Abstract
Spike (S) protein is the most important membrane protein on the surface of severe acute respiratory syndrome coronavirus (SARS-CoV). It associates with cellular receptors to mediate infection of their target cells. Inspired by such a mechanism, an in-depth investigation into the genome sequences of S protein of SARS-CoV and its receptor are conducted thru a mathematical transformation and graphic approach. As an outcome, a novel method for visualizing the characteristic of SARS-CoV is suggested. An extensive comparison among a large number of genome sequences has proved that the characteristic thus revealed is unique for SARS-CoV. As such, the characteristic can be regarded as the fingerprint map of SARS-CoV for diagnostic usage. Moreover, the conclusion has been further supported in a real case in Guangdong province of China. The fingerprint map proposed here has the merits of clear visibility and reliability that can serve as a complementary clinical tool for detecting SARS-CoV, particularly for the cases where the results obtained by the conventional methods are uncertain or conflicted with each other.
Abbreviations: SARS-CoV, severe acute respiratory syndrome coronavirus; ACE2, angiotensin-converting enzyme 2; S protein, spike protein
Keywords: SARS-CoV, Fingerprint map, Angiotensin-converting enzyme 2, Spike protein, Z-curve, Graphical approach
1. Introduction
Severe acute respiratory syndrome (SARS), which was first recognized in Guangdong province of China in November 2002, emerged as a life-threatening disease associated with pneumonia. A novel coronavirus, called SARS-coronavirus or SARS-CoV, has been proved to be the cause of SARS [1], [2], [3]. Since then, progresses in finding inhibitors against SARA from different angles have been reported (see, e.g. [4], [5], [6], [7], [8], [9]). It is also important to timely and accurately diagnose SARS-CoV, which can help understand the mechanism of causing the disease and optimize the healing treatment. Unfortunately, it is quite slow to use the routine clinical procedures to diagnose SARS-CoV, as illustrated below.
SARS-CoV-specific RNA can be detected in various clinical specimens such as blood, stool, respiratory secretions, and body tissues by the polymerase chain reaction (PCR) [3]. Although the existing PCR tests were made available during the outbreak, their sensitivity and specificity were unknown because there were no “gold standard” laboratory or clinical definitions for the diagnosis of SARS [10], [11], [12]. For instance, the SARS-CoV can be detected by inoculating suitable cell cultures (e.g. Vero cells) with patient specimens (such as respiratory secretions) and propagating the virus in vitro, but the problem is: although the positive cell culture results indicate the existence of SARS-CoV in the sample tested; the negative results do not exclude SARS. Again, although the enzyme-linked immunosorbent assay (ELISA), immunofluorescence assay (IFA), and neutralization test (NT) have the strongpoint of convenience, reliability and repeatability, they are unsuitable for the acute illness [12]. Therefore, the existing diagnostic tests lack sufficient sensitivity for clinical usage in timely ruling out SARS.
From the emergence of SARS-CoV, many studies have been done on its biological medicine aspects, such as pathogeny characteristics, mechanisms of causing disease, clinic diagnosis and treatment, bacterin development and the spreading rules. At the level of molecule biology, studies have been conducted on its genome sequences’ characteristic, structures and functions of the translated proteins, and the evolution relationships in sequences. SARS-CoV genomes are distinguished by the presence of a single stranded plus-sense RNA genome about 30 kb in length. Eleven open reading frames (ORF) of the viral genome are translated into 23 proteins, including four main structural proteins: Spike (S) protein, Membrane (M) protein, Envelope (E) protein, and Nucleocapsid (N) protein [13], [14], [15]. Research on other known coronaviruses has proved that among the structural proteins of coronaviruses, S protein plays a very important role in virus entry, virus–receptor interactions and their relationship to tropism [16], [17]. The S proteins of coronaviruses are large type-I transmembrane glycoproteins that are responsible for receptor building and membrane fusion. On 26 November 2003, angiotensin-converting enzyme 2 (ACE2) was identified as a functional receptor for the SARS-CoV, and S protein associate with cellular receptors to mediate infection of their target cells. Recently, in virtue of giant human antibody libraries, a human monoclonal antibody, which potently neutralizes SARS-CoV and inhibits syncytia formation between cells expressing the S protein and those expressing the SARS-CoV receptor ACE2, was identified [18]. Research on molecular evolution of the SARS-CoV reveals that the highest rate of mutation is seen in the part of the virus genome coding for the S protein. Research data suggests that through the change of S protein, the early SARS-CoV was under significant pressure to mutate in order to become an efficient human virus [19]. In addition, the research on S protein of SARS-CoV will do great favor to clinic diagnosis and bacterin development [20], [21], [22].
In view of the important roles of S protein in SARS-CoV's infection and evolution, in the current study we will regard S protein as the primary research object, investigating it at a deeper level. The previous studies on virus gene sequences only concern about the structure or property of the virus gene sequence by itself, seldom relating the investigation to the other objects such as the receptor for the virus. Here we shall take into account the interaction of SARS-CoV and its receptor. It has been observed that that after a certain kind of mathematic transformation, there is an obvious symmetric characteristic shown in gene sequences between SARS-CoV S protein segment and human ACE2; however, no such a characteristic shown in gene sequences between non-SARS-CoV S protein segment and human ACE2. The characteristic can be regarded as a fingerprint map and be applied to SARS virus detection, thus providing a simple and intuitive lab detection method to complement the traditional clinical and epidemiological methods.
2. Methods
Graphical approaches have been successfully used to deal with many biologically interesting problems, such as enzyme kinetics [23], [24], [25], [26], protein folding kinetics [27], analysis of codon usage in E. coli protein coding sequences [28] and HIV protein sequences [29], among many others. The advantages of graphical approach are in providing an intuitive picture, helping investigators visualize a very complicated or abstract problem and catch its essence or signature (see, e.g. [30], [31], [32], [33], [34], [35], [36], [37], [38]). Here let us use graphical approach to study the interaction of SARS-CoV and its receptor ACE2, and see if there is any special feature in the embodiment of their sequences. The mathematic transformation method presented in this paper is based on the graphic approach developed by the previous investigators [28], [29], [39], [40]. Using the graphic approach, or Z-curve method [41], we can obtain the transformed graph of the sequences and find the visualization characteristic between SARS-CoV and its receptor ACE2. Also, we do the same analysis on the S protein of non-SARS-CoV.
The Z-curve is a novel method for mapping the DNA or RNA sequence into a folding curve in a three-dimensional space. Originally, the DNA (or RNA) sequences are expressed in terms of a series of four letters A, C, G, and T(U) that may be called the letter sequence representation (LSR) of the DNA (or RNA) sequences. Z-curve representation is one-to-one correspondence; it is a geometrical approach to express the LSR of DNA (or RNA) sequences. Such a geometric form of DNA (or RNA) sequence displays the new characters of sequence like symmetry, periodicity and local motif, thus providing a brand-new method for decryption of DNA or RNA sequence. The method is presented briefly as follows. Consider one strand DNA or RNA sequence with N bases. Suppose that the cumulative numbers of bases A, C, G, and T(U) occurring in this subsequence from the 1st base to the nth base in the sequence are denoted by A n, C n, G n, and T n, respectively. The Z-curve is composed of a series of nodes P 0, P 1, P 2, …, P N whose coordinates x n, y n and z n (n = 0, 1, 2, …, N) are uniquely determined by the so-called Z-transform of DNA (RNA) sequence:
| (1) |
Let us define A 0 = C 0 = G 0 = T 0 = 0. The Z curve is defined as the connection of the nodes P 0, P 1, P 2, …, P N one by one sequentially with straight lines. Define x 0 = y 0 = z 0 = 0 so that the Z-curve always starts from the origin of the three-dimensional coordinate system.
3. Results and discussion
The gene sequences in our experiment with the annotation information were downloaded from the website of NCBI RefSeq project (http://www.ncbi.nih.gov/RefSeq). We have conducted experiments on the following three kinds of sequences: (1) the S protein segments of more than 80 SARS complete gene sequences; (2) human ACE2 sequences (with the accession numbers of AB046569, NM_021804, AF291820, and AY358714); (3) non-SARS coronavirus sequences including house mouse, cotton aphid, Norway rat (with the accession numbers of BC026801, AB053181, AB053182, YSCACE2, XM_228924, XM_136130, AF502082, and AB122152).
We analyzed the visualization characteristics of the above sequences. The result, derived from the comparisons among a large number of gene sequences, shows a regular quadrilateral between SARS-CoV S protein segment and human ACE2 (cf. Fig. 1 ). However, there is no such feature between non-SARS coronavirus S protein segment and human ACE2 (cf. Fig. 2 ). Such a unique feature represents a fingerprint map that can be used to identify and distinguish SARS from other infectious diseases, helping the diagnosis of some ambiguous SARS cases.
Fig. 1.
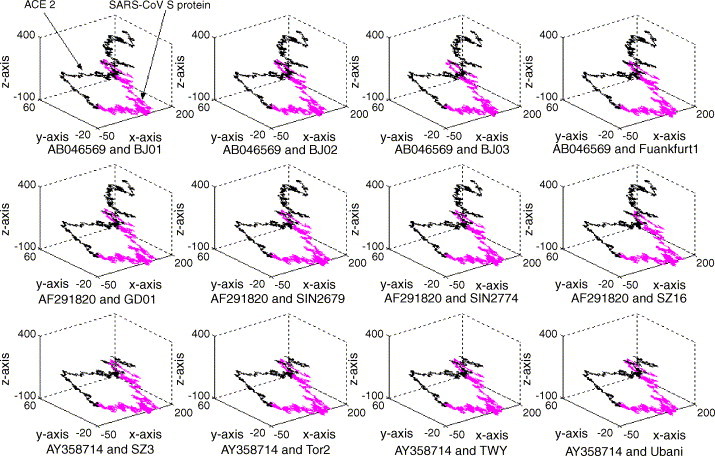
The fingerprint map of the SARS-CoV S protein segment formed by the mathematic transformation of Eq. (1). In each sub-graph, the black part in the left is formed by human ACE2 gene sequence while the purplish part in the right formed by S protein segment.
Fig. 2.
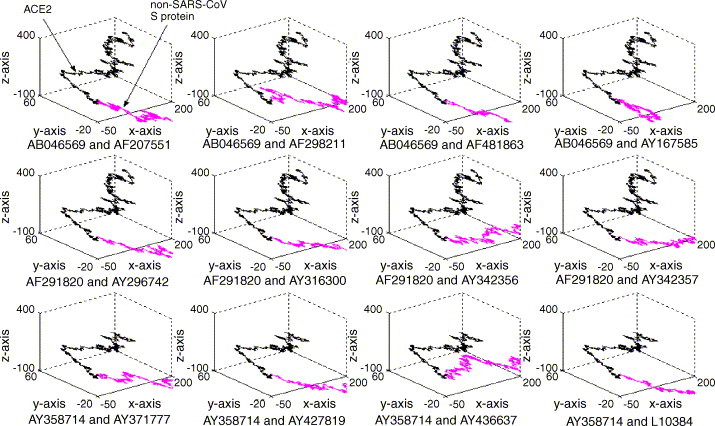
The representation generated by the same procedure as that of Fig. 1 for the non-SARS coronavirus S protein segment. See the legend of Fig. 1 for further explanation.
Using the fingerprint map, we have analyzed a suspected case in Guangdong province of China. It was quickly found by our method that the S protein of suspected virus did display a regular quadrilateral between the S protein of suspected case and the four kinds of human ACE2 sequences, as shown in Fig. 3 . Such character can help us draw the conclusion that this is a SARS-CoV infected case, which has been confirmed by a series of conventional methods performed later.
Fig. 3.
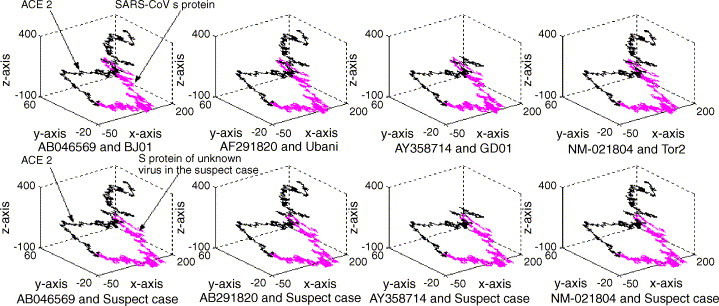
The representation generated for the corresponding segment from a diagnose SARS case in Guangdong province of China. See the legend of Fig. 1 for further explanation.
4. Conclusions
Gene sequence carries all the information of an organism. Since the sequence is a one-dimensional, it is not easy to get the genetic information directly from the sequence itself. The transformation from gene sequence to the visualized graph with a unique feature is an effective way to observe and study the characteristics of gene sequences. Taking the interaction of SARS-CoV and its receptor into consideration, we have obtained a unique characteristic of SARS-CoV using the graphic approach. A visualization interactive relationship has been revealed between SARS-CoV and its receptor human ACE2, which does not exist between the non-SARS-CoV and human ACE2. Such a unique feature can be deemed as the fingerprint map of SARS-CoV that can be used to diagnose suspected cases against SARS. The outcome diagnosed by the fingerprint method has been confirmed by a real SARS case in Guangdong province of China. It has not escaped our notice that the proposed fingerprint map method, along with the method based on cellular automata images reported earlier [42], can also be used to the in-depth research for revealing the molecular mechanism of the disease.
Acknowledgments
This work was supported in part by the National Nature Science Foundation of China (Nos. 60474037 and 60004006), Program for New Century Excellent Talents in University, and Specialized Research Fund for the Doctoral Program of Higher Education from Educational Committee of China (No. 20030255009).
References
- 1.Ruan Y.J., Wei C.L., Ee A.L., Vega V.B., Thoreau H., Su S.T., Chia J.M., Ng P., Chiu K.P., Lim L., Zhang T., Peng C.K., Lin E.O., Lee N.M., Yee S.L., Ng L.F., Chee R.E., Stanton L.W., Long P.M., Liu E.T. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerberding J.L. N. Engl. J. Med. 2003;348:2030–2031. doi: 10.1056/NEJMe030067. [DOI] [PubMed] [Google Scholar]
- 3.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Chou K.C., Wei D.Q., Zhong W.Z. Biochem. Biophys. Res. Commun. 2003;308:148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du Q.S., Wang S.Q., Wei D.Q., Zhu Y., Guo H., Sirois S., Chou K.C. Peptides. 2004;25:1857–1864. doi: 10.1016/j.peptides.2004.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sirois S., Wei D.Q., Du Q.S., Chou K.C. J. Chem. Inf. Comput. Sci. 2004;44:1111–1122. doi: 10.1021/ci034270n. [DOI] [PubMed] [Google Scholar]
- 7.Du Q.S., Wang S., Wei D.Q., Sirois S., Chou K.C. Anal. Biochem. 2005;337:262–270. doi: 10.1016/j.ab.2004.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du Q.S., Wang S.Q., Jiang Z.Q., Gao W.N., Li Y.D., Wei D.Q., Chou K.C. Med. Chem. 2005;1:209–213. doi: 10.2174/1573406053765468. [DOI] [PubMed] [Google Scholar]
- 9.Chou K.C. Curr. Med. Chem. 2004;11:2105–2134. doi: 10.2174/0929867043364667. [DOI] [PubMed] [Google Scholar]
- 10.McIntosh K. Clin. Chem. 2003;49:845–846. doi: 10.1373/49.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang P., Louie M., Richardson S.E., Smieja M., Simor A.E., Jamieson F., Fearon M., Poutanen S.M., Mazzulli T., Tellier R., Mahony J., Loeb M., Petrich A., Chernesky M., McGeer A., Low D.E., Phillips E., Jones S., Bastien N., Li Y., Dick D., Grolla A., Fernando L., Booth T.F., Henry B., Rachlis A.R., Matukas L.M., Rose D.B., Lovinsky R., Walmsley S., Gold W.L., Krajden S. Cmaj. 2004;170:47–54. [PMC free article] [PubMed] [Google Scholar]
- 13.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 14.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 15.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 16.Bonavia A., Zelus B.D., Wentworth D.E., Talbot P.J., Holmes K.V. J. Virol. 2003;77:2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breslin J.J., Mork I., Smith M.K., Vogel L.K., Hemmila E.M., Bonavia A., Talbot P.J., Sjoestrom H., Noren O., Holmes K.V. J. Virol. 2003;77:4435–4438. doi: 10.1128/JVI.77.7.4435-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinese SARS Molecular Epidemiology Consortium Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 20.Pohl-Koppe A., Raabe T., Siddell S.G., ter Meulen V. J. Virol. Met. 1995;55:175–183. doi: 10.1016/0166-0934(95)00041-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C.H., Hong C.C., Seak J.C. Vet. Microbiol. 2002;85:333–342. doi: 10.1016/s0378-1135(01)00525-9. [DOI] [PubMed] [Google Scholar]
- 22.Streatfield S.J., Jilka J.M., Hood E.E., Turner D.D., Bailey M.R., Mayor J.M., Woodard S.L., Beifuss K.K., Horn M.E., Delaney D.E., Tizard I.R., Howard J.A. Vaccine. 2001;19:2742–2748. doi: 10.1016/S0264-410X(00)00512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou G.P., Deng M.H. J. Biochem. 1984;222:169–176. doi: 10.1042/bj2220169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou K.C., Jiang S.P., Liu W.M., Fee C.H. Sci. Sin. 1979;22:341–358. [Google Scholar]
- 25.Chou K.C., Forsen S. J. Biochem. 1980;187:829–835. doi: 10.1042/bj1870829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou K.C. J. Biol. Chem. 1989;264:12074–12079. [PubMed] [Google Scholar]
- 27.Chou K.C. Biophys. Chem. 1990;35:1–24. doi: 10.1016/0301-4622(90)80056-d. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C.T., Chou K.C. J. Mol. Biol. 1994;238:1–8. doi: 10.1006/jmbi.1994.1263. [DOI] [PubMed] [Google Scholar]
- 29.Chou K.C., Zhang C.T. AIDS Res. Hum. Retroviruses. 1992;8:1967–1976. doi: 10.1089/aid.1992.8.1967. [DOI] [PubMed] [Google Scholar]
- 30.Chou K.C., Kezdy F.J., Reusser F. Anal. Biochem. 1994;221:217–230. doi: 10.1006/abio.1994.1405. [DOI] [PubMed] [Google Scholar]
- 31.Althaus I.W., Gonzales A.J., Chou J.J., Diebel M.R., Chou K.C., Kezdy F.J., Romero D.L., Aristoff P.A., Tarpley W.G., Reusser F. J. Biol. Chem. 1993;268:14875–14880. [PubMed] [Google Scholar]
- 32.Althaus I.W., Chou J.J., Gonzales A.J., Diebel M.R., Chou K.C., Kezdy F.J., Romero D.L., Aristoff P.A., Tarpley W.G., Reusser F. Biochemistry. 1993;32:6548–6554. doi: 10.1021/bi00077a008. [DOI] [PubMed] [Google Scholar]
- 33.Althaus I.W., Chou J.J., Gonzales A.J., Diebel M.R., Chou K.C., Kezdy F.J., Romero D.L., Aristoff P.A., Tarpley W.G., Reusser F. J. Biol. Chem. 1993;268:6119–6124. [PubMed] [Google Scholar]
- 34.Lin S.X., Neet K.E. J. Biol. Chem. 1990;265:9670–9675. [PubMed] [Google Scholar]
- 35.Kuzmic P., Ng K.Y., Heath T.D. Anal. Biochem. 1992;200:68–73. doi: 10.1016/0003-2697(92)90278-f. [DOI] [PubMed] [Google Scholar]
- 36.Althaus I.W., Chou J.J., Gonzales A.J., Diebel M.R., Chou K.C., Kezdy F.J., Romero D.L., Aristoff P.A., Tarpley W.G., Reusser F. Experientia. 1994;50:23–28. doi: 10.1007/BF01992044. [DOI] [PubMed] [Google Scholar]
- 37.Althaus I.W., Chou J.J., Gonzales A.J., Diebel M.R., Chou K.C., Kezdy F.J., Romero D.L., Thomas R.C., Aristoff P.A., Tarpley W.G., Reusser F. Biochem. Pharmacol. 1994;47:2017–2028. doi: 10.1016/0006-2952(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 38.Althaus I.W., Chou K.C., Franks K.M., Diebel M.R., Kezdy F.J., Romero D.L., Thomas R.C., Aristoff P.A., Tarpley W.G., Reusser F. Biochem. Pharmacol. 1996;51:743–750. doi: 10.1016/0006-2952(95)02390-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhang C.T., Chou K.C. J. Protein Chem. 1993;12:329–335. doi: 10.1007/BF01028195. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C.T., Chou K.C. Amino Acids. 1996;10:253–262. doi: 10.1007/BF00807327. [DOI] [PubMed] [Google Scholar]
- 41.Chen L.L., Ou H.Y., Zhang R., Zhang C.T. Biochem. Biophys. Res. Commun. 2003;307:382–388. doi: 10.1016/S0006-291X(03)01192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M., Yao J.S., Huang Z.D., Xu Z.J., Liu G.P., Zhao H.Y., Wang X.Y., Yang J., Zhu Y.S., Chou K.C. Med. Chem. 2005;1:39–47. doi: 10.2174/1573406053402505. [DOI] [PubMed] [Google Scholar]


