellular adhesion molecule; (LeX), Lewis-x; (ManLAM), mannosylated lipoarabinomannan; (LPS), lipopolysaccharide; (SEA), soluble egg antigen; (TLR), Toll-like receptor; (Pak), p21-activated kinases; (PRR), pathogen recognition receptor
Keywords: Dendritic cells, Pathogen recognition receptor, Adhesion, Migration, Endocytic receptor, Immune modulation
1. Introduction
Dendritic cells (DCs) are a heterogeneous population and the most potent antigen-presenting cells (APCs) known so far [1]. The functional characteristics of DCs are unique in that they are responsible for generating strong Ag-specific immune responses, as well as for inducing tolerance and maintenance of immune homeostasis. Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) is a type II transmembrane lectin receptor. Attention on DC-SIGN has increased greatly in recent years due to important discoveries documenting its immune-related functions and cell-specific expression that is mostly restricted to DCs. DC-SIGN is abundantly expressed on immature DCs (iDCs) that are present in peripheral tissues, but is also found, albeit down-regulated, on mature or activated DCs (mDCs) in lymphoid tissues such as lymph nodes, tonsils and spleen [2]. However, it is not expressed on certain DC-types such as follicular DCs or on skin-resident Langerhans DCs.
Although C-type lectin receptors (CLRs) were initially thought to function as scavenger receptors that bind various pathogens upon recognition of particular carbohydrate profiles, it has become clear that some C-type lectins may function as adhesion, signalling or antigen receptors. CLRs are well known for their function of serving as antigen-uptake receptors, and this is consistent with the fact that most CLRs are present on APCs [3]. In addition, several CLRs have been shown to contribute to loading of endocytosed Ags on MHC class I and class II molecules, thereby facilitating effective Ag-specific CD4 and CD8 T-cell responses [4], [5]. Besides foreign Ags, DC-SIGN binds to a number of endogenous ligands, particularly to intracellular adhesion molecules (ICAM)-2 on endothelial cells and ICAM-3 on T lymphocytes, contributing to transendothelial migration of DCs and the formation of the DC–T cell synapse, respectively [2], [6]. Apart from supporting the initial immune response between DCs and T cells, DC-SIGN also recognizes several bacterial pathogens [7], contributing to generation of pathogen-tailored immune responses. Furthermore, DC-SIGN can capture HIV-1 at entry sites and transport the virus into lymphoid tissues, where HIV-1 can be transmitted to CD4+ T cells. Recently, it has been demonstrated that, besides adhesive and Ag-recognition properties, engagement of DC-SIGN on DCs results in activation of signal transduction pathways that can cause extensive modulation of immune responses, particularly when co-activated with Toll-like receptor (TLR)-induced signalling [8]. Whereas simultaneous signalling through TLRs is present during pathogen-specific immune responses during infection [9], both in vitro and in vivo targeting to CLRs on immature DCs leads to tolerance by default. The physiological function of DC-SIGN may thus be induction of tolerance by immature DC, after recognition of glycosylated self-antigens, for homeostatic control [10]. Several pathogens that target DC-SIGN appear to exploit this signalling and subvert its functions, either by inhibition of antigen presentation or by alteration of TLR mediated signalling, resulting in modification of T cell responses [11]. In this review, we will discuss the structural, functional and signalling characteristics of DC-SIGN in the context of its functions in immune regulation, tumour immunity and potential therapy.
2. DC-SIGN structure and expression on the DC surface
2.1. DC-SIGN expression
DC-SIGN is preferentially expressed on myeloid DCs and is found on dermal DCs, interstitial DCs, a subset of blood DCs, and on in vitro prepared, monocyte-derived DCs [12], [13]. Due to its highly restricted expression, DC-SIGN is considered a DC-specific phenotypic marker. During in vitro differentiation of DCs from monocytes, DC-SIGN expression is dependent on IL-4 signalling [14]. Furthermore, IL-4 and IL-13 (both of which act through the STAT6 signalling pathway) can also cause de novo expression of DC-SIGN on the THP-1 monocytic cell line, primary monocytes and, alternatively, activated macrophages [15]. Pre-treatment of THP-1 cells with differentiation-inducing agents such as phorbol esters and bryostatin further conveys the DC-SIGN-inducing ability of IL-4. In accordance with the study of Relloso et al. [15], our group recently demonstrated that the surface expression of DC-SIGN on monocytes appears as early as 24 h after GM-CSF and IL-4 induced DC differentiation, and reaches peak levels on day four (Svajger et al., unpublished observations). The JAK-STAT signalling route is involved in IL-4-dependent induction of DC-SIGN. The use of tryphostin AG490, a specific inhibitor of JAK2 and JAK3, results in complete abrogation of DC-SIGN induction [14]. Thus, it can be speculated that STAT6, as a major IL-4 signalling element, is directly responsible for binding to promoter regions of the DC-SIGN gene and induction of transcription. Furthermore, IFN-α and IFN-γ inhibit the up-regulation of DC-SIGN by IL-4, and both cytokines have been demonstrated to suppress IL-4-dependent gene expression by inhibiting tyrosine phosphorylation and nuclear translocation of STAT6, most probably via induced expression of suppressors of cytokine signalling [16], [17]. Considering the negative regulation of DC-SIGN expression by myeloid DCs, DC-SIGN was shown to be negatively regulated by type I and II interferons, as well as by the anti-inflammatory drug dexamethasone and TGF-β [14]. Dexamethasone blocks in vitro DC differentiation at the monocyte stage, resulting in CD14+ macrophage-like cells [18]. It has been demonstrated that dexamethasone can interfere with the JAK-STAT pathway [19] indicating that prevention of DC-SIGN expression could be achieved by direct inhibition of IL-4 signalling. However, although TGF-β can inhibit the Jak-STAT activation in certain systems, it has been demonstrated that it fails to suppress STAT6 activation by IL-4 in monocytes [16]. This tells us that factors other than STAT6 are probably also involved in the regulation of DC-SIGN expression. The group of Corbi recently demonstrated that the transcription factor PU.1 regulates basal and tissue-specific expression of DC-SIGN through occupancy of two DNA elements within the proximal regulatory region of the DC-SIGN gene [20]. The expression of DC-SIGN correlated with nuclear levels of PU.1 transcription factor during DC maturation as well as during classical and alternative macrophage activation [20]. This finding further supports the connection between proper DC differentiation and DC-SIGN expression, since high PU.1 activity directs the differentiation of bone marrow progenitors and blood monocytes towards DCs and suppresses macrophage development [21].
2.2. DC-SIGN structure
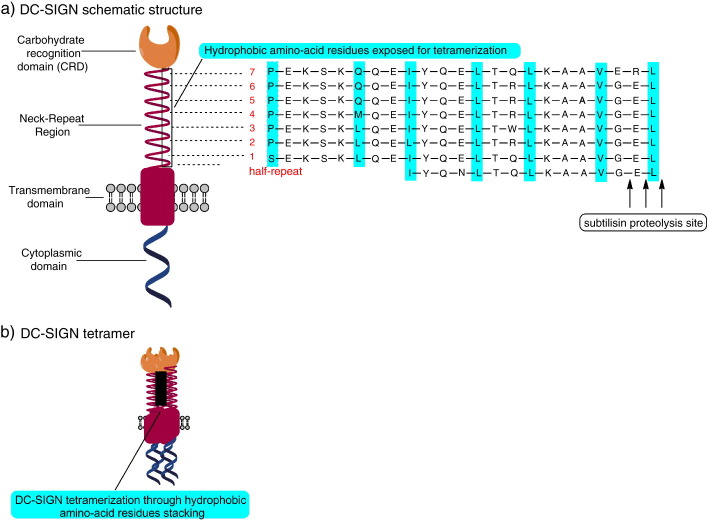
DC-SIGN contains a carbohydrate recognition domain (CRD), a neck region composed of 7 and a half repeats containing 23 amino-acid residue repeats, and a transmembrane region followed by a cytoplasmic tail containing recycling and internalization motifs [2], [22]. DC-SIGN ligation can result in transmission of intracellular signalling and this has been associated with the presence of a di-leucine motif and a tyrosine residue in the cytoplasmic tail [23].
The formation of multimeric complexes or, alternatively, conformational changes of their receptor is a possible way of increasing binding affinity/avidity of ligands containing repetitive sugar moieties. DC-SIGN tetramerization is thought to have the main impact on binding affinity, and occurs through the DC-SIGN neck-repeat domain [24]. The hydrophobic necks are believed to stabilize the DC-SIGN oligomers and project the CRDs away from the cell surface, which positions the CRDs for appropriate multivalent interaction with glycan ligands. Moreover, the carbohydrate recognition domain (CRD) of DC-SIGN when tetramerized, provides a means of amplifying specificity for multiple repetitive units on host molecules [25]. The repeats form extended stalks, stabilized largely by lateral interactions of α-helical regions in the 23-amino-acid repeats. This helical neck shape presents hydrophobic residues in recurring intervals of 3–5 that stack spontaneously to form dimers or tetramers (Fig. 1 ) [26]. Feinberg et al. [26] have created a series of truncated forms of DC-SIGN and demonstrated that oligomerization status depends on the number of helical repeats in the neck region; at least 6 repeats are needed for tetramerization, the neck region with 5.5 repeats provokes equilibrium between tetramer and dimer, while equilibrium between dimer and monomer takes place at a neck region of two repeats only. The organization into tetramers also amplifies the specificity and defines the set of pathogens that are recognized by DC-SIGN [25]. In this way, DC-SIGN binds particularly well to closely spaced (approx. 5 nm between sugar binding sites) oligosaccharides on the envelopes of viruses and membranes of parasites [27], [28]. However, some adjustments of this stringent binding model have been demonstrated very recently. Menon et al., by force-distance measurements, have shown a certain flexibility, allowing for conformational changes in DC-SIGN upon ligand binding [29]. This allows it to adapt to the arrangement of target monosaccharides and thus enables all CDRs to interact with their ligands. The multimeric organization and conformational flexibility of DC-SIGN molecules on the DC surface is therefore necessary for effective and selective binding of various mannose- and fucose-containing oligosaccharide patterns. Furthermore, due to the nature of receptor-mediated intercellular signalling, oligomerization of DC-SIGN could contribute to signal transduction after ligand binding.
Fig. 1.
a. Schematic structure of DC-SIGN and amino-acid sequence alignment of the neck-repeat domain. The repeated hydrophobic amino-acid residues (hydrophobic heptad), crucial for tetramerization, is highlighted. Arrows point to the subtilisin site of digestion. b. DC-SIGN tetramerization through hydrophobic residues stacking in the neck-repeat domain (modified from Feinberg et al. [26]).
Studies using transmission electronic microscopy and near-field scanning optical microscopy have revealed that, on the surface of immature DCs, DC-SIGN is arranged into distinct molecular clusters [30], [31]. This has been shown both on live DCs as well as in cells that ectopically express DC-SIGN. Such clustering of DC-SIGN is thought to improve binding to viral particles or bacteria with multivalent binding sites, by providing high-avidity binding platforms. The binding of oligosaccharides by the CRD is spatially constricted. When binding both mannose- and fucose-containing oligosaccharides, the CRD forms a 1-to-1 complex that consists of multiple interactions with the constituent monosaccharides, which provides specificity based on spatial constraints [24], [32], [33].
Its localization in lipid microdomains may further create a scaffold that favours ligand binding as well as interaction of DC-SIGN with signalling molecules that are also recruited into the same membrane domains.
3. Involvement of DC-SIGN in various DC functions: differentiation/migration/antigen capture/T cell priming
3.1. Differentiation
DC-SIGN greatly affects IL-4 guided DC differentiation from monocytes (Svajger et al., unpublished observations). Furthermore, the results point to a relationship between DC-SIGN and IL-4 signalling, emphasizing their inseparable role for DC differentiation. Since DC-SIGN regularly recognizes self-antigens, precise and coordinate interactions in vivo might also play an important part in maintaining DC homeostasis and proper immune function. In addition to the tolerogenic characteristics displayed by such DCs, enhanced apoptosis of early DC progenitors following DC-SIGN engagement could be yet another mechanism in the complex and finely tuned process of peripheral tolerance to self.
3.2. Migration
The capacity to migrate and exert continuous surveillance is a fundamental aspect of DC function. Transendothelial migration is a multistep process and DC-SIGN has been acknowledged to mediate the tethering and rolling along ICAM-2-expressing surfaces, as well as the adhesion of DC to endothelium and their subsequent migration [12]. Although DC-SIGN binds to both ICAM-2 and -3 under static conditions, only the former interaction resists shear stresses, and cells tether to and roll on ICAM-2, in contrast to ICAM-3, surfaces [12]. In particular, DC-SIGN mediates adhesion and rolling of dendritic cells on primary human umbilical vein endothelial cells through LeY antigen expressed on ICAM-2 [34].
3.3. Antigen capture
In contrast to TLRs, the main function of CLRs is to internalize antigens for degradation in order to enhance antigen processing and presentation on MHC class I and II molecules [3]. For this reason, CLRs are also called endocytic receptors or antigen-uptake receptors. DC-SIGN has also been demonstrated to mediate antigen uptake [13], [35], [36]. Binding of soluble ligand to DC-SIGN induces rapid internalization from the cell surface, mediated by a di-leucine motif present in the cytoplasmic domain [13]. DC-SIGN contains an additional tri-acidic cluster (EEE or DDD) in its cytoplasmic tail, important for targeting to proteolytic compartments. Moreover, a dual role has been indicated for the EEE motif as a sorting signal in the secretory pathway and a lysosomal targeting signal in the endocytic pathway [37]. Accordingly, DC-SIGN-ligand complexes are targeted to late endosomal or lysosomal compartments [38] where ligands are processed for MHC class II presentation to T cells, indicating an important role for DC-SIGN as an antigen receptor. Both the tyrosine and the di-leucine motif are required for the association of the cytoplasmic tail of DC-SIGN with leukocyte specific protein (LSP-1), an F-actin binding protein that mediates DC-SIGN dependent transport of HIV-1 to proteasomes [39].
Cambi et al. recently showed that DC-SIGN-mediated internalization occurs via clathrin-coated pits [40]. On the plasma membrane of DC, DC-SIGN is organized in nanoclusters, some of which co-localize with lipid rafts that specifically confer to the receptor its capacity for binding and internalization [30], [41]. The nanoclusters are enriched near the leading edge of living DC, but are preferentially endocytosed at lamellar sites posterior to the leading edge, suggesting a mobility of DC-SIGN from areas of concentration at the front to rearward sites of internalization [22], [42].
3.4. T cell priming
DC-SIGN, by binding ICAM-3, mediates transient adhesion of DCs with T cells, allowing screening of the MHC-peptide complexes. In particular, DC-SIGN supports early, antigen nonspecific contact between T cells and DCs, enabling T cell receptor engagement by stabilization of the DC–T cell contact zone and effective T cell receptor engagement [2]. The importance of DC-SIGN-ICAM-3 interactions in the initial DC–T cell contact is emphasized by the ability of anti-DC-SIGN antibodies to inhibit DC–T cell clustering and DC-induced proliferation of resting T cells. Apparently the transient nature of the DC-SIGN–ICAM-3 interactions enables screening of a large number of resting T cells until a productive TCR engagement is achieved.
4. Various ligands and signalling through DC-SIGN
The recognition of its ligands by DC-SIGN is highly regulated [2]. As noted above, DC-SIGN mediates the contact between DC and T lymphocytes by binding to ICAM-3 [2], mediates rolling of DCs on endothelium by interacting with ICAM-2 [6], and recognizes a variety of microorganisms, including viruses (HIV-1, HCV, CMV, Dengue, Ebola, SARS-CoV, HSV, coronaviruses, H5N1, West Nile virus, measles virus) [2], [43], [44], [45], [46], [47], [48], [49], [50], bacteria (Helicobacter pylori, Mycobacterium tuberculosis, and Leptospira interrogans) [11], [51], fungi (Candida albicans and Aspergillus fumigatus) [35] and several parasites (Leishmania, and Schistosoma mansoni) [52], [53].
4.1. Specificity in binding of endogenous and exogenous ligands
DC-SIGN was discovered by the observation that DCs bind the intercellular adhesion molecule (ICAM)-3 (CD50) with very high affinity. ICAM-3 is N-linked glycosylated by high mannose-type oligosaccharides and, depending on the peripheral blood cell population, Lewis-x residues [54]. DC-SIGN also binds ICAM-2, but not ICAM-1. Enzymatic removal of the N-linked carbohydrates from ICAM-2 and ICAM-3 completely abrogates the binding of DC-SIGN [55]. DC-SIGN interacts primarily with the Ig-like second domains of ICAM-2 and ICAM-3. However, although the interactions exhibit similar features, DC-SIGN interacts with ICAM-2 differently from that with ICAM-3. The distinct carbohydrate structure and/or the different size of the ICAM molecules may determine the manner of interaction. DC-SIGN binding to ICAM-3 is calcium dependent, and DC-SIGN CRD binds two Ca2+ ions, one essential for the tertiary structure and the other for coordinating ligand binding [32].
DC-SIGN is capable of binding to the gp120 envelope protein of HIV-1. Site-directed mutagenesis demonstrated that DC-SIGN binding to both gp120 and ICAM-3is mediated by Ca2+ at site 2 and by nearby amino-acid residues, and that interaction with HIV-1, ICAM-2 and ICAM-3 is blocked by the polycarbohydrate mannan. However, the binding of DC-SIGN to gp120 differs from that to ICAM-3. In particular, DC-SIGN has a distinct binding site for HIV-1 gp120.
DC-SIGN interacts with high mannose-type oligosaccharides but not with single terminal mannose residues [2], [24]. It recognizes high mannose, with a minimum of three mannose residues located more internally within a glycan structure [32], and the external saccharides also interact with the surface of DC-SIGN, as well as terminal di-mannoses. DC-SIGN has a higher affinity for more complex mannose residues in specific arrangements [24], [32], [55] and the recognition of specific carbohydrate structures appears to depend on their spacing on a glycoprotein [24]. Apart from interacting with several less complex mannose-containing glycoconjugates, i.e. mannose and α1 → 3, α1 → 6 mannotriose, DC-SIGN also demonstrates a high affinity for Lewis blood group antigens, that contain fucose residues. The cysteine-rich domain (CRD) of DC-SIGN recognizes the unsialated forms of Lewis-x (LeX), Lewis-y (Ley) and Lewis-a (Lea), Lewis-b (Leb) [33], [52], that contain fucose residues in different anomeric linkages. Notably, DC-SIGN has a much higher affinity for the fucose-containing carbohydrate Lex than for mannotriose. In addition, DC-SIGN binds the strongly sulfated LeX. Guo et al. showed that DC-SIGN reacts with a wider range of glycans, including Lewis blood group antigens, than hitherto realized [33]. The DC-SIGN crystal structure reported by the same group reveals that two distinct ligand groups bind to the Ca2+-binding pocket (designated the principal Ca2+ site) that accommodates either mannose or fucose moieties. Apart from this mutual starting binding pocket however, mannose and fucose-based oligosaccharides lie in opposite directions, confirming the existence of two distinct binding sites for mannose- and fucose-based oligosaccharides.
DC-SIGN interacts with pathogens through either mannose or fucose-containing glycans. It binds strongly to H. pylori lipopolysaccharide (LPS) and S. mansoni soluble egg antigen (SEA). LeX is expressed on surface located LPS in H. pylori and on surface located LPS, including SEA, at all stages of the parasite S. mansoni. Mannose-capped surface lipophosphoglycan (LPG) expressed by Leishmania mexicana and the mannose-capped cell-wall component of M. tuberculosis ManLAM (lipoarabinomannan) also interact with DC-SIGN. DC-SIGN binds specifically to the dimeric and trimeric mannose residues in ManLAM and does not recognize ManLAM capped with single mannose residues in Mycobacterium avium [56], which correlates with the specificity of DC-SIGN for di- and tri-mannose structures. No binding of DC-SIGN was observed to Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa or Staphylococcus aureus [52].
4.2. Signalling through DC-SIGN
DC-SIGN possesses several motifs in its cytoplasmic tail that could allow the induction of intracellular signalling pathways, however specific mechanisms of signal transduction are still highly speculative. By studying the accumulation of proteins containing phosphotyrosine and phosphoserine after DC-SIGN ligation with H200 anti-DC-SIGN Ab, Hodges et al. suggested the presence of the tyrosine residue in the cytoplasmic tail of DC-SIGN to be crucially important [23]. However, a tyrosine residue does not appear to be relevant in the DC-SIGN-induced activation of the serine/threonine kinase Raf-1 [8], shown to be important in DC-SIGN signalling by studies of various kinases potentially involved in DC-SIGN signalling. Blockade of Raf-1 activity by specific chemical inhibitors, as well as by silencing through RNAi, led to the inhibition of IL-10 induction by DC-SIGN triggering [8]. Activation of Raf-1 is still speculative and so far explained mainly by known direct upstream mediators. However, it has been shown that ManLAM binding to DC-SIGN activates the small GTPase Ras. During activation, Raf-1 translocates to the membrane through interaction with the active form of Ras [57]. This induces a conformational change in Raf-1 and is required for its activation. However, the conformational change in Raf-1 alone is not sufficient for its activation and requires the phosphorylation of serine 338 (Ser338) and tyrosines 340 and 341 (Tyr 340/341) [58] (Fig. 2 ). Gringhuis et al. demonstrated that ManLAM activation of DC-SIGN results in induced phosphorylation of Raf-1 on Ser338 and Tyr340/341 [8]. The phosphorylation of Ser338 is carried out by p21-activated kinases (Pak), while the phosphorylation of Tyr340/341 depends on a yet unidentified member of the Src family of tyrosine kinases. However, in imunoprecipitation studies, DC-SIGN from lipid rafts of DCs was found to co-precipitate with Lyn, a member of the Src family kinases, as well as with Syk tyrosine kinase, indicating their possible involvement in DC-SIGN signalling [59]. As shown by others, Raf-1 does not appear to be the sole important factor in DC-SIGN signalling. Stimulation of DC-SIGN with the MR-1 Ab has been shown to result in phosphorylation of ERK1/2 and Akt [59]. The MEK–ERK kinase cascade is the most highly characterized pathway down-stream of Raf proteins. However, the activation of Raf-1 by ManLAM does not lead to activation of ERK1/2 or MEK1/2 [8]. In addition, it has not been shown whether MR-1 Ab is able to activate Raf-1. Therefore, the activation of Raf-1 by ManLAM must employ a signalling route distinct from that of ERK and MEK. In this manner, signal transduction after DC-SIGN ligation appears to be dependent on the specific DC-SIGN ligand at hand. In addition, its engagement by MR-1 Ab in DC-SIGN-transfected Jurkat cells triggers PLC-γ phosphorylation [59]. Also, it has been demonstrated very recently that hepatitis C virus E2 can induce p38MAPK activation in DC-SIGN transfected HEK293T cells [60].
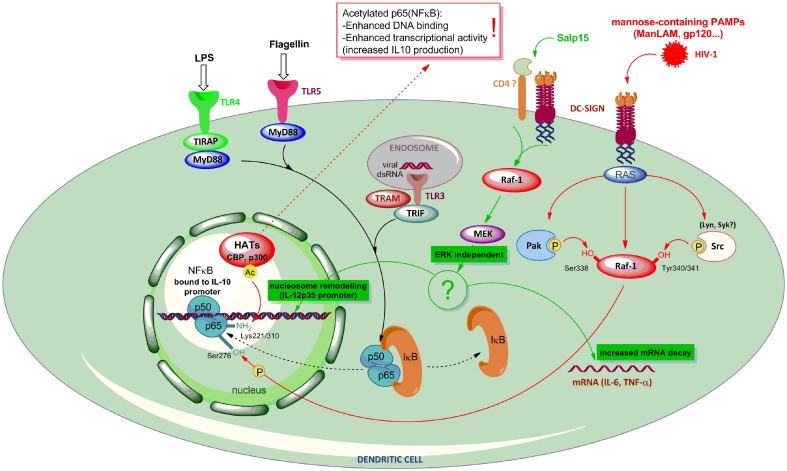
Fig. 2.
DC-SIGN signalling is ligand-dependent, activates Raf and down-stream mediators and contributes to modulation of signalling involved in DC activation. Activation of DC-SIGN with ManLam or gp120 leads to increased phosphorylation of Raf-1. This process requires a conformational change in Raf-1, that is mediated by small GTPase Ras. Raf-1 can then be phosphorylated at Ser338 by p21-activated kinases (Pak) and at Tyr340 and Tyr341 by kinases belonging to Src family, most probably Lyn and Syk. Activated Raf-1 is neccessary for phosphorylation of p50/p65 Nf-κB dimer in the nucleus, which is a prerequisite for later Nf-κB modification by histone acetyltransferases (HATs). The acetylation of Nf-κB is carried out at lysine residues 221 and 310 and this enables increased DNA binding and transcriptional activity by Nf-κB, leading to up-regulated IL-10 production. Salp15 binding to DC-SIGN leads to Raf-1 activation and modulated cytokine production without modulation of Nf-κB. In this manner, Raf-1 activates MEK, which without the activity of ERK, leads to increased degradation of IL-6 and TNF-α mRNA, as well as nucleosome remodelling at the IL-12p35 promoter.
Down-stream mediators from Raf-1 are not well defined and appear to be dependent on specific ligands. Nevertheless, it is clear that modulation of TLR signalling by DC-SIGN engagement depends on modification of Nf-κB activity. Activation of DCs by TLR agonists and subsequent production of pro-inflammatory cytokines, such as IL-12p70, TNF-α, IL-6, and immunosuppressive IL-10, is dependent on activation and nuclear translocation of transcription factor Nf-κB. In immature DCs, The family of Nf-κB proteins, c-Rel, RelA (p65), RelB, NF-κB1 (p50), and NF-κB2 (p52), reside in the cytoplasm and cannot translocate into the nucleus because of their association with the inhibitory proteins IκBα, IκBβ and IκBε [61]. The most studied and frequently present form of active Nf-κB is the heterodimer between p65 and p50 subunits. After DC activation, the p65-p50 dimer translocates to the nucleus and influences the expression of many genes by binding to target DNA sequences. Besides inhibition by IκB proteins, NF-κB activity is also regulated by covalent modifications that alter the ability of Nf-κB dimers to bind DNA, leading to differential gene expression. Regulation of p65 activity involves several post-translational modifications, such as phosphorylation and acetylation [62]. ManLAM engagement of DC-SIGN has been shown to result in Raf-1 activation and subsequent phosphorylation of Ser276 on the p65 subunit [8]. Importantly, phosphorylation of p65 subunit is required for the subsequent interaction of Nf-κB with histone acetyltransferases (HATs), such as CREB-binding protein (CBP) and p300, and histone deacetylases (HDACs), such as HDAC1 and HDAC2 [63]. Signalling through DC-SIGN leads to modification of p65-p50 dimer, resulting in increased activity as well as prolonged retention at the IL-10 promoter. The latter has been shown to be associated with increased acetylation of the p65 subunit [8]. Previous reports have demonstrated the crucial importance of direct acetylation of Nf-κB, in terms of either positive or negative regulation [64], [65]. As demonstrated by Chen et al. [65], acetylation of various lysine residues, namely Lys221 and Lys310, orchestrates distinct cellular functions of Nf-κB, thus making the acetylation-dependent regulation of Nf-κB site-specific. The acetylation of Lys310 is necessary for full transcriptional activity of p65 subunit which could explain the enhanced Nf-κB activity observed by Gringhuis et al. [8] Furthermore, acetylation at Lys221 of p65 leads to enhancement of DNA binding by Nf-κB and impairs its assembly with IκBα [65], which would consequently result in prolonged Nf-κB activity. Such an explanation could explain the results obtained by Gringhuis et al., where DC-SIGN induced signalling, together with LPS stimulation in DCs, led to prolonged transcriptional activity of Nf-κB and enhanced transcription rate from the IL-10 gene [8]. Thus, in the context of DC-SIGN ligation by ManLAM, it can be said that DC-SIGN activation of Raf-1 leads to enhanced acetylation of Nf-κB , which results in its enhanced activity and prolonged nuclear retention.
Further, we must stress that mechanisms besides modification of Nf-κB activity might well play an important role in immunoregulation of TLR signalling by DC-SIGN. Salp15 is a salivary tick protein, produced by the Ixodes scapularis tick species, with reported immunomodulatory properties [66], [67]. Similarly to ManLAM, binding of Salp15 to DC-SIGN results in activation of Raf-1 kinase and modulation of cytokine production by DCs when added together with LPS [68]. Salp15 reduces the production of IL-12p70, TNF-α and IL-6, and increases the production of IL-10. However, inhibition of CBP/p300-dependent acetylation of Nf-κB does not abrogate pro-inflammatory cytokine suppression by Salp15. Instead, Salp15-induced Raf-1 activation leads to activation of MEK. Furthermore, inhibition of MEK by inhibitor U0126 completely abrogates modulation of cytokine production by Salp15 [68]. It was demonstrated that Salp15 modulates DC-cytokine production by increasing IL-6 and TNF-α mRNA decay and by impairing nucleosome remodelling at the IL-12p35 promoter [68]. In this context, it should be noted that Salp15 also binds to CD4 [66] and DC-SIGN has been shown to co-localize with CD4 on the cell surface [69]. It is possible that the distinct signalling observed with Salp15 could be due to concomitant engagement of both CD4 and DC-SIGN, which would affect Raf-1 activation on down-stream mediators. Like Salp15, an anti-DC-SIGN Ab MR-1 has been shown to up-regulate IL-10 production when present in cultures of LPS-stimulated DCs. Additionally, MR-1-stimulation of DC-SIGN results in increased activation of ERK and PI3K (67). It is difficult to speculate whether MR-1 ligation of DC-SIGN correlates well with that of Salp15, since activation of Raf-1 was not determined for MR-1 and, although MEK kinases are well known to activate ERK, its phosphorylation was not seen with Salp15 [68]. Interestingly, H. pylori engagement of DC-SIGN through fucose-elements present on their Lewis Ags leads to Raf-1-independent immune responses with low IL-12 and IL-6 and increased IL-10 production [70]. Taken together, many DC-SIGN ligands lead to activation of Raf-1, but further activation of down-stream mediators appears to depend on specific binding of a particular ligand. So far, despite incomplete information, signalling through DC-SIGN could probably activate all elements of the classical Raf–MEK–ERK pathway, however detailed studies on the importance of individual signalling elements in association with particular ligands are necessary to fully understand DC-SIGN-induced regulation of immune responses.
5. Immune modulation through concomitant signalling of DC-SIGN with other pathogen recognition receptors (PRRs)
DCs have evolved unique ways in which they regulate immunity and contribute to tolerance induction [1]. DC-SIGN functions not only as an independent PRR, but is also implicated in immunoregulation of DCs. Recently, DC-SIGN has emerged as a key player in the induction of immune responses by modulating TLR-induced activation of DCs. However, the specific modulation of immune responses is dependent on the pathogen involved. Binding of DC-SIGN by particular pathogens can lead to the inhibition or promotion of T helper type 1 (Th1) polarization, Th2 responses and/or induction of regulatory T cell differentiation.
Gringhuis et al. [70] have recently demonstrated carbohydrate-specific signalling by DC-SIGN, i.e. mannose- and fucose-expressing pathogens induce distinct DC-SIGN signalling pathways. Carbohydrate-specific ligation of DC-SIGN leads to a switch in the proximal DC-SIGN signalling complex (signalosome) consisting of LSP1, KSR1 and CNK, which is required for the constitutive recruitment of Raf-1 to DC-SIGN. After binding of DC-SIGN by mannose-expressing pathogens such as M. tuberculosis and HIV-1, Raf-1 became activated by recruitment of the ‘upstream’ effectors LARG and RhoA to the DC-SIGN signalosome. This Raf-1-dependent signalling modulates TLR4 signalling and enhances the expression of IL-10, IL-12 and IL-6. In contrast, fucose-expressing pathogens, such as H. pylori, actively dissociate the KSR1–CNK–Raf-1 complex from the signalosome and enhance the expression of IL-10, but downregulate the expression of IL-12 and IL-6 in a Raf-1-independent but LSP1-dependent manner. The association of LSP1 with DC-SIGN was a prerequisite for cytokine modulation by mannose and fucose-containing ligands.
In particular, M. tuberculosis binds to DC-SIGN mainly via ManLAM, which is abundantly expressed in mycobacterial cell walls and also secreted by M. tuberculosis-infected cells [71], [72], [73]. Other mycobacterial ligands have also been documented for DC-SIGN, i.e. lipomannan [74], mannose-capped arabinomannan [74], two mannosylated glycoproteins [74], the phosphatidylinositol mannosides [75] and, recently, β-glucan [76]. The binding of ManLAM, in addition to TLR activation, enhances the production of different TLR-induced cytokines such as IL-12p70 and IL-10 [11], [70] where the increased production of IL-10 leads to down-regulation of DC function and, subsequently, more successful infection by M. tuberculosis. This double stimulus, from both DC-SIGN and TLR, is necessary for modulation of cytokine production, since the binding of ManLAM on its own does not induce cytokine production. Upon binding of ManLAM, DC-SIGN triggers an intracellular signalling cascade independently of other recognition receptors [8], and inhibition of Raf-1 completely blocks DC-SIGN-mediated immune responses to mycobacteria. As for ManLAM, binding of β-glucan to DC-SIGN stimulates the production of the immunosuppressive IL-10 by LPS-activated, monocyte-derived DCs [76].
Similarly, Enterobacter sakazakii targeting of DC-SIGN prevents the maturation of DCs by triggering the production of high levels of IL-10 and TGF-β, and by suppressing the activation of MAPKs, for which outer membrane protein A (OmpA) expression is critical, although it is not required for uptake [77]. The major S layer protein, SlpA, of Lactobacillus acidophilus NCFM is also a DC-SIGN ligand that is functionally involved in the modulation of DCs and T cell functions, by inducing concentration-dependent production of IL-10 and low IL-12p70 [78]. Other probiotic bacteria (Lactobacillus reuteri and Lactobacillus casei) also exert immune suppression through engagement of DC-SIGN on DCs by instructing DCs to induce IL-10 producing regulatory T cells that suppress T cell responses [79].
The central feature of pathogens that interact with DC-SIGN is that they cause chronic infections and that manipulation of the Th1 versus Th2 cell balance by these pathogens is central to their persistence [80]. A Th1 toTh2 shift is crucial for the virulence and persistence of L. mexicana. Similarly, the Th2-tvpe immune response to infection with S. mansoni is associated with persistence of the pathogen, and SEA and its major glycan antigen Lex can cause a switch towards a Th2-cell mediated immune response [81]. H. pylori expresses several Le blood group antigens in their LPS, such as Lex and Ley. However, the expression of Lex/y is not stable but subjected to reversible on and off switching of LPS epitopes, resulting in Lex/y+ and Lex/y– bacteria within a single strain [82]. Bergman et al. [83] have shown that H. pylori can modulate the Th1/Th2 balance through interaction of phase-variable LPS with DC-SIGN on DCs. In this manner, the binding of Lex/y+ antigens results in enhanced IL-10 production by DCs and inhibition of Th1 responses. However, mutant strains with truncations of the LPS outer core of Neisseria meningitides have been shown to induce strong Th1 responses after interaction with DC-SIGN [84].
Modulation of TLR activity by DC-SIGN is not limited to TLR4 signalling, but has also been demonstrated when the DCs were stimulated with TLR3 ligand, poly I:C, and TLR5 ligand, flagellin. Concomitant stimulation of DC-SIGN and TLR3 or TLR5 resulted in increased expression of IL-10 protein in both cases [8]. Furthermore, measles virus induces the production of IL-10 by DCs via concomitant binding of DC-SIGN and TLR2 [85], [86]. DC-SIGN also plays an important role in capturing HIV-1 [87]. Since HIV-1 does not activate TLRs, it does not modulate the cytokine profile of DCs on its own. A recent report demonstrated that AIDS patients have elevated amounts of microbial LPS in their serum [88] and HIV-1 can induce IL-10 production, with concomitant TLR4 triggering by LPS [8]. Furthermore, some immunosuppressive effects exerted by HIV-1 could be attributed to IL-10 production by DCs, since elevated amounts of IL-10 have consistently been found in AIDS patients [89]. As discussed above the modulation of TLR signalling by DC-SIGN appears to depend mainly on Raf-1 induced acetylation of the p65 Nf-κB subunit. HIV-1 binding also mediates effects independently of TLR activation, namely by inducing the negative regulator of TLR4, ATF3 [90], and by inducing LARG-mediated activation of the GTPase RhoA, which is required for viral synapse formation [23].
The convergence of DC-SIGN and TLR pathways is thus an important mode of immune regulation and presents an important mechanism by which several pathogens reduce immune responses and escape immune surveillance.
6. DC-SIGN as a mechanism to escape immune surveillance
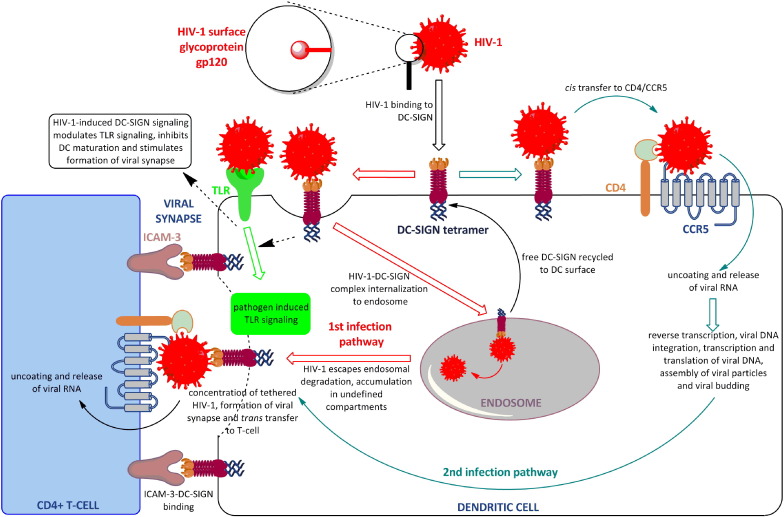
HIV-1 is a perfect example of a virus that exploits native DC-SIGN functions for infection (Fig. 3 ). To develop a thoroughly disseminated infection, HIV-1 has to be transported from mucosal surfaces and/or blood to lymphoid tissues, where it most importantly infects CD4+ T cells. DCs make a perfect host for this assignment as they are abundant at mucosal surfaces and, upon maturation, migrate to lymphoid tissues. HIV-1 binds to DCs through interaction with its envelope glycoprotein gp120 and DC-SIGN. The complex HIV-1–DC-SIGN is promptly internalized via clathrin-coated pits to DCs' endosomes [40], where the acidic endosomal media cause ligands to dissociate from DC-SIGN. Free DC-SIGN is then recycled to the DC surface while bound ligands are lysed and processed [91]. Indeed, a large part of the HIV-1 that enters the DCs is destroyed by this mechanism. In contrast with these findings, HIV-1 bound to DC-SIGN is astonishingly stable and a small quantity of HIV-1 that enters DCs remains protected from the host immune system, while retaining its infectiveness [2], [22]. HIV-1 stays in DCs (and the THP monocyte cell line) in a highly infectious state for days, hidden in multivesicular bodies that are different from endosomes and/or lysosomes [92]. Alternatively, after interaction with DC-SIGN, HIV-1 may be transferred laterally to bind CD4 and CCR5 receptors expressed on immature DCs, followed by fusion of the viral envelope with the plasma membrane and infection of DCs [93]. In either way, HIV-1 exploits DCs as a Trojan horse to escape the host immune system [94].
Fig. 3.
HIV-1 interaction with DC-SIGN and subsequent events. The first CD4+ T-cell infection pathway through an HIV-1 escape mechanism is depicted with red arrows; an alternative 2nd pathway through DC infection is depicted with green arrows. Both pathways merge in HIV-1 trans transfer to CD4+ cells. DC-SIGN signalling modulation is presented in acid green.
HIV-1-infected DCs are able to mediate transmission of the virus to T cells through the formation of a so-called infectious synapse. Hodges et al. demonstrated that DC-SIGN signalling is responsible for viral synapse formation between DCs and T cells [23]. Thus, HIV-1-induced DC-SIGN signalling triggers two paradoxical conditions, inhibiting DC maturation while inducing formation of viral synapse, a process previously attributed exclusively to mature DCs. This process may be facilitated by the DC-SIGN function as an adhesion molecule for ICAM-3 at the surface of CD4+ T cells. Namely, Sol-Foulon et al. [95] have demonstrated that HIV-1 protein Nef induces DC-SIGN up-regulation in HIV-1 infected DCs and this markedly stimulates DC–T cell clustering which facilitates HIV-1 transmission and dissemination. The intracellular trafficking of DC-SIGN is affected by the Nef. Nef can interact with the cell sorting machinery to downregulate expression levels of CD4 and MHC class I and thus facilitate immune evasion. Furthermore, Trumpfheller et al. [92] speculated that DC-SIGN, once expressed in high quantity on infected DCs, could serve to concentrate and tether infective HIV-1 virions on the DC surface, which could facilitate HIV-1 transfer to CD4 and CCR5 proteins on CD4+ T cells in a manner similar to that for cis transfer. At the end, the two separate pathways depicted in Fig. 2 merge in trans infection of CD4+ T cells, in which DC-SIGN is of vital importance, as demonstrated by Geijtenbeek et al. [2]. To summarize, DC-SIGN dictates the mode of HIV-1 infection, from DC infection and immune system modulation to HIV-1 transmission and dissemination, and HIV-1 clearly benefits from DC-SIGN-mediated signalling. The recent studies discussed above demonstrate that other pathogens may share a similar mechanism of host infection. From this point of view, inhibition of pathogen interaction with DC-SIGN is a plausible concept for new anti-infectives, preventing not only localized infection of DCs, but also pathogen dissemination.
Acknowledgments
The authors thank Professor Roger Pain for critical reading of the manuscript. This work was supported by a grant from the Research Agency of the Republic of Slovenia (grant P1-0208 to MA) and by the Women in Science UNESCO award (NO).
References
- 1.Banchereau J., Briere F., Caux C., Davoust J., Lebecque S., Liu Y.J., Pulendran B., Palucka K. Annu. Rev. Immunol. 2000;18:767. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 2.Geijtenbeek T.B., Torensma R., van Vliet S.J., van Duijnhoven G.C., Adema G.J., van Kooyk Y., Figdor C.G. Cell. 2000;100(5):575. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 3.Figdor C.G., van Kooyk Y., Adema G.J. Nat. Rev. Immunol. 2002;2(2):77. doi: 10.1038/nri723. [DOI] [PubMed] [Google Scholar]
- 4.Tacken P.J., de Vries I.J., Gijzen K., Joosten B., Wu D., Rother R.P., Faas S.J., Punt C.J., Torensma R., Adema G.J., Figdor C.G. Blood. 2005;106(4):1278. doi: 10.1182/blood-2005-01-0318. [DOI] [PubMed] [Google Scholar]
- 5.Bonifaz L.C., Bonnyay D.P., Charalambous A., Darguste D.I., Fujii S., Soares H., Brimnes M.K., Moltedo B., Moran T.M., Steinman R.M. J. Exp. Med. 2004;199(6):815. doi: 10.1084/jem.20032220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J., Cornelissen I.L., Nottet H.S., KewalRamani V.N., Littman D.R., Figdor C.G., van Kooyk Y. Cell. 2000;100(5):587. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 7.Geijtenbeek T.B., den Dunnen J., Gringhuis S.I. Future Microbiol. 2009;4:879. doi: 10.2217/fmb.09.51. [DOI] [PubMed] [Google Scholar]
- 8.Gringhuis S.I., den Dunnen J., Litjens M., van Het Hof B., van Kooyk Y., Geijtenbeek T.B. Immunity. 2007;26(5):605. doi: 10.1016/j.immuni.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Hawiger D., Inaba K., Dorsett Y., Guo M., Mahnke K., Rivera M., Ravetch J.V., Steinman R.M., Nussenzweig M.C. J. Exp. Med. 2001;194(6):769. doi: 10.1084/jem.194.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geijtenbeek T.B., van Vliet S.J., Engering A., t Hart B.A., van Kooyk Y. Annu. Rev. Immunol. 2004;22:33. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- 11.Geijtenbeek T.B., Van Vliet S.J., Koppel E.A., Sanchez-Hernandez M., Vandenbroucke-Grauls C.M., Appelmelk B., Van Kooyk Y. J. Exp. Med. 2003;197(1):7. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geijtenbeek T.B., Krooshoop D.J., Bleijs D.A., van Vliet S.J., van Duijnhoven G.C., Grabovsky V., Alon R., Figdor C.G., van Kooyk Y. Nat. Immunol. 2000;1(4):353. doi: 10.1038/79815. [DOI] [PubMed] [Google Scholar]
- 13.Engering A., Van Vliet S.J., Geijtenbeek T.B., Van Kooyk Y. Blood. 2002;100(5):1780. doi: 10.1182/blood-2001-12-0179. [DOI] [PubMed] [Google Scholar]
- 14.Relloso M., Puig-Kroger A., Pello O.M., Rodriguez-Fernandez J.L., de la Rosa G., Longo N., Navarro J., Munoz-Fernandez M.A., Sanchez-Mateos P., Corbi A.L. J. Immunol. 2002;168(6):2634. doi: 10.4049/jimmunol.168.6.2634. [DOI] [PubMed] [Google Scholar]
- 15.Puig-Kroger A., Serrano-Gomez D., Caparros E., Dominguez-Soto A., Relloso M., Colmenares M., Martinez-Munoz L., Longo N., Sanchez-Sanchez N., Rincon M., Rivas L., Sanchez-Mateos P., Fernandez-Ruiz E., Corbi A.L. J. Biol. Chem. 2004;279(24):25680. doi: 10.1074/jbc.M311516200. [DOI] [PubMed] [Google Scholar]
- 16.Dickensheets H.L., Venkataraman C., Schindler U., Donnelly R.P. Proc. Natl. Acad. Sci. U. S. A. 1999;96(19):10800. doi: 10.1073/pnas.96.19.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickensheets H.L., Donnelly R.P. J. Leukoc. Biol. 1999;65(3):307. doi: 10.1002/jlb.65.3.307. [DOI] [PubMed] [Google Scholar]
- 18.Piemonti L., Monti P., Allavena P., Sironi M., Soldini L., Leone B.E., Socci C., Di Carlo V. J. Immunol. 1999;162(11):6473. [PubMed] [Google Scholar]
- 19.Bianchi M., Meng C., Ivashkiv L.B. Proc. Natl. Acad. Sci. U. S. A. 2000;97(17):9573. doi: 10.1073/pnas.160099797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez-Soto A., Puig-Kroger A., Vega M.A., Corbi A.L. J. Biol. Chem. 2005;280(39):33123. doi: 10.1074/jbc.M503401200. [DOI] [PubMed] [Google Scholar]
- 21.Bakri Y., Sarrazin S., Mayer U.P., Tillmanns S., Nerlov C., Boned A., Sieweke M.H. Blood. 2005;105(7):2707. doi: 10.1182/blood-2004-04-1448. [DOI] [PubMed] [Google Scholar]
- 22.Kwon D.S., Gregorio G., Bitton N., Hendrickson W.A., Littman D.R. Immunity. 2002;16(1):135. doi: 10.1016/s1074-7613(02)00259-5. [DOI] [PubMed] [Google Scholar]
- 23.Hodges A., Sharrocks K., Edelmann M., Baban D., Moris A., Schwartz O., Drakesmith H., Davies K., Kessler B., McMichael A., Simmons A. Nat. Immunol. 2007;8(6):569. doi: 10.1038/ni1470. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell D.A., Fadden A.J., Drickamer K. J. Biol. Chem. 2001;276(31):28939. doi: 10.1074/jbc.M104565200. [DOI] [PubMed] [Google Scholar]
- 25.Frison N., Taylor M.E., Soilleux E., Bousser M.T., Mayer R., Monsigny M., Drickamer K., Roche A.C. J. Biol. Chem. 2003;278(26):23922. doi: 10.1074/jbc.M302483200. [DOI] [PubMed] [Google Scholar]
- 26.Feinberg H., Guo Y., Mitchell D.A., Drickamer K., Weis W.I. J. Biol. Chem. 2005;280(2):1327. doi: 10.1074/jbc.M409925200. [DOI] [PubMed] [Google Scholar]
- 27.Cambi A., Figdor C.G. Curr. Opin. Immunol. 2005;17(4):345. doi: 10.1016/j.coi.2005.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmskov U., Thiel S., Jensenius J.C. Annu. Rev. Immunol. 2003;21:547. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 29.Menon S., Rosenberg K., Graham S.A., Ward E.M., Taylor M.E., Drickamer K., Leckband D.E. Proc. Natl. Acad. Sci. U. S. A. 2009;106(28):11524. doi: 10.1073/pnas.0901783106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cambi A., de Lange F., van Maarseveen N.M., Nijhuis M., Joosten B., van Dijk E.M., de Bakker B.I., Fransen J.A., Bovee-Geurts P.H., van Leeuwen F.N., Van Hulst N.F., Figdor C.G. J. Cell. Biol. 2004;164(1):145. doi: 10.1083/jcb.200306112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koopman M., Cambi A., de Bakker B.I., Joosten B., Figdor C.G., van Hulst N.F., Garcia-Parajo M.F. FEBS Lett. 2004;573(1–3):6. doi: 10.1016/j.febslet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 32.Feinberg H., Mitchell D.A., Drickamer K., Weis W.I. Science. 2001;294(5549):2163. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 33.Guo Y., Feinberg H., Conroy E., Mitchell D.A., Alvarez R., Blixt O., Taylor M.E., Weis W.I., Drickamer K. Nat. Struct. Mol. Biol. 2004;11(7):591. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Vallejo J.J., van Liempt E., da Costa Martins P., Beckers C., van het Hof B., Gringhuis S.I., Zwaginga J.J., van Dijk W., Geijtenbeek T.B., van Kooyk Y., van Die I. Mol. Immunol. 2008;45(8):2359. doi: 10.1016/j.molimm.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Cambi A., Gijzen K., de Vries J.M., Torensma R., Joosten B., Adema G.J., Netea M.G., Kullberg B.J., Romani L., Figdor C.G. Eur. J. Immunol. 2003;33(2):532. doi: 10.1002/immu.200310029. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig I.S., Lekkerkerker A.N., Depla E., Bosman F., Musters R.J., Depraetere S., van Kooyk Y., Geijtenbeek T.B. J. Virol. 2004;78(15):8322. doi: 10.1128/JVI.78.15.8322-8332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azad A.K., Torrelles J.B., Schlesinger L.S. J. Leukoc. Biol. 2008;84(6):1594. doi: 10.1189/jlb.0308192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahnke K., Guo M., Lee S., Sepulveda H., Swain S.L., Nussenzweig M., Steinman R.M. J. Cell. Biol. 2000;151(3):673. doi: 10.1083/jcb.151.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith A.L., Ganesh L., Leung K., Jongstra-Bilen J., Jongstra J., Nabel G.J. J. Exp. Med. 2007;204(2):421. doi: 10.1084/jem.20061604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cambi A., Beeren I., Joosten B., Fransen J.A., Figdor C.G. Eur. J. Immunol. 2009;39(7):1923. doi: 10.1002/eji.200939351. [DOI] [PubMed] [Google Scholar]
- 41.Cambi A., Lidke D.S., Arndt-Jovin D.J., Figdor C.G., Jovin T.M. Nano Lett. 2007;7(4):970. doi: 10.1021/nl0700503. [DOI] [PubMed] [Google Scholar]
- 42.Neumann A.K., Thompson N.L., Jacobson K. J. Cell. Sci. 2008;121(Pt 5):634. doi: 10.1242/jcs.022418. [DOI] [PubMed] [Google Scholar]
- 43.Klimstra W.B., Nangle E.M., Smith M.S., Yurochko A.D., Ryman K.D. J. Virol. 2003;77(22):12022. doi: 10.1128/JVI.77.22.12022-12032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lozach P.Y., Lortat-Jacob H., de Lacroix de Lavalette A., Staropoli I., Foung S., Amara A., Houles C., Fieschi F., Schwartz O., Virelizier J.L., Arenzana-Seisdedos F., Altmeyer R. J. Biol. Chem. 2003;278(22):20358. doi: 10.1074/jbc.M301284200. [DOI] [PubMed] [Google Scholar]
- 45.Tassaneetrithep B., Burgess T.H., Granelli-Piperno A., Trumpfheller C., Finke J., Sun W., Eller M.A., Pattanapanyasat K., Sarasombath S., Birx D.L., Steinman R.M., Schlesinger S., Marovich M.A. J. Exp. Med. 2003;197(7):823. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin G., Simmons G., Pohlmann S., Baribaud F., Ni H., Leslie G.J., Haggarty B.S., Bates P., Weissman D., Hoxie J.A., Doms R.W. J. Virol. 2003;77(2):1337. doi: 10.1128/JVI.77.2.1337-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., Steinkasserer A., Becker S., Bates P., Hofmann H., Pohlmann S. J. Virol. 2004;78(21):12090. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. J. Virol. 2004;78(11):5642. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Regan A.D., Whittaker G.R. J. Virol. 2008;82(23):11992. doi: 10.1128/JVI.01094-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S.F., Huang J.C., Lee Y.M., Liu S.J., Chan Y.J., Chau Y.P., Chong P., Chen Y.M. Biochem. Biophys. Res. Commun. 2008;373(4):561. doi: 10.1016/j.bbrc.2008.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaudart N., Ekpo P., Pattanapanyasat K., van Kooyk Y., Engering A. FEMS Immunol. Med. Microbiol. 2008;53(3):359. doi: 10.1111/j.1574-695X.2008.00437.x. [DOI] [PubMed] [Google Scholar]
- 52.Appelmelk B.J., van Die I., van Vliet S.J., Vandenbroucke-Grauls C.M., Geijtenbeek T.B., van Kooyk Y. J. Immunol. 2003;170(4):1635. doi: 10.4049/jimmunol.170.4.1635. [DOI] [PubMed] [Google Scholar]
- 53.Colmenares M., Constant S.L., Kima P.E., McMahon-Pratt D. Infect. Immun. 2002;70(12):6597. doi: 10.1128/IAI.70.12.6597-6605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogoevska V., Nollau P., Lucka L., Grunow D., Klampe B., Uotila L.M., Samsen A., Gahmberg C.G., Wagener C. Glycobiology. 2007;17(3):324. doi: 10.1093/glycob/cwl073. [DOI] [PubMed] [Google Scholar]
- 55.Geijtenbeek T.B., van Duijnhoven G.C., van Vliet S.J., Krieger E., Vriend G., Figdor C.G., van Kooyk Y. J. Biol. Chem. 2002;277(13):11314. doi: 10.1074/jbc.M111532200. [DOI] [PubMed] [Google Scholar]
- 56.Maeda N., Nigou J., Herrmann J.L., Jackson M., Amara A., Lagrange P.H., Puzo G., Gicquel B., Neyrolles O. J. Biol. Chem. 2003;278(8):5513. doi: 10.1074/jbc.C200586200. [DOI] [PubMed] [Google Scholar]
- 57.Baccarini M. FEBS Lett. 2005;579(15):3271. doi: 10.1016/j.febslet.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 58.Wellbrock C., Karasarides M., Marais R. Nat. Rev. Mol. Cell. Biol. 2004;5(11):875. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 59.Caparros E., Munoz P., Sierra-Filardi E., Serrano-Gomez D., Puig-Kroger A., Rodriguez-Fernandez J.L., Mellado M., Sancho J., Zubiaur M., Corbi A.L. Blood. 2006;107(10):3950. doi: 10.1182/blood-2005-03-1252. [DOI] [PubMed] [Google Scholar]
- 60.Chen Q.L., Zhu S.Y., Bian Z.Q., Zhao L.J., Cao J., Pan W., Qi Z.T. Cell Biochem. Biophys. 2010;56(1):49. doi: 10.1007/s12013-009-9069-0. [DOI] [PubMed] [Google Scholar]
- 61.Vallabhapurapu S., Karin M. Annu. Rev. Immunol. 2009;27:693. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 62.Hayden M.S., Ghosh S. Genes Dev. 2004;18(18):2195. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 63.Li Q., Verma I.M. Nat. Rev. Immunol. 2002;2(10):725. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 64.Ashburner B.P., Westerheide S.D., Baldwin A.S., Jr. Mol. Cell. Biol. 2001;21(20):7065. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen L.F., Mu Y., Greene W.C. EMBO J. 2002;21(23):6539. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anguita J., Ramamoorthi N., Hovius J.W., Das S., Thomas V., Persinski R., Conze D., Askenase P.W., Rincon M., Kantor F.S., Fikrig E. Immunity. 2002;16(6):849. doi: 10.1016/s1074-7613(02)00325-4. [DOI] [PubMed] [Google Scholar]
- 67.Paveglio S.A., Allard J., Mayette J., Whittaker L.A., Juncadella I., Anguita J., Poynter M.E. J. Immunol. 2007;178(11):7064. doi: 10.4049/jimmunol.178.11.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hovius J.W., de Jong M.A., den Dunnen J., Litjens M., Fikrig E., van der Poll T., Gringhuis S.I., Geijtenbeek T.B. PLoS Pathog. 2008;4(2):e31. doi: 10.1371/journal.ppat.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee B., Leslie G., Soilleux E., O'Doherty U., Baik S., Levroney E., Flummerfelt K., Swiggard W., Coleman N., Malim M., Doms R.W. J. Virol. 2001;75(24):12028. doi: 10.1128/JVI.75.24.12028-12038.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gringhuis S.I., den Dunnen J., Litjens M., van der Vlist M., Geijtenbeek T.B. Nat. Immunol. 2009;10(10):1081. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- 71.Chatterjee D., Khoo K.H. Glycobiology. 1998;8(2):113. doi: 10.1093/glycob/8.2.113. [DOI] [PubMed] [Google Scholar]
- 72.Xu S., Cooper A., Sturgill-Koszycki S., van Heyningen T., Chatterjee D., Orme I., Allen P., Russell D.G. J. Immunol. 1994;153(6):2568. [PubMed] [Google Scholar]
- 73.Sturgill-Koszycki S., Schlesinger P.H., Chakraborty P., Haddix P.L., Collins H.L., Fok A.K., Allen R.D., Gluck S.L., Heuser J., Russell D.G. Science. 1994;263(5147):678. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 74.Pitarque S., Herrmann J.L., Duteyrat J.L., Jackson M., Stewart G.R., Lecointe F., Payre B., Schwartz O., Young D.B., Marchal G., Lagrange P.H., Puzo G., Gicquel B., Nigou J., Neyrolles O. Biochem. J. 2005;392(Pt 3):615. doi: 10.1042/BJ20050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torrelles J.B., Azad A.K., Schlesinger L.S. J. Immunol. 2006;177(3):1805. doi: 10.4049/jimmunol.177.3.1805. [DOI] [PubMed] [Google Scholar]
- 76.Geurtsen J., Chedammi S., Mesters J., Cot M., Driessen N.N., Sambou T., Kakutani R., Ummels R., Maaskant J., Takata H., Baba O., Terashima T., Bovin N., Vandenbroucke-Grauls C.M., Nigou J., Puzo G., Lemassu A., Daffe M., Appelmelk B.J. J. Immunol. 2009;183(8):5221. doi: 10.4049/jimmunol.0900768. [DOI] [PubMed] [Google Scholar]
- 77.Mittal R., Bulgheresi S., Emami C., Prasadarao N.V. J. Immunol. 2009;183(10):6588. doi: 10.4049/jimmunol.0902029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Konstantinov S.R., Smidt H., de Vos W.M., Bruijns S.C., Singh S.K., Valence F., Molle D., Lortal S., Altermann E., Klaenhammer T.R., van Kooyk Y. Proc. Natl. Acad. Sci. U. S. A. 2008;105(49):19474. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smits H.H., Engering A., van der Kleij D., de Jong E.C., Schipper K., van Capel T.M., Zaat B.A., Yazdanbakhsh M., Wierenga E.A., van Kooyk Y., Kapsenberg M.L. J. Allergy Clin. Immunol. 2005;115(6):1260. doi: 10.1016/j.jaci.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 80.van Kooyk Y., Geijtenbeek T.B. Nat. Rev. Immunol. 2003;3(9):697. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- 81.Okano M., Satoskar A.R., Nishizaki K., Abe M., Harn D.A., Jr. J. Immunol. 1999;163(12):6712. [PubMed] [Google Scholar]
- 82.Appelmelk B.J., Shiberu B., Trinks C., Tapsi N., Zheng P.Y., Verboom T., Maaskant J., Hokke C.H., Schiphorst W.E., Blanchard D., Simoons-Smit I.M., van den Eijnden D.H., Vandenbroucke-Grauls C.M. Infect. Immun. 1998;66(1):70. doi: 10.1128/iai.66.1.70-76.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bergman M.P., Engering A., Smits H.H., van Vliet S.J., van Bodegraven A.A., Wirth H.P., Kapsenberg M.L., Vandenbroucke-Grauls C.M., van Kooyk Y., Appelmelk B.J. J. Exp. Med. 2004;200(8):979. doi: 10.1084/jem.20041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steeghs L., van Vliet S.J., Uronen-Hansson H., van Mourik A., Engering A., Sanchez-Hernandez M., Klein N., Callard R., van Putten J.P., van der Ley P., van Kooyk Y., van de Winkel J.G. Cell Microbiol. 2006;8(2):316. doi: 10.1111/j.1462-5822.2005.00623.x. [DOI] [PubMed] [Google Scholar]
- 85.de Witte L., Abt M., Schneider-Schaulies S., van Kooyk Y., Geijtenbeek T.B. J. Virol. 2006;80(7):3477. doi: 10.1128/JVI.80.7.3477-3486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bieback K., Lien E., Klagge I.M., Avota E., Schneider-Schaulies J., Duprex W.P., Wagner H., Kirschning C.J., Ter Meulen V., Schneider-Schaulies S. J. Virol. 2002;76(17):8729. doi: 10.1128/JVI.76.17.8729-8736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hong P.W., Flummerfelt K.B., de Parseval A., Gurney K., Elder J.H., Lee B. J. Virol. 2002;76(24):12855. doi: 10.1128/JVI.76.24.12855-12865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brenchley J.M., Price D.A., Schacker T.W., Asher T.E., Silvestri G., Rao S., Kazzaz Z., Bornstein E., Lambotte O., Altmann D., Blazar B.R., Rodriguez B., Teixeira-Johnson L., Landay A., Martin J.N., Hecht F.M., Picker L.J., Lederman M.M., Deeks S.G., Douek D.C. Nat. Med. 2006;12(12):1365. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 89.Redpath S., Ghazal P., Gascoigne N.R. Trends Microbiol. 2001;9(2):86. doi: 10.1016/s0966-842x(00)01919-3. [DOI] [PubMed] [Google Scholar]
- 90.Gilchrist M., Thorsson V., Li B., Rust A.G., Korb M., Roach J.C., Kennedy K., Hai T., Bolouri H., Aderem A. Nature. 2006;441(7090):173. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 91.Bernhard O.K., Lai J., Wilkinson J., Sheil M.M., Cunningham A.L. J. Biol. Chem. 2004;279(50):51828. doi: 10.1074/jbc.M402741200. [DOI] [PubMed] [Google Scholar]
- 92.Trumpfheller C., Park C.G., Finke J., Steinman R.M., Granelli-Piperno A. Int. Immunol. 2003;15(2):289. doi: 10.1093/intimm/dxg030. [DOI] [PubMed] [Google Scholar]
- 93.Sodhi A., Montaner S., Gutkind J.S. Nat. Rev. Mol. Cell. Biol. 2004;5(12):998. doi: 10.1038/nrm1529. [DOI] [PubMed] [Google Scholar]
- 94.van Kooyk Y., Appelmelk B., Geijtenbeek T.B. Trends Mol. Med. 2003;9(4):153. doi: 10.1016/s1471-4914(03)00027-3. [DOI] [PubMed] [Google Scholar]
- 95.Sol-Foulon N., Moris A., Nobile C., Boccaccio C., Engering A., Abastado J.P., Heard J.M., van Kooyk Y., Schwartz O. Immunity. 2002;16(1):145. doi: 10.1016/s1074-7613(02)00260-1. [DOI] [PubMed] [Google Scholar]