Abstract
Drug discovery and development are long and financially taxing processes. On an average it takes 12–15 years and costs 1.2 billion USD for successful drug discovery and approval for clinical use. Many lead molecules are not developed further and their potential is not tapped to the fullest due to lack of resources or time constraints. In order for a drug to be approved by FDA for clinical use, it must have excellent therapeutic potential in the desired area of target with minimal toxicities as supported by both pre-clinical and clinical studies. The targeted clinical evaluations fail to explore other potential therapeutic applications of the candidate drug. Drug repurposing or repositioning is a fast and relatively cheap alternative to the lengthy and expensive de novo drug discovery and development. Drug repositioning utilizes the already available clinical trials data for toxicity and adverse effects, at the same time explores the drug's therapeutic potential for a different disease. This review addresses recent developments and future scope of drug repositioning strategy.
Keywords: Drug repurposing, Drug repositioning, Clinical trials, Lead molecule
Graphical abstract

1. Introduction
The discovery process of a potential new drug is time-consuming [1]. The development and approval of active pharmaceutical ingredient can take 10–15 years. Pharmaceutical companies spend nearly 1.2 billion U.S. dollars for bringing the new drug to the market. Researchers identify particular cellular and genetic factors that play a major role in each disease condition [2]. They target these biological markers with newly developed biological and chemical agents. Sometimes the developed molecules show drug-like effects. During the discovery process, nearly 5000 new compounds are identified and undergo various preclinical evaluations [3]. Out of these compounds, on an average approximately 5 are selected for further clinical development. After several years of clinical evaluations only one of these compounds would finally receive approval for human use [4]. Fig. 1 shows the steps involved in the drug development and drug repositioning. The discovery and developmental stages of new drug molecules include target identification (targeting cellular and genetic markers associated with a particular disease), target prioritization/validation (identification of developed molecule that have an effect on selected target), lead identification (molecules capable of treating the disease), and lead optimization (comparison of properties of various lead compounds and selection of lead compound with the greatest potential) [5,6]. Preclinical studies which follow drug discovery ensure the safety of the drug by laboratory and animal testing. After the preclinical research, investigational new drug (IND) application is submitted to FDA for permission to conduct clinical testing [7]. Clinical research phase includes phase I, II and III studies and is mainly to ensure safety and efficacy of drug in humans [8,9]. US Food and Drug Administration (US-FDA) review team examines all the submitted documents related to the drug and makes a final decision. After approval of drug by FDA, manufacturing companies conduct post-market safety monitoring (Phase IV) and post approval studies.
Fig. 1.
Various stages of drug development and drug repositioning.
Despite the huge investments and time-consuming processes, the chances of the new drug molecule clearing all the drug approval processes are often negligible [10]. So, the drug manufacturing companies focus on other viable options for drug research. National centre for advancing translational sciences (NCATS), an initiative from of national institute of health (NIH) in the united states of america (USA), defined drug repurposing as “studying the drugs that are already approved to treat one disease or condition to see if they are safe and effective for treating other diseases” [11]. In May 2012, NCATS introduced the program “Discovery of New Therapeutic Uses for Existing Molecules” to focus on new therapeutic indications for the existing drugs, drugs currently in clinical development as well as shelved and discontinued drugs [12]. This approach is also known as repositioning or drug reprofiling. Drug repurposing is a promising approach and is mainly applied for the treatment of rare genetic diseases, and it offers significant benefits to the pharmaceutical industries [13,14]. The usual drug development process may delay the translation of discovery from bench to bedside. This alternate approach overcomes most of the cost and time-consuming hurdles during the drug development process [15]. Drug repurposing was initiated with expanding interest from pharmaceutical organizations and the recognizable proof of different cheminformatics and bioinformatics findings [16]. Out of these, nearly 10% of repurposed drugs were approved by regulatory bodies and 70% are in various stages of clinical development [17].
2. History of drug repurposing
In mid 2000s sildenafil for angina was repositioned to erectile dysfunction and thalidomide for morning sickness was repositioned to multiple myeloma. The success created a big interest in repurposing which resulted in the formation of many repurposing focused startup companies [18]. Many reviews as well as reports of market research confirms repurposing has an important share in the life cycle management of products with an R&D spending of 10–50% [19]. In order to understand the history on the scope of practice of drug repurposing Nancy et al. reported a bibliometric analysis by examining in depth a few drugs. Few examples include chlorpromazine synthesized in 1950 that was indicated for use in controlling mental disorders and as a preoperative medication which was later tried for various diseases like treating whooping cough and symptoms developed in radiation therapy for cancer patients in 1972 [[20], [21], [22]]. Chloroquine was a well-known antimalarial compound synthesized in the year 1934 and was later targeted towards many other diseases including parasitic diseases (before 1960), fever, lupus skin rashes [23,24]. Table 1 summarizes various repurposed drugs which are categorized based on biological activity.
Table 1.
List of repurposed drugs categorized based on biological activity.
| Sl.no | Name of drug | Original indication | Novel indication | Reference |
|---|---|---|---|---|
| Drug Repurposing targeting CNS | ||||
| 1 | Amphetamine | Stimulant | ADHD, Hyperkinesis in children | [25,26] |
| 2 | Atomoxetine | Parkinson's disease | ADHD | [25,[27], [28], [29]] |
| 3 | Milnacipran | Depression | Fibromyalgia syndrome | [25] |
| 4 | Mifepristone | Pregnancy termination | Psychotic major depression, Cushing's syndrome | [25] |
| 5 | Duloxetine | Depression, Diabetic Neuropathies | Urinary incontinence, fibromyalgia, musculoskeletal pain, shoulder pain, back pain, osteoarthritis, Diabetic peripheral neuropathy | [30,31] |
| 6 | Chlorpromazine | Anti-emetic, antihistamine | Non-sedating tranquillizer | [32] |
| 7 | Galantamine | Polio, paralysis and anaesthesia | Alzheimer's disease | [32,33] |
| 8 | Ropinirole | Hypertension | Parkinson's disease and idiopathic restless leg syndrome | [25,32,34] |
| 9 | Mecamylamine | Hypertension and uncomplicated cases of malignant hypertension | ADHD | [32] |
| 10 | Amantadine | Influenza | Parkinson's disease | [35] |
| 11 | Infliximab | Crohn's disease | Alzheimer's disease, Different arthritis forms | [30] |
| 12 | Perindopril | Hypertension | Alzheimer's disease | [36] |
| 13 | Gabapentin | Epilepsy | Neuropathic pain | [37] |
| Drug Repurposing targeting CVS | ||||
| 14 | Paclitaxel | Cancer | Restenosis | [25,38] |
| 15 | Lidocaine | Local anesthetic | Arrhythmia | [25] |
| 16 | Aspirin | Inflammation, pain | Antiplatelet | [39] |
| 17 | Lomitapide | Lipidemia | Familial hypercholesterolemia | [40] |
| 18 | Drospirenone | Oral contraceptive | Hypertension | [36] |
| 19 | Fludrocortisone | Cerebral salt wasting syndrome | Hypertension | [36] |
| 20 | Pegvisomant | Acromegaly | Hypercholesterolemia | [36] |
| Drug Repurposing targeting cancer | ||||
| 21 | Thalidomide | Morning sickness | Multiple myeloma | [25,41] |
| 22 | Celecoxib | adult rheumatoid arthritis, osteoarthritis | Colon and breast cancer, familial adenomatous polyposis | [32,42] |
| 23 | Arsenic | Syphilis | Leukemia | [43] |
| 24 | Aspirin | Analgesic, antipyretic | Colorectal cancer | [44] |
| 25 | Disulfiram | Alcoholism | Melanoma | [45] |
| 26 | Gemcitabine | Antiviral | Cancer | [46] |
| 27 | Premetrexed | Mesothelioma | Lung cancer | [47,48] |
| 28 | Tretinoin | Acne | Leukemia | [49] |
| 29 | Crizotinib | Clinical trials for anaplastic large-cell lymphoma | NSCLC | [50] |
| 30 | Imidapril | Hypertension | Cancer cachexia | [30] |
| 31 | Leflunomide | Rheumatoid arthritis | Prostate cancer | [44,51] |
| 32 | Metformin | Diabetes mellitus | Colorectal cancer, prostate, breast, adenocarcinoma | [44] |
| 33 | Methotrexate | Acute leukemia | Hodgkin lymphoma, breast cancer, osteosarcoma | [25] |
| 34 | Minocycline | Acne | Glioma, ovarian cancer | [44] |
| 35 | Nelfinavir | AIDS | Clinical trials for multiple cancer | [50,52] |
| 36 | Nitroxoline | Antibiotic | Bladder, breast cancer | [44] |
| 37 | Noscapine | Antitussive, antimalarial,analgesic | Multiple cancer types | [44] |
| 38 | Rapamycin | Immunosuppressant | Lymphoma, leukemia colorectal cancer | [44,53] |
| 39 | Retinoic acid | Acne | Acute promyelocytic leukemia | [30] |
| 40 | Statins | Myocardial infarction | Leukemia, cancer | [44,52] |
| 41 | Sunitinib | Renal cell carcinoma, GIST | Gastrointestinal tumor, pancreatic tumors | [50,54] |
| 42 | Trastuzumab | HER2-positive breast cancer | HER2-positive metastatic gastric cancer | [50] |
| 43 | Valproic acid | Antiepileptic | Solid tumors, leukemia | [44] |
| 44 | Vesnarinone | Cardioprotective | Lymphoma, leukemia, oral cancer | [44] |
| 45 | Wortmannin | Antifungal | Leukemia | [44] |
| 46 | Zoledronic acid | Anti-bone resorption | Breast cancer, prostate cancer, multiple myeloma. | [44] |
| Drug Repurposing targeting respiratory system | ||||
| 47 | Etanercept | Rheumatoid arthritis | Asthma | [36] |
| Drug Repurposing targeting GIT | ||||
| 48 | Imatinib | BCR-ABL | GIST | [50] |
| Drug Repurposing targeting genetic, immunological response | ||||
| 49 | Zidovudine | Cancer | HIV/AIDS | [25] |
| 50 | Methotrexate | Cancer | Rheumatoid arthritis | [44] |
| 51 | Everolimus | Immunosuppressant | Pancreatic neuroendocrine tumors | [50,55] |
| 52 | Furosemide | Edema associated with congestive heart failure | Bartter syndrome | [36] |
| 53 | Hydroxychloroquine | Antiparasitic | Anti-arthritic systemic lupus erythematosus | [30] |
| 54 | Mycophenolate/Mycophenolate mofetil | Transplanted organ rejection | Renal symptoms/lupus nephritis of systemic lupus erythematosus | [30] |
| Drug Repurposing targeting pain and inflammatory diseases | ||||
| 55 | Allopurinol | Tumor lysis syndrome | Gout | [25] |
| 56 | Tofisopam | Anxiety-related conditions | Irritable bowel syndrome | [32] |
| 57 | Colchicine | Gout | Recurrent pericarditis | [[56], [57], [58]] |
| 58 | Propranolol | Hypertension | Infantile hemangioma, Migraine prophylaxis | [[59], [61], [60]] |
| 59 | Budesonide | Asthma | Ulcerative collitis | [32,62] |
| Drug Repurposing targeting metabolic disorders and complications | ||||
| 60 | Sibutramine | Depression | Obesity | [25,30] |
| 61 | Bromocriptine | Parkinson's disease | Diabetes mellitus | [63,64] |
| 62 | Colesevelam | Hyperlipidemia | Type 2 diabetes mellitus | [65,66] |
| 63 | Nortriptyline | Depression | Neuropathic pain | [67,68] |
| 64 | Pioglitazone | Type 2 diabetes mellitus | Nonalcoholic steatohepatitis | [69,70] |
| Drug Repurposing targeting reproductive system | ||||
| 65 | Fluoxetine | Depression | Premenstrual dysphoric disorder | [25,52] |
| 66 | Sildenafil | Angina | Erectile dysfunction | [25,30] |
| 67 | Dapoxetine | Analgesia and depression | Premature ejaculation | [25] |
| 68 | Tadalafil | Cardiovascular disease, Inflammation | Male erectile dysfunction | [32] |
| 69 | Apomorphine | Parkinson's disease | Erectile dysfunction | [44] |
| Drug Repurposing targeting infectious diseases | ||||
| 70 | Thalidomide | Sedation, nausea and insomnia, | ||
| Anti-emetic | Erythema nodosum leprosum in leprosy | [25,30,32,54] | ||
| 71 | Amphotericin | Antifungal | Leishmaniasis | [71,72] |
| 72 | Dapsone | Leprosy | Malaria | [73,74] |
| 73 | Miltefosine | Cancer | Cutaneous and visceral leishmaniasis | [[75], [76], [77], [78], [79], [80]] |
| Drug Repurposing targeting miscellaneous categories of diseases | ||||
| 74 | Minoxidil | Hypertension | Alopecia | [59,[81], [82], [83], [84]] |
| 75 | Bupropion | Depression | Smoking Cessation | [25,30] |
| 76 | Finasteride | Benign prostatic hyperplasia | Alopecia | [25,32] |
| 77 | Lumigan | Glaucoma | Hypotrichosis simplex | [25] |
| 78 | Phentolamine | Hypertension | Dental anesthesia reversal agent | [25,85] |
| 79 | Eflornithine | Anti-infective | Reduction of unwanted facial hair in women | [32,86] |
| 80 | Raloxifene | Prostate and breast cancer | Osteoporosis | [30] |
| 81 | Bimatoprost | Glaucoma | Promotes growth of eyelash | [[87], [88], [89], [90], [91]] |
| 82 | Doxepin | Depressive disorder | Antipruritic | [30] |
| 83 | Naltrexone | Opioid addiction | Alcohol withdrawal | [[92], [93], [94]] |
| 84 | Zileuton | Asthma | Acne | [95,96] |
CNS: central nervous system.
ADHD: Attention-Deficit/Hyperactivity Disorder.
CVS: Cardiovascular system.
NSCLC: Non-small-cell lung carcinoma.
AIDS: acquired immune deficiency syndrome.
GIST: Gastrointestinal stromal tumors.
HER2: Human epidermal growth factor receptor 2.
HIV: Human immunodeficiency virus.
3. Approaches to drug repurposing
Finding out novel disease-drug relationship is a very important issue in drug repurposing. There are various approaches to address the issue such as in silico approaches, biological approaches, experimental, mixed approaches and knowledge based approach. Various approaches to drug repurposing is summarized in Table 2 .
Table 2.
Various approaches to drug repurposing.
| Sl.no | Approaches to drug repurposing | Description |
|---|---|---|
| 1 | In silico drug repurposing | Database consists of information gathered from research, clinical trials, reports of label uses, and other published datas |
| 1.1. | Text mining approach | Mine the information and identify the connections or relationships |
| 1.2 | Cluster approaches | Can discover novel drug-target or drug-disease relationships based on the concept that biological entities and network of same module have identical characters. |
| 1.3 | Propagation approach | Useful in discovering disease-genes, disease-disease, and target- disease relationships based on the concept that information transferred from source node to network nodes and to various sub network nodes. |
| 1.4 | Semantics approach | Biological entity relationships are found out from data in medical databases and a semantic network is built also based on existing ontology network and algorithms are developed to discover relationship in the network. |
| 1.5 | Databases and resources | Maintained with the purpose of storing, managing and retrieving information. Different types of databases used include pharmacological databases, chemical databases, proteomics databases |
| 2 | Biological approaches | Models are developed based on understanding of molecular level pathways and simulate physiological environment of target proteins |
| 3 | Experimental approaches | Based on experimentation including targets screening, cell assay, animal model and clinical trials |
| 4 | Mixed approaches | It is a mixture of computational observation, biological experiments and clinical testing. |
| 5 | Knowledge-based repurposing | Based on the knowledge of researchers and doctors and their capability and skill to interpret observations. |
3.1. In silico drug repurposing
In this approach, various public databases are utilised and information is gathered from research, clinical trials, reports of label uses, and other published datas. Then, with the help of bioinformatics tools as well as artificial intelligence, interaction networks between drug targets and drugs are identified [97,98]. There are various disease-drug knowledge databases such as ChemBank [99], DrugBank [100], KEGG [101], Pubmed [102], OMIA [103] and genomic databases such as PDB [104], GenBank [105], GEO [106], MIPS [107,108].
The information helps in development of various computational approaches rapidly which are cost effective and have much less barriers [109]. In silico approaches are based on merging and analysing a plethora of information regarding drug-disease relationship [110]. Some are based on gene expression analysis after drug treatment of relevant cell lines. There are various viewpoints [[111], [112], [113]] such as repurposing based on biological networks employed or hypothesis driven and data driven [113]. Based on methodologies repurposing can be done by text mining [[114], [115], [116], [117], [118], [119], [120], [121], [122], [123], [124], [125], [126]], network-based [[127], [128], [129], [130], [131], [132], [133], [134], [135], [136], [137]], and semantic approaches [[138], [139], [140], [141]].
3.1.1. Text mining approach
In addition to a lot of exploitative information regarding drug repurposing, a large data of biological concept relationships are available from publications. Many are novel and text mining can be employed to mine the information and identify the connections or relationships. Text mining discovers novel information by extracting from several published resources with the help of a computer [142]. Text mining in biology includes, retrieval of relevant information filtered and extracted from literature; biological name entity recognition with the use of controlled vocabularies; biological information extraction and knowledge discovery with the extracted concepts. Relationships between drug-target and drug-disease can also be discovered. Natural language processing techiniques increases the development of tools for text mining and helps finding out repurposable drugs [120,121].
3.1.2. Cluster approaches
Cluster approaches can discover novel drug-target or drug-disease relationships. It is based on the concept, biological entities (protein, drug, disease etc) and network of same module have identical characters. Various modules or groups or sub networks show drug-drug or drug-target or drug-disease relations and are found using cluster algorithms. Few examples are CLIQUE [143], DBSCAN [144], OPTICS [145], STING [146].
3.1.3. Propagation approach
This method can be useful in discovering disease-genes, disease-disease, and target-disease relationships [147,148]. It is based on the concept that information transmit from source node to network nodes and to various sub network nodes. There are local and global approaches. Local approaches extract small information from network, so the prediction may not be successful in some instance [149]. Global approaches extract data from complete network and are better and currently focussed.
3.1.4. Semantics approach
It is extensively used for extracting information and images which can be applied to drug repurposing. In this approach biological entity relationships are found out from datas in medical databases and a semantic network is built also based on existing ontology network and algorithms are developed to discover relationship in the network [108].
3.1.5. Databases and resources
Advancements in biotechnology, bioinformatics and omics techniques (proteomics, genomics, metabolomics, etc) resulted in the development of several databases in biology, chemistry, medicine and pharmacology. These can be utilised for drug repurposing. A list of the various resources are listed out in Table 3 .
Table 3.
List of various resources for drug repurposing.
| Sl.no | Resources | Repurposing Approaches | Website link |
|---|---|---|---|
| 1 | Pubmed | Text Mining approach | https://www.ncbi.nlm.nih.gov/pubmed/ |
| 2 | Online Mendelian Inheritance in Animals (OMIA) | Text Mining approach | https://omia.org/home/ |
| 3 | Drugbank | Pharmacological databases | http://www.drugbank.ca/ |
| 4 | Drug versus Disease (DvD) | Pharmacological databases | www.ebi.ac.uk/saezrodriguez/dvd |
| 5 | Drug Combination Database (DCDB) | Pharmacological databases | http://www.cls.zju.edu.cn/dcdb |
| 6 | Drug Map Central (DMC) | Pharmacological databases | http://r2d2drug.org/index.html |
| 7 | Side Effect Resource (SIDER) | Pharmacological databases | http://sideeffects.embl.de |
| 8 | DailyMed (US FDA) | Medical database | http://dailymed.nlm.nih.gov/dailymed/about.cfm |
| 9 | Drugs@FDA Database | Medical database | http://www.fda.gov/Drugs/InformationOnDrugs/ |
| 10 | FAERS (US FDA) | Medical database | http://www.fda.gov/Drugs/ |
| 11 | FDALABEL (US FDA) | Medical database | https://labels.fda.gov/ |
| 12 | ChemBank | Chemical databases | http://chembank.broad.harvard.edu |
| 13 | ChEMBL | Chemical databases | https://www.ebi.ac.uk/chembl/ |
| 14 | ChemDB | Chemical databases |
http://www.chemdb.com (Service is currently not available) |
| 15 | ChemDB | Chemical databases | http://www.chemdb.com |
| 16 | PubChem | Chemical databases | https://pubchem.ncbi.nlm.nih.gov/ |
| 17 | BindingDB | Chemical databases | http://www.bindingdb.org/bind/index.jsp |
| 18 | ChemFrog | Chemical databases |
http://www.chemfrog.com (Service is currently not available) |
| 19 | Chemicalize (ChemAxon) | Chemical databases | http://www.chemicalize.org |
| 20 | ChemSpider | Chemical databases | http://www.chemspider.com |
| 21 | Protein Data Bank (PDB) | Proteomics databases | http://www.rcsb.org/pdb/home/home.do |
| 22 | Database of Interacting Proteins (DIP) | Proteomics databases | http://dip.doe-mbi.ucla.edu/dip/Main.cgi |
| 23 | Human Protein Reference Database (HPRD) | Proteomics databases | http://www.hprd.org/ |
| 24 | IntAct Molecular Interaction Database | Proteomics databases | http://www.ebi.ac.uk/intact/ |
| 25 | OCA | Proteomics databases | http://oca.weizmann.ac.il/oca-bin/ocamain |
| 26 | Sequence Read Archive (SRA) | Proteomics databases | http://www.ncbi.nlm.nih.gov/Traces/sra/ |
| 27 | Oncomine | Genomics database | https://www.oncomine.org |
| 28 | Online Mendelian Inheritance in Man (OMIM) | Genomics database | http://www.omim.org |
| 29 | Pharmacogenomics Knowledge Base (PharmGKB) | Genomics database | http://www.pharmgkb.org |
| 30 | ArrayExpress | Genomics database | http://www.ebi.ac.uk/arrayexpress/ |
| 31 | Kyoto Encyclopedia of Genes and Genomes (KEGG) | Genomics database | http://www.genome.jp/kegg |
| 32 | CellMiner | Genomics database | http://discover.nci.nih.gov/cellminer/ |
| 33 | Database for Annotation Visualization and Integrated Discovery (DAVID) | Genomics database | http://david.abcc.ncifcrf.gov |
| 34 | DbSNP | Genomics database | http://www.ncbi.nlm.nih.gov/projects/SNP/ |
| 35 | Gene Expression Atlas | Genomics database | http://www.ebi.ac.uk/gxa |
| 36 | Gene Expression Omnibus (GEO) | Genomics database | http://www.ncbi.nlm.nih.gov/geo |
| 37 | Gene Set Enrichment Analysis(GSEA) | Genomics database | http://www.broadinstitute.org/gsea |
| 38 | GeneCards Databases | Genomics database | http://www.genecards.org/ |
| 39 | International Cancer Genome Consortium | Genomics database | https://icgc.org |
| 40 | Molecular Signature Database(MsigDB) | Genomics database | http://www.broadinstitute.org/gsea/msigdb |
| 41 | Mammalian protein-protein interaction database (MIPS) | Heterogenous networks | http://mips.helmholtz-muenchen.de/proj/ppi/ |
| 42 | Search tool for interactions of chemicals (STRING) | Biological database/Proteomics databases | http://string-db.org/ |
| 43 | Biological General Repository for Interaction Datasets (BioGRID) | Biological database | http://thebiogrid.org/ |
| 44 | Cancer Cell Line Encyclopedia (CCLE) | Biological database | http://www.broadinstitute.org/ccle |
| 45 | GPCR-Ligand Database (GLIDA) | Biological approach | http://pharminfo.pharm.kyoto-u.ac.jp/services/glida/ |
| 46 | NCI Pathway Interaction Database (NCI-PID) | Biological approaches |
http://pid.nci.nih.gov/ (Service is currently not available) |
| 47 | OPM(membrane proteins) | Biological approaches | http://opm.phar.umich.edu |
| 48 | Pathway Commons | Biological approaches | http://www.pathwaycommons.org/about/ |
| 49 | Psychoactive Drug Screening Program Ki (PDSP Ki) | Biological approaches | https://pdspdb.unc.edu/pdspWeb/ |
| 50 | Reactome | Biological approaches | http://www.reactome.org |
| 51 | Human Metabolome Database (HMDB) | Metabolomics databases | http://www.hmdb.ca |
| 52 | Clinicaltrial.gov | Experimental approaches | http://clinicaltrials.gov |
| 53 | Library of Integrated Network based Cellular Signatures(LINCS) | Experimental approaches | http://www.lincsproject.org |
| 54 | NCGC Database | Experimental approaches | https://tripod.nih.gov/npc/ |
| 55 | Princeton University MicroArray database (PUMAdb) | Experimental approaches | http://puma.princeton.edu |
| 56 | Stanford Microarray Database | Experimental approaches |
http://smd.princeton.edu (Service is currently not available) |
| 57 | Collaborative Drug Discovery Vault | Mixed approaches | https://www.collaborativedrug.com |
| 58 | STITCH (Chemical-Protein Interactions) | Mixed approaches | http://stitch.embl.de/ |
| 59 | SWEETLEAD | Mixed approaches | https://simtk.org/home/sweetlead |
| 60 | The Cancer Genome Atlas (TCGA) | Mixed approaches | http://cancergenome.nih.gov |
| 61 | The Connectivity Map (CMap) | Mixed approaches | http://www.broadinstitute.org/cmap |
| 62 | The NCGC Pharmaceutical Collection (NPC) | Mixed approaches | http://tripod.nih.gov/npc/ |
| 63 | TOPSAN | Mixed approaches |
http://www.topsan.org (Service is currently not available) |
| 64 | DistilBio | Knowledge based approach | http://distilbio.com |
| 65 | Proteopedia | Knowledge-based repurposing | http://proteopedia.org |
| 66 | Pharmacogenomics Knowledge Base (PharmGKB) | Knowledge based approach | http://www.pharmgkb.org |
| 67 | Therapeutic Target Database (TTD) | Knowledge based approach | http://bidd.nus.edu.sg/group/cjttd/ |
3.1.5.1. Pharmacological databases such as DrugBank, DvD, DCDB, CDD, DrugMap central, network medicine, PharmGKB are very important in repurposing
They extract datas regarding drug properties, drug-biological entities interaction and provides the use of various computational as well as network based tools [100,[149], [150], [151], [152], [153], [154]].
3.1.5.2. Chemical databases such as chembank, ChEMBL, Chem2Bio2RDF, ChemDB, PubChem provide data regarding chemical structures, 2 D topological fingerprints and 3D conformations
It can predict and find novel drug structures for new indications based on drugs with similar structures and can construct chemical networks [99,[155], [156], [157], [158]]. PubChem is widely used for chemical structures [159].
3.1.5.3. Proteomics databases such as MIPS, PDB, human proteinpedia and HPRD explore protein-protein interactions and can be employed for network-based repurposing [33,36,37,83].
Heterogenous networks such as drug-protein- disease network can be constructed. MIPS is a popular database [107,154,160,161].
Analysing huge amount of literature is essential for drug repurposing, therefore medical literature databases are also important [102,162,163]. PubMed is the most employed with enormous amount of data with over 27 million citations. Properly utilising the data to mine novel information is still evolving.
Various machine learning algorithms along with computational and experimental approaches can improve the potential for repurposing.
3.2. Biological approaches
Systems and network biology help drug discovery by the development of various models which simulate the physiological environment of target proteins and the consequences of modulating them. This approach is very important in targeting multi factorial complex diseases. Polypharmacology approach is also useful as it modulates entire pathways instead of a single target protein. In systems and network biology, models are developed understanding molecular level pathways without losing key details. A multi target approach is also gaining importance to target complex illnesses. Various disease pathway based interaction maps and the effects of modulating various target proteins in model organisms will increase the utilization and influence of systems biology in drug repurposing as well as the drug development process [164].
3.3. Experimental approaches
Experimental approaches include screening of targets [[165], [166], [167], [168], [169]], cell assay [[170], [171], [172], [173]], animal model [[174], [175], [176], [177]] and clinical aspects [55].
3.4. Mixed approaches
There are mixed approaches which confirms computational observation, biological experiments and clinical testing and is very effective for drug repurposing [[178], [179], [180]].
Considering the market, huge number of diseases are in need for new drugs for its treatment. Even in the case of rare diseases more than 6000 to be still explored with research being limited to only 5% of them.
3.5. Knowledge-based repurposing
Drug repurposing can be based on the knowledge of researchers and doctors and their capability and skill to interpret observations. There are chances of serendipity while doing research in some other cases. The first cases of repurposing where mainly by serendipity. Then similar disease conditions which shared altered pathways were explored for combination treatment and to repurpose the drug for its treatment, instead of the originally targeted disease [181].
4. Drug repurposing in various diseases
4.1. Drug repurposing for CNS disorders
Recently Gaspar (2019) reported a novel and promising in silico approach for drug repurposing based on the 44 risk variants published by genome-wide association study (GWAS) for major depressive disorder (MDD). The authors used the online tool Drug Targetor (drugtargetor.com) to evaluate the integration of interactions between drugs and their targets, genome-wide association statistics, and also for genetically predicted expression levels in different tissues. Outside the major histocompatibility complex (MHC) region, 153 protein-coding genes are significantly associated with MDD in MAGMA (which is used to perform pathway analyses) after multiple testing correction. The authors suggests that among these, five are predicted to be down or upregulated in brain regions and 24 are known druggable genes [182].
Alzheimer's disease (AD) and Parkinson's disease (PD) are the two major hotspots of CNS drug research due to the debilitating nature of the disease and dearth of effective treatment options. Lack of better understanding of the underlying mechanisms and involvement of multiple pathophysiological pathways are the major challenges associated with these two neurological diseases. Therefore, drug repurposing gives the hope for finding a successful drug candidate for the treatment of such situations. Tau, a microtubule-associated protein (MAP) functions in maintaining the assembly as well as disassembly of the microtubules [183]. An aberration in tau phosphorylation disrupting its normal physiology is known to be the cause of neurodegenerative disorders such as AD. Conditions such as hyperphosphorylation or abnormal phosphorylation of tau is known to be caused due to several kinases such as cyclin-dependent kinase 5 (CDK5), microtubule-affinity-regulating kinase (MARK) and glycogen synthase kinase 3 (GSK3) and various signaling pathways such as MAPK and CAMP- dependent protein kinase A respectively, resulting in β,γ-secretases regulation and neuronal apoptosis [[184], [185], [186], [187], [188], [189], [190], [191]].
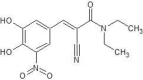
Silva (2019) reported the anti-aggregating activity of tau proteins by nitrocatechol scaffold, which can be further enhanced by the introduction of bulky substituents at the side chain. A remarkable increase in activity was observed for α-cyanocarboxamide derivative. The authors used docking studies to confirm the activity of these compounds and demonstrated 5-nitro-α-cyanocarboxamide derivatives of caffeic acid and caffeic acid phenethyl ester can effectively inhibit aggregation of tau-derived hexapeptide AcPHF6. which is illustrated in Fig. 2 [192].
Fig. 2.
Anti-aggregating activity of tau proteins by 5-Nitro-α-cyanocarboxamide.
Similarly Bourque (2018) provided a detailed review on the protective effect of sex hormones, particularly estrogens, progesterone, androgens and dehydroepiandrosterone on PD through animal models and also in human studies. They suggest that drugs affecting estrogen neurotransmission such as selective estrogen receptor modulators (SERM) or steroid metabolism inhibitors such as 5α-reductase inhibitors could be repositioned for treatment of PD. Sex steroids are also modulator of neurotransmission. This information can be vital in repurposing of drugs for PD [193].
Johnston (2018) provided the scope of drug repurposing in l-DOPA-induceddyskinesia (LID) in Parkinson's disease. The authors approached the subject in three ways such as in vivo phenotypic screening in a hypothesis-free manner; based on analogy to a related disorder; and a hypothesis-driven evaluation of candidates in vivo and in silico screening selection by artificial intelligence. They also provided a case study using IBM-Watson where -a training set of compounds, with demonstrated ability to reduce LID, were employed to identify novel repurposing candidates [194].
Zhu (2019) reported the opportunity for repurposing of omeprazole for oligodendrocyte differentiation and remyelination [195]. Multiple sclerosis is a disease which is immune mediated, hence causes dysregulation of several inflammatory cells and damages the myelin sheath. Proper molecular mechanism is not known so far [196]. The authors identifed omeprazole as a potential, applicable pro-remyelinating drug candidate. The finding was supported by public microarray results from the GEO database, which was analyzed using Connectivity-Map, a database connecting diseases, genes and drugs. This study revealed that the proportion of myelinated axons was found to be increased after omeprazole administration. Thus, it can be concluded that omeprazole is a promising drug candidate for remyelination associated with multiple sclerosis.
Tranfaglia (2018) provided a detailed investigation on the possibilities of drug repurposing for the treatment of Fragile X syndrome (FXS). It is one of the most common inherited causes of intellectual impairment and the major monogenic cause of autism. The authors used Disease-Gene Expression Matching (DGEM) for the repurposing analysis which is a novel and promising approach for drug repurposing. This DGEM study predicted that sulindac could be a potent candidate for fragile X treatment, and subsequent preclinical validation studies have shown promising results. The authored suggest that the use of combinations of available drugs and nutraceuticals has the potential to greatly expand the options for repurposing [197].
4.2. Drug repurposing for cancer
Overcoming the self developed drug resistance of tumours is a huge obstacle in cancer therapy and management. Suitable strategies need to be developed in order to minimise drug resistance and maximize anticancer activity. Discovery and development of anticancer therapeutic agents are deterred by multiple factors including the pharmacology, pharmacokinetics, pathophysiology of the type of cancer and drug metabolism. This major challenge can be successfully resolved by repurposing the drugs since most of the aspects discussed earlier is already known to a greater extent and the drugs are already in existing therapeutic use.
Cyclin-dependent kinase-5(Cdk-5), a threonine or serine kinaseis overexpressed in glioma cells in comparison to the healthy human astrocytes is responsible for neuronal dysfunction and death [198]. Pandey (2019) reported both in vitro and in vivo enhancement of the antitumour activity of temozolomide (TMZ) by the drug Roscovitine (RSV) known to inhibit Cdk-5 [199,200]. This is an interesting result since use of TMZ alone is not effective for gliomas after a while due to the development of drug resistance by malignant cells. The authors observed that TMZ treatment followed by a pre-treatment with RSV significantly enhanced chemo-sensitivity and suppressed the growth of glioma cells by reducing Cdk-5 activity along with simultaneous induction of autophagy and Caspase-3 mediated apoptosis. This study also revealed the reduced expression of Ki67, GFAP and markers of angiogenesis (CD31, VEGF) in case of TMZ + RSV therapy in glioma treatment. CD31, a platelet endothelial cell adhesion molecule-1 (PECAM-1) and vascular endothelial growth factor (VEGF), a cytokine with multiple functions play a key role in endothelial migration as well as proliferation and is highly expressed in glioma [[201], [202], [203], [204], [205], [206]]. The presence of reactive astrocytes in peri-tumoral areas and in areas around blood vessels was completely diminished in TMZ + RSV treated brain sections.
Drug resistance affects many other drug pathways in cancer therapy. Imatinib is a perfect example for this scenario, which is used in the therapy of gastrointestinal stromal tumors (GISTs), one of the most common mesenchymal tumors of the GI tract. Imatinib loses its desired activity due to development of resistance by tumor cells. Second line therapies involving sunitinib and regorafenib are also not effective due to multiple reasons including cytotoxicity and limited clinical response. Lu (2019) attempted to solve this problem by repurposing cabozantinib for GIST treatment. Authors reported that cabozantinib exhibited higher potency than imatinib against primary gain-of-function mutations of cKIT, a tyrosine kinase receptor which plays the major role in the pathophysiology of GIST by the downstream signaling of MEK/ERK/RAS/RAF, STAT 5 and PI3K/AKT pathways [[207], [208], [209], [210]]. In this study, cabozantinib showed good in vitro and in vivo efficacy in the cKIT mutant-driven preclinical models of GISTs and also displayed a prolonged and sustained effect in the watchful waiting period after the treatment withdrawal. This study increases the scope of carbozantinib as a potent candidate for GIST treatment in future.
Sunitinib is another important chemotherapeutic drug which is a multi-targeted tyrosine kinase inhibitor. It is approved by the FDA for the treatment of renal cell carcinoma (RCC) and imatinib resistant GIST. Recently, Chatziathanasiadou (2019) studied the activity of sunitinib against glioblastoma multiforme (GBM), one of the most dangerous form of malignancy. Treatment of GBM has many limitations due to the unknown pathophysiology of the disease and also due to the variety of physiological factors like poor blood brain barrier (BBB) permeability. The authors successfully developed a validated LC-MS/MS method for the in vitro and in vivo quantitation of sunitinib in glioblastoma cells. There is no report of other study for the estimation of sunitinib uptake in GBM, therefore this finding is quite significant. This method was validated and accredited according to ISO 17025:2005 guidelinein human plasma and successfully applied to cancer patient plasma. The method was successfully adopted to establish a protocol for the evaluation of sunitinib accumulation into M095K glioma cell lines. This work definitely gives some additional momentum in repurposing studies of cancer drugs [211].
Kuenzi (2019) analysed the drug repurposing opportunities for tivantinib, a c-MET inhibitor used in the treatment of acute myeloid leukemia (AML) through a target based approach. The authors repored that tivantinib is a novel glycogen synthase kinase 3 (GSK3α/β) inhibitor that potently kills AML cells. Tivantinib as single agent or combination therapy with ABT-199 may represent attractive new therapeutic opportunities for AML [212]. GSK3α has been already reported as a new target in the treatment of acute myeloid leukemia (AML) by its inactivation via activation of several pathways such as PKB/AKT, S6K, RAS-MAPK and Wnt and hedgehog pathways [[212], [213], [214]]. However, most GSK3 inhibitors lack specificity for GSK3α over GSK3β and other kinases. This work revealed that tivantinib alone or in combination with ABT-199 can effectively inhibit the colony forming capacity of AML bone marrow mononuclear cells. This study gives the possibilities of developing treatment for AML based on tivantinib and further in vivo studies can shed more light on the anticancer potential of the same [212].
There are previous reports of anti-alcoholic drug disulfiram(DSF) being used for cancer treatment. But the mechanism of action of disulfiram remained unknown for a long time and has been explored only recently. Spillier (2019) reported the structure-activity relationships (SAR) of phosphoglycerate dehydrogenase (PHGDH) inhibition by disulfram analogues. This study provided significant insight into the mechanism of anticancer activity of disulfiram by PHGDH inhibition which is illustrated in Fig. 3 . This is a result of the oxidation of mutated cysteine 116 residue present in PHGDH which further disrupts the tetrameric state of PHGDH. PHGDH inhibition causes inhibition in proliferation of tumor cells [45].
Fig. 3.
Disulfiram inhibition of tumor cell proliferation via PHGDH inhibition.
4.3. Drug repurposing for infectious diseases
Mihai (2019) explored the repurposing of antidepressants consisting of dibenzo[b,e]thiepin scaffold against dengue virus replication. The authors synthesized a series ofdihydrodibenzo[b,e]thiepin derivatives and analysed at 10 μg/mL in HEK293 cells infected with DENV2. In this work, molecular docking studies were also conducted to confirm the tested compounds to DENV2 NS3 helicase and also on dopamine D4 receptor. The objective of the study is to develop a correlation between in vitro and in silico results. It is observed that majority of compounds appear to inhibit only the viral helicase, while others inhibit both helicase and D4 receptors but the mechanism is very complex [215].
Lv (2018) studied target proteins of Japanese encephalitis virus(JEV) for drug repurposing since there is no drugs available for successful treatment. The authors adopted systems biology methods which are a combination of both gene expression data and the data from phenome-wide association studies (PheWAS). By using this efficient approach the authors identified 286 genes which are involved in mechanism of JEV infection [216]. Based on in vivo studies in mice model, it is observed that bortezomib, an anticancer drug indicated for multiple myeloma and lymphoma can lower JEV-induced death in mice, along with alleviating the suffering in JEV infection and also reduce the brain damage possibly by proteasome inhibition [217,218]. This study highlights the possibility of a new drug candidate for JEV treatment.
Another very serious and major zoonotic pathogenic disease is tuberculosis (TB) which is responsible for large scale death and disaster in this class. Thus, identifying a novel and efficient drug is very essential regarding the aggressiveness of TB. Passi (2018) reports a very effective and unique repurposing method based on molecular function correlations among known drug-target pairs to predict novel drug-target interactions. This approach is called ‘RepTB’ and not much explored anywhere else for drug repurposing strategies. The authors used Network based inference (NBI) method for analyzing a gene Ontology based network containing 26,404 edges, 6630 drug and 4083 target nodes. The study identified four TB targets; FolP1 (Dihydropteroate synthase), Tmk (Thymidylate kinase), Dut (Deoxyuridine 5′-triphosphate nucleotidohydrolase) and MenB (1,4-dihydroxy-2-naphthoyl-CoA synthase) and revealed the similarity in these targets based on examining the drugs. This study is very significant since it introduces a new method for the repurposing giving emphasis on the target proteins of the pathogens [219]. Another study by Singh, A et al., 2019 revealed the stable binding of terlipressin, a vasopressin analogue used for hypotension management with that of EmbC responsible for arabinogalactan synthesis within mycobacterial cell wall [220,221].
Recently Keighobadi (2019) focused the application of 3-(1,2,4-triazol-1-yl)flavanones (TF) against promastigote and amastigote forms of Leishmania major (L. major). The authors identified 4-chloro derivative (TF-2) having potency about 13 times more than fluconazole against promastigotes. Further analysis using in silico tools showed that TF-2 can be properly incorporated in the active site of parasitic CYP51 and coordinated to the heme. The SAR analysis revealed that the introduction of 4-chloro on 2- phenyl moiety results in the best profile of activity and selectivity. Accordingly, the compound TF-2 prototype can be considered as promising candidate for development of new antileishmanial agents [222].
Staphylococcus aureus has gained significant research interest in drug repurposing studies. The major feature that makes this pathogen for investigation is that it exploits the metabolic pathways of the host organism for the survival which we can investigate with more clarity. Santano (2019) demonstated a host based approach for drug repurposing in S. aureus infection. The authors screened 133 host-targeting drugs and found three tyrosine kinase inhibitors; ibrutinib, dasatinib and crizotinib can effectively impair intracellular bacterial survival. Further, ibrutinib significantly increased host cell viability after infection through inhibition of cell invasion and intracellular bacterial proliferation. This study also proposes a putative mechanism of action of ibrutinib based on phosphoproteomics data which involves several host factors, including EPHA2, C-JUN and NWASP [223]. Additionally, Li (2019) also reviewed host based therapeutics were effective against corona virus and influenza virus infections [224].
Similarly, Oleivera (2019) reports the repurposing of non-steroidal anti-inflammatory drug (NSAID) ibuprofen against S. aureus infection. The authors used control of adhered cells (2 h) and 24 h old biofilms of Staphylococcus aureus for the study and observed that ibuprofen causes metabolic reductions up to 80% and total loss of culturability of adhered cells and 24 h old biofilm. But the drug is not found to be active against antibiotic resistant strains. This aspect requires future investigations for developing another potent drug molecule which can overcome the antibiotic resistant feature of the pathogen [225].
Anticancer drugs are also reported against pathogenic diseases through repurposing. Cisplatin is a major anticancer drug used in the treatment of different kinds of malignancies. Yuan (2018) demonstrated the repurposing of cisplatin against Pseudomonas aeruginosa. This study confirms the efficiency of the drug as an antimicrobial agent and kills the pathogens by acting through inhibiting the DNA replication [226,227]. In vitro and in vivo studies also confirm this feature and additionally the drug repressed the type III secretion system (T3SS) of the microbe which is important for the secretion of exotoxins.
Pic (2018) used anthelminthic drug salicylanilide oxyclozanide against Candida albicans strains which is found to be clinically resistant. This study found that the drug is active in both sensitive and resistant strains and the antifungal activity of oxyclozanide was enhanced when C. albicans grew in nonfermentable carbon sources. The authors reported the possible mechanism of action as disruption of the mitochondrial membrane potential [228]. A list of the various recently repurposed drugs with their chemical structure and efficacy are listed out in Table 4 .
Table 4.
Recently repurposed drugs in various diseases.
| Repurposed Drug | Novel activity/efficacy |
|---|---|
| CNS disorders | |
 5-nitro-α-cyanocarboxamide derivatives |
Anti-aggregating activity of tau proteins (AD) |
 Omeprazole |
Oligodendrocyte differentiation and remyelination associated with multiple sclerosis |
| Cancer | |
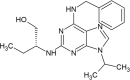 Roscovitine |
Individually or in combination with drugs like temozolomide inhibits Cyclin-dependent kinase-5(Cdk-5) there by enhanced chemo-sensitivity and suppressed the growth of glioma cells |
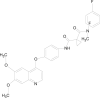 Cabozantinib |
Gastrointestinal stromal tumors (GIST) treatment |
 Sunitinib |
Multi-targeted tyrosine kinase inhibitor (renal cell carcinoma), Treatment of glioblastoma multiforme (GBM) |
 Tivantinib |
Treatment of acute myeloid leukemia (AML) |
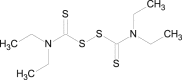 Disulfiram |
PHGDH inhibition causes anti- proliferation |
| Infectious diseases | |
 Dibenzo[b,e]thiepin |
Inhibit dengue virus replication |
| Miscellaneous diseases | |
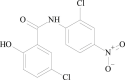 Niclosamide ` |
Treat pulmonary fibrosis (PF) |
4.4. Drug repurposing in other diseases
Patrick (2018) introduced an efficient and successful bioinformatics approach for drug repurposing by using word embedding to summarize drug information from more than 20 million articles and applied machine learning to model the drug-disease relationship. The authors applied drug repurposing approach separately on nine cutaneous diseases including psoriasis, atopic dermatitis, and alopecia areata and eight other immune mediated diseases and obtained a mean area under the receiver operating characteristics of 0.93 in cross validation. They gave emphasis on psoriasis, which is a chronic inflammatory condition of skin that affects more than 100 million people worldwide and using this approach the authors were able to confirm drugs that are known to be effective for psoriasis and to identify potential candidates used to treat other diseases. This work provided an effective algorithm that can be used to identify repurposed drugs for immune disrupted dermatological diseases [229].
Boyapally (2018) reported the repurposing of niclosamide which is an antihelminthic drug for treating pulmonary fibrosis (PF) one of the most deadly lung disease. The authors demonstated the anti-fibrotic potential of niclosamide in TGF-β1 induced in vitro model of pulmonary fibrosis and 21-day in vivo model of bleomycin induced PF respectively. This study showed that niclosamide holds the potential for anti-fibrotic effect by various mechanisms as illustrated in Fig. 4 which includes blockade of epithelial to mesenchymal transition (EMT)as well inhibiting the deposition of extracellular matrix, downregulation of platelet-derived growth factor (PDGF) expression. This leads to reduction in angiogenesis and blockade of WNT/β-catenin signaling thereby inhibiting β-catenin translocation into the nucleus, TGF-β1 production, fibroblasts proliferation and collagen fibers secretion which gives scope for developing successful drug candidate for PF treatment [[230], [231], [232], [233]].
Fig. 4.
Mechanisms involved in antifibrotic activity produced by niclosamide.
Fang (2018) focuses on drug repurposing for coronary artery disease (CAD) using computational tools. The authors employed in silico approaches by integrating known drug-target interactions, CAD genes derived from the genetic and genomic studies and also through human protein interactomics. This work gives some interesting and novel findings based on reported case studies in which various approved drugs such as Fasudil, Parecoxib, and Dexamethasone and some natural products including Resveratrol, Luteolin, Daidzein and Caffeic acid are efficient in developing drugs for CAD [234].
5. Conclusion and future perspectives
This review focuses mainly on the most recent research advancements in drug repurposing. We have done an analysis of repurposing works and found that most of the recent repurposing falls under anticancer, CNS and pathogenic diseases. Since drug development involves multistage process which is time and money intense, this type of innovative ideas and novel concepts definitely will expedite the drug development process. The repurposed drug is already qualified initial phases of screening and this makes the new objective more feasible and significantly reduces the cost of development.
The process of drug development starts from finding a lead molecule to implementation in the market. But this process is a big scenario which utilizes lot of cost including money and facility and takes very long time of around fifteen years. Repurposing is a vital concept which can overcome these tedious tasks and can boost the drug development process to a significant level. The current repurposing methods include statistical screening of the approved drug and to find the good binding target using in silico methods. Since this requires the use of wide library of compounds and data more feasible techniques for repurposing have to be developed. The influence of target is another possibility which is not much explored. The original and repurposed target may have a significant similarity owing to the good binding drug. This aspect has to be exploited at molecular level which can further make the drug repurposing process more efficient and feasible.
References
- 1.DiMasi J.A., Feldman L., Seckler A., Wilson A. Trends in risks associated with new drug development: success rates for investigational drugs. Clin. Pharmacol. Ther. 2010;87(3):272–277. doi: 10.1038/clpt.2009.295. [DOI] [PubMed] [Google Scholar]
- 2.DiMasi J.A., Grabowski H.G., Hansen R.W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., Schacht A.L. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat. Rev. Drug Discov. 2010;9(3):203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 4.Mohs R.C., Greig N.H. Drug discovery and development: role of basic biological research. Alzheimer's Dementia: Transl. Res. Clin. Interv. 2017;3(4):651–657. doi: 10.1016/j.trci.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owens P.K., Raddad E., Miller J.W., Stille J.R., Olovich K.G., Smith N.V., et al. A decade of innovation in pharmaceutical R&D: the Chorus model. Nat. Rev. Drug Discov. 2015;14(1):17. doi: 10.1038/nrd4497. [DOI] [PubMed] [Google Scholar]
- 6.Drews J. Drug discovery: a historical perspective. Science. 2000;287(5460):1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 7.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discov. Today. 2005;10(2):139–147. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 8.Arrowsmith J., Miller P. 2013. Trial Watch: Phase II and Phase III Attrition Rates 2011–2012. [DOI] [PubMed] [Google Scholar]
- 9.Lipinski C.A. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 10.GNS H.S., Saraswathy G.R., Murahari M., Krishnamurthy M. An update on Drug Repurposing: Re-written saga of the drug's fate. Biomed. Pharmacother. 2019;110:700–716. doi: 10.1016/j.biopha.2018.11.127. [DOI] [PubMed] [Google Scholar]
- 11.Allison M. 2012. NCATS Launches Drug Repurposing Program. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins A.L. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008;4(11):682. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 13.Talevi A., Carrillo C., Comini M. Current Medicinal Chemistry. 2019. The thiol-polyamine metabolism of trypanosoma cruzi: molecular targets and drug repurposing strategies. [DOI] [PubMed] [Google Scholar]
- 14.Konreddy A.K., Rani G.U., Lee K., Choi Y. Recent drug-repurposing-driven advances in the discovery of novel antibiotics. Curr. Med. Chem. 2019 doi: 10.2174/0929867325666180706101404. [DOI] [PubMed] [Google Scholar]
- 15.Dandu K., Kallamadi P.R., Thakur S.S., Rao C.M. Drug repurposing for retinoblastoma: recent advances. Curr. Top. Med. Chem. 2019 doi: 10.2174/1568026619666190119152706. [DOI] [PubMed] [Google Scholar]
- 16.Martorana A., Perricone U., Lauria A. The repurposing of old drugs or unsuccessful lead compounds by in silico approaches: new advances and perspectives. Curr. Top. Med. Chem. 2016;16(19):2088–2106. doi: 10.2174/1568026616666160216153457. [DOI] [PubMed] [Google Scholar]
- 17.Olgen S., Kotra L.P. Drug repurposing in the development of anticancer agents. Curr. Med. Chem. 2019 doi: 10.2174/0929867325666180713155702. [DOI] [PubMed] [Google Scholar]
- 18.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol. Sci. 2013;34(5):267–272. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Arrowhead conferences . 2011. Drug Repositioning: New Technologies, Business Models and Strategies for Successful Development Arrowhead Conferences. [Google Scholar]
- 20.López-Muñoz F., Alamo C., Cuenca E., Shen W.W., Clervoy P., Rubio G. History of the discovery and clinical introduction of chlorpromazine. Ann. Clin. Psychiatr. 2005;17(3):113–135. doi: 10.1080/10401230591002002. [DOI] [PubMed] [Google Scholar]
- 21.Frankenburg F.R., Baldessarini R.J. Neurosyphilis, malaria, and the discovery of antipsychotic agents. Harv. Rev. Psychiatry. 2008;16(5):299–307. doi: 10.1080/10673220802432350. [DOI] [PubMed] [Google Scholar]
- 22.Gordon R.A., Campbell M. The use of chlorpromazine in intractable pain associated with terminal carcinoma. Can. Med. Assoc. J. 1956;75(5):420. [PMC free article] [PubMed] [Google Scholar]
- 23.Schlitzer M. Malaria chemotherapeutics part I: history of antimalarial drug development, currently used therapeutics, and drugs in clinical development. ChemMedChem. 2007;2(7):944–986. doi: 10.1002/cmdc.200600240. [DOI] [PubMed] [Google Scholar]
- 24.Wallace D.J. The history of antimalarials. Lupus. 1996;5(1_suppl):2–3. [PubMed] [Google Scholar]
- 25.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol. Sci. 2013;34(5):267–272. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Mullen J., Cockell S.J., Tipney H., Woollard P.M., Wipat A. Mining integrated semantic networks for drug repositioning opportunities. PeerJ. 2016;4:e1558. doi: 10.7717/peerj.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeu Y., Yoon Y., Park S. Protein localization vector propagation: a method for improving the accuracy of drug repositioning. Mol. Biosyst. 2015;11(7):2096–2102. doi: 10.1039/c5mb00306g. [DOI] [PubMed] [Google Scholar]
- 28.Kehagia A.A., Housden C.R., Regenthal R., Barker R.A., Müller U., Rowe J., et al. Targeting impulsivity in Parkinson's disease using atomoxetine. Brain. 2014;137(7):1986–1997. doi: 10.1093/brain/awu117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haynes V., Lopez-Romero P., Anand E. Attention-deficit/hyperactivity disorder under Treatment Outcomes Research (AUTOR): a European observational study in pediatric subjects. ADHD Atten. Deficit Hyperactivity Disord. 2015;7(4):295–311. doi: 10.1007/s12402-015-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henriksen K., Christiansen C., Karsdal M.A. Serological biochemical markers of surrogate efficacy and safety as a novel approach to drug repositioning. Drug Discov. Today. 2011;16(21–22):967–975. doi: 10.1016/j.drudis.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Maciukiewicz M., Marshe V.S., Hauschild A.C., Foster J.A., Rotzinger S., Kennedy J.L., et al. GWAS-based machine learning approach to predict duloxetine response in major depressive disorder. J. Psychiatr. Res. 2018;99:62–68. doi: 10.1016/j.jpsychires.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3(8):673. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 33.Luo H., Zhang P., Huang H., Huang J., Kao E., Shi L., et al. DDI-CPI, a server that predicts drug–drug interactions through implementing the chemical–protein interactome. Nucleic Acids Res. 2014;42(W1):W46–W52. doi: 10.1093/nar/gku433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke C.E., Deane K.H. Ropinirole for levodopa-induced complications in Parkinson's disease. Cochrane Database Syst. Rev. 2001;(1) doi: 10.1002/14651858.CD001516. [DOI] [PubMed] [Google Scholar]
- 35.Schwab R.S., England A.C., Poskanzer D.C., Young R.R. Amantadine in the treatment of Parkinson's disease. Jama. 1969;208(7):1168–1170. [PubMed] [Google Scholar]
- 36.Wang Z.Y., Zhang H.Y. Rational drug repositioning by medical genetics. Nat. Biotechnol. 2013;31(12):1080. doi: 10.1038/nbt.2758. [DOI] [PubMed] [Google Scholar]
- 37.Moore R.A., Wiffen P.J., Derry S., Rice A.S. Gabapentin for chronic neuropathic pain and fibromyalgia in adults. Cochrane Database Syst. Rev. 2014;(4) doi: 10.1002/14651858.CD007938.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H., Choo S., Park J., Jung J., Kang Y., Lee D. vol. 10. 2016, December. Prediction of drugs having opposite effects on disease genes in a directed network. (BMC Systems Biology). 1, p. S2). (BioMed Central) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paez Espinosa E.V., Murad J.P., Khasawneh F.T. Aspirin: pharmacology and clinical applications. Thrombosis. 2012:173124. doi: 10.1155/2012/173124. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.deGoma E.M. Lomitapide for the management of homozygous familial hypercholesterolemia. Rev. Cardiovasc. Med. 2014;15(2):109–118. doi: 10.3909/ricm0735. [DOI] [PubMed] [Google Scholar]
- 41.Dudley J.T., Deshpande T., Butte A.J. Exploiting drug–disease relationships for computational drug repositioning. Briefings Bioinf. 2011;12(4):303–311. doi: 10.1093/bib/bbr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagaraj A.B., Wang Q.Q., Joseph P., Zheng C., Chen Y., Kovalenko O., et al. Using a novel computational drug-repositioning approach (DrugPredict) to rapidly identify potent drug candidates for cancer treatment. Oncogene. 2018;37(3):403. doi: 10.1038/onc.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andresen V., Gjertsen B.T. Drug repurposing for the treatment of acute myeloid leukemia. Front. Med. 2017;4:211. doi: 10.3389/fmed.2017.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta S.C., Sung B., Prasad S., Webb L.J., Aggarwal B.B. Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends Pharmacol. Sci. 2013;34(9):508–517. doi: 10.1016/j.tips.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Spillier Q., Vertommen D., Ravez S., Marteau R., Thémans Q., Corbet C., et al. Anti-alcohol abuse drug disulfiram inhibits human PHGDH via disruption of its active tetrameric form through a specific cysteine oxidation. Sci. Rep. 2019;9(1):4737. doi: 10.1038/s41598-019-41187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toschi L., Finocchiaro G., Bartolini S., Gioia V., Cappuzzo F. 2005. Role of Gemcitabine in Cancer Therapy. [DOI] [PubMed] [Google Scholar]
- 47.Manegold C. vol. 30. WB Saunders; 2003. Pemetrexed (Alimta, MTA, multitargeted antifolate, LY231514) for malignant pleural mesothelioma; pp. 32–36. (Seminars in Oncology). [DOI] [PubMed] [Google Scholar]
- 48.Cohen M.H., Justice R., Pazdur R. Approval summary: pemetrexed in the initial treatment of advanced/metastatic non-small cell lung cancer. The Oncologist. 2009;14(9):930–935. doi: 10.1634/theoncologist.2009-0092. [DOI] [PubMed] [Google Scholar]
- 49.Gillis J.C., Goa K.L. Tretinoin. Drugs. 1995;50(5):897–923. doi: 10.2165/00003495-199550050-00008. [DOI] [PubMed] [Google Scholar]
- 50.Deotarse P.P., Jain A.S., Baile M.B., Kolhe N.S., Kulkarni A.A. Drug repositioning: a review. Int. J. Pharm. Rev. Res. 2015;4:51–58. [Google Scholar]
- 51.Lu J., Chen L., Yin J., Huang T., Bi Y., Kong X.,., Cai Y.D. Identification of new candidate drugs for lung cancer using chemical–chemical interactions, chemical–protein interactions and a K-means clustering algorithm. J. Biomol. Struct. Dyn. 2016;34(4):906–917. doi: 10.1080/07391102.2015.1060161. [DOI] [PubMed] [Google Scholar]
- 52.Cheng F., Liu C., Jiang J., Lu W., Li W., Liu G.,., Tang Y. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput. Biol. 2012;8(5) doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei G., Twomey D., Lamb J., Schlis K., Agarwal J., Stam R.W., et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10(4):331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 54.Abbruzzese C., Matteoni S., Signore M., Cardone L., Nath K., Glickson J.D., Paggi M.G. Drug repurposing for the treatment of glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2017;36(1):169. doi: 10.1186/s13046-017-0642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Q., Tao C., Shen F., Chute C.G. Exploring the pharmacogenomics knowledge base (PharmGKB) for repositioning breast cancer drugs by leveraging Web ontology language (OWL) and cheminformatics approaches. Biocomputing. 2014;2014:172–182. [PMC free article] [PubMed] [Google Scholar]
- 56.Hong I.S., Ipema H.J., Gabay M.P., Lodolce A.E. Medication repurposing: new uses for old drugs. J. Pharm. Technol. 2011;27(3):132–140. [Google Scholar]
- 57.Kuo I.F., Pearson G.J., Koshman S.L. Colchicine for the primary and secondary prevention of pericarditis: an update. Ann. Pharmacother. 2009;43(12):2075–2081. doi: 10.1345/aph.1M234. [DOI] [PubMed] [Google Scholar]
- 58.Imazio M., Bobbio M., Cecchi E., Demarie D., Pomari F., Moratti M., et al. Colchicine as first-choice therapy for recurrent pericarditis: results of the CORE (COlchicine for REcurrent pericarditis) trial. Arch. Intern. Med. 2005;165(17):1987–1991. doi: 10.1001/archinte.165.17.1987. [DOI] [PubMed] [Google Scholar]
- 59.Ishida J., Konishi M., Ebner N., Springer J. Repurposing of approved cardiovascular drugs. J. Transl. Med. 2016;14(1):269. doi: 10.1186/s12967-016-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leaute-Labreze C., Hoeger P., Mazereeuw-Hautier J., Guibaud L., Baselga E., Posiunas G.,., Friedlander S.F. A randomized, controlled trial of oral propranolol in infantile hemangioma. N. Engl. J. Med. 2015;372(8):735–746. doi: 10.1056/NEJMoa1404710. [DOI] [PubMed] [Google Scholar]
- 61.Rao B.S., Das D.G., Taraknath V.R., Sarma Y. A double blind controlled study of propranolol and cyproheptadine in migraine prophylaxis. Neurol. India. 2000;48(3):223. [PubMed] [Google Scholar]
- 62.Ross M.K., Yoon J., van der Schaar A., van der Schaar M. Discovering pediatric asthma phenotypes on the basis of response to controller medication using machine learning. Ann. Am. Thorac. Soc. 2018;15(1):49–58. doi: 10.1513/AnnalsATS.201702-101OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parkes D. Bromocriptine. N. Engl. J. Med. 1979;301(16):873–878. doi: 10.1056/NEJM197910183011606. [DOI] [PubMed] [Google Scholar]
- 64.Pijl H., Ohashi S., Matsuda M., Miyazaki Y., Mahankali A., Kumar V.,., DeFronzo R.A. Bromocriptine: a novel approach to the treatment of type 2 diabetes. Diabetes Care. 2000;23(8):1154–1161. doi: 10.2337/diacare.23.8.1154. [DOI] [PubMed] [Google Scholar]
- 65.Steinmetz K.L. Colesevelam hydrochloride. Am. J. Health Syst. Pharm. 2002;59(10):932–939. doi: 10.1093/ajhp/59.10.932. [DOI] [PubMed] [Google Scholar]
- 66.Fonseca V.A., Rosenstock J., Wang A.C., Truitt K.E., Jones M.R. Colesevelam HCl improves glycemic control and reduces LDL cholesterol in patients with inadequately controlled type 2 diabetes on sulfonylurea-based therapy. Diabetes Care. 2008;31(8):1479–1484. doi: 10.2337/dc08-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yella J., Yaddanapudi S., Wang Y., Jegga A. Changing trends in computational drug repositioning. Pharmaceuticals. 2018;11(2):57–77. doi: 10.3390/ph11020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiang M.C., Tseng M.T., Pan C.L., Chao C.C., Hsieh S.T. Progress in the treatment of small fiber peripheral neuropathy. Expert Rev. Neurother. 2015;15(3):305–313. doi: 10.1586/14737175.2015.1013097. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z.Y., Quan Y., Zhang H.Y. Medical genetic inspirations for anticancer drug repurposing. Trends Pharmacol. Sci. 2014;35(1):1–3. doi: 10.1016/j.tips.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 70.Fukuoka Y., Takei D., Ogawa H. A two-step drug repositioning method based on a protein-protein interaction network of genes shared by two diseases and the similarity of drugs. Bioinformation. 2013;9(2):89. doi: 10.6026/97320630009089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemke A., Kiderlen A.F., Kayser O. Amphotericin B. Appl. Microbiol. Biotechnol. 2005;68(2):151–162. doi: 10.1007/s00253-005-1955-9. [DOI] [PubMed] [Google Scholar]
- 72.Saravolatz L.D., Bern C., Adler-Moore J., Berenguer J., Boelaert M., den Boer M.,., Ritmeijer K. Liposomal amphotericin B for the treatment of visceral leishmaniasis. Clin. Infect. Dis. 2006;43(7):917–924. doi: 10.1086/507530. [DOI] [PubMed] [Google Scholar]
- 73.Wolf R., Matz H., Orion E., Tuzun B., Tuzun Y. Dapsone. Dermatol. Online J. 2002;8(1) [PubMed] [Google Scholar]
- 74.Brabin B.J., Eggelte T.A., Parise M., Verhoeff F. Dapsone therapy for malaria during pregnancy. Drug Saf. 2004;27(9):633–648. doi: 10.2165/00002018-200427090-00002. [DOI] [PubMed] [Google Scholar]
- 75.Croft S.L., Engel J. Miltefosine—discovery of the antileishmanial activity of phospholipid derivatives. Trans. R. Soc. Trop. Med. Hyg. 2006;100(Supplement_1):S4–S8. doi: 10.1016/j.trstmh.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 76.Smorenburg C.H., Seynaeve C., Bontenbal M., Planting A.S., Sindermann H., Verweij J. Phase II study of miltefosine 6% solution as topical treatment of skin metastases in breast cancer patients. Anti Canccer Drugs. 2000;11(10):825–828. doi: 10.1097/00001813-200011000-00006. [DOI] [PubMed] [Google Scholar]
- 77.Verweij J., Planting A., van der Burg M., Stoter G. A dose-finding study of miltefosine (hexadecylphosphocholine) in patients with metastatic solid tumours. J. Cancer Res. Clin. Oncol. 1992;118(8):606–608. doi: 10.1007/BF01211805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verweij J., Planting A.T., Stoter G., Gandia D., Armand J.P. Phase II study of oral miltefosine in patients with squamous cell head and neck cancer. Eur. J. Cancer. 1993;29(5):778–779. doi: 10.1016/s0959-8049(05)80369-7. [DOI] [PubMed] [Google Scholar]
- 79.Planting A.S.T., Stoter G., Verweij J. Phase II study of daily oral miltefosine (hexadecylphosphocholine) in advanced colorectal cancer. Eur. J. Cancer. 1993;29(4):518–519. doi: 10.1016/s0959-8049(05)80142-x. [DOI] [PubMed] [Google Scholar]
- 80.Soto J., Arana B.A., Toledo J., Rizzo N., Vega J.C., Diaz A.,., Junge K. Miltefosine for new world cutaneous leishmaniasis. Clin. Infect. Dis. 2004;38(9):1266–1272. doi: 10.1086/383321. [DOI] [PubMed] [Google Scholar]
- 81.LINAS S.L., NIES A.S. Minoxidil. Ann. Intern. Med. 1981;94(1):61–65. doi: 10.7326/0003-4819-94-1-61. [DOI] [PubMed] [Google Scholar]
- 82.Olsen E.A., Weiner M.S., Delong E.R., Pinnell S.R. Topical minoxidil in early male pattern baldness. J. Am. Acad. Dermatol. 1985;13(2):185–192. doi: 10.1016/s0190-9622(85)70157-0. [DOI] [PubMed] [Google Scholar]
- 83.Olsen E.A., Dunlap F.E., Funicella T., Koperski J.A., Swinehart J.M., Tschen E.H., Trancik R.J. A randomized clinical trial of 5% topical minoxidil versus 2% topical minoxidil and placebo in the treatment of androgenetic alopecia in men. J. Am. Acad. Dermatol. 2002;47(3):377–385. doi: 10.1067/mjd.2002.124088. [DOI] [PubMed] [Google Scholar]
- 84.Weiss V.C., West D.P., Fu T.S., Robinson L.A., Cook B., Cohen R.L., Chambers D.A. Alopecia areata treated with topical minoxidil. Arch. Dermatol. 1984;120(4):457–463. [PubMed] [Google Scholar]
- 85.Yan X.Y., Zhang S.W., Zhang S.Y. Prediction of drug–target interaction by label propagation with mutual interaction information derived from heterogeneous network. Mol. Biosyst. 2016;12(2):520–531. doi: 10.1039/c5mb00615e. [DOI] [PubMed] [Google Scholar]
- 86.Mimoto M.S., Oyler J.L., Davis A.M. Evaluation and treatment of hirsutism in premenopausal women. Jama. 2018;319(15):1613–1614. doi: 10.1001/jama.2018.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Easthope S.E., Perry C.M. Topical bimatoprost. Drugs Aging. 2002;19(3):231–248. doi: 10.2165/00002512-200219030-00008. [DOI] [PubMed] [Google Scholar]
- 88.Woodward D.F., Krauss A.P., Chen J., Lai R.K., Spada C.S., Burk R.M., et al. The pharmacology of bimatoprost (Lumigan™) Surv. Ophthalmol. 2001;45:S337–S345. doi: 10.1016/s0039-6257(01)00224-7. [DOI] [PubMed] [Google Scholar]
- 89.Woodward D.F., Krauss A.P., Chen J., Liang Y., Li C., Protzman C.E., et al. Pharmacological characterization of a novel antiglaucoma agent, Bimatoprost (AGN 192024) J. Pharmacol. Exp. Ther. 2003;305(2):772–785. doi: 10.1124/jpet.102.047837. [DOI] [PubMed] [Google Scholar]
- 90.Law S.K. Bimatoprost in the treatment of eyelash hypotrichosis. Clin. Ophthalmol. 2010;4:349. doi: 10.2147/opth.s6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tosti A., Pazzaglia M., Voudouris S., Tosti G. Hypertrichosis of the eyelashes caused by bimatoprost. J. Am. Acad. Dermatol. 2004;51(5):S149–S150. doi: 10.1016/j.jaad.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 92.Gonzalez J.P., Brogden R.N. Naltrexone. Drugs. 1988;35(3):192–213. doi: 10.2165/00003495-198835030-00002. [DOI] [PubMed] [Google Scholar]
- 93.Minozzi S., Amato L., Vecchi S., Davoli M., Kirchmayer U., Verster A. Oral naltrexone maintenance treatment for opioid dependence. Cochrane Database Syst. Rev. 2011;(4) doi: 10.1002/14651858.CD001333.pub2. [DOI] [PubMed] [Google Scholar]
- 94.Volpicelli J.R., Alterman A.I., Hayashida M., O'Brien C.P. Naltrexone in the treatment of alcohol dependence. Arch. Gen. Psychiatr. 1992;49(11):876–880. doi: 10.1001/archpsyc.1992.01820110040006. [DOI] [PubMed] [Google Scholar]
- 95.McGill K.A., Busse W.W. Zileuton. The Lancet. 1996;348(9026):519–524. doi: 10.1016/S0140-6736(95)12297-4. [DOI] [PubMed] [Google Scholar]
- 96.Zouboulis C.C. Zileuton, a new efficient and safe systemic anti-acne drug. Derm. Endocrinol. 2009;1(3):188–192. doi: 10.4161/derm.1.3.8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Z., Fang H., Reagan K., Xu X., Mendrick D.L., Slikker W., Jr., Tong W. In silico drug repositioning–what we need to know. Drug Discov. Today. 2013;18(3–4):110–115. doi: 10.1016/j.drudis.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 98.Patel M.N., Halling-Brown M.D., Tym J.E., Workman P., Al-Lazikani B. Objective assessment of cancer genes for drug discovery. Nat. Rev. Drug Discov. 2013;12(1):35. doi: 10.1038/nrd3913. [DOI] [PubMed] [Google Scholar]
- 99.Seiler K.P., George G.A., Happ M.P., Bodycombe N.E., Carrinski H.A., Norton S., et al. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2007;36(suppl_1):D351–D359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(suppl_1):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Training N.L.M. US National Library of Medicine, National Institutes of Health; 2010. PubMed. [Google Scholar]
- 103.Lenffer J., Nicholas F.W., Castle K., Rao A., Gregory S., Poidinger M., et al. OMIA (Online Mendelian Inheritance in Animals): an enhanced platform and integration into the Entrez search interface at NCBI. Nucleic Acids Res. 2006;34(suppl_1):D599–D601. doi: 10.1093/nar/gkj152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bernstein F.C., Koetzle T.F., Williams G.J., Meyer E.F., Jr., Brice M.D., Rodgers J.R., et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. Arch. Biochem. Biophys. 1978;185(2):584–591. doi: 10.1016/0003-9861(78)90204-7. [DOI] [PubMed] [Google Scholar]
- 105.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., et al. GenBank. Nucleic Acids Res. 2012;41:36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Yefanov A. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012;41(D1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mewes H.W., Hani J., Pfeiffer F., Frishman D. MIPS: a database for protein sequences and complete genomes. Nucleic Acids Res. 1998;26(1):33–37. doi: 10.1093/nar/26.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xue H., Li J., Xie H., Wang Y. Review of drug repositioning approaches and resources. Int. J. Biol. Sci. 2018;14(10):1232. doi: 10.7150/ijbs.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oprea T.I., Overington J.P. Computational and practical aspects of drug repositioning. Assay Drug Dev. Technol. 2015;13(6):299–306. doi: 10.1089/adt.2015.29011.tiodrrr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Napolitano F., Zhao Y., Moreira V.M., Tagliaferri R., Kere J., D'Amato M., Greco D. Drug repositioning: a machine-learning approach through data integration. J. Cheminf. 2013;5(1):30. doi: 10.1186/1758-2946-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lotfi Shahreza M., Ghadiri N., Mousavi S.R., Varshosaz J., Green J.R. A review of network-based approaches to drug repositioning. Briefings Bioinf. 2017;19(5):878–892. doi: 10.1093/bib/bbx017. [DOI] [PubMed] [Google Scholar]
- 112.Zou J., Zheng M.W., Li G., Su Z.G. Advanced systems biology methods in drug discovery and translational biomedicine. BioMed Res. Int. 2013;2013 doi: 10.1155/2013/742835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gönen M. Predicting drug–target interactions from chemical and genomic kernels using Bayesian matrix factorization. Bioinformatics. 2012;28(18):2304–2310. doi: 10.1093/bioinformatics/bts360. [DOI] [PubMed] [Google Scholar]
- 114.Li J., Zhu X., Chen J.Y. Building disease-specific drug-protein connectivity maps from molecular interaction networks and PubMed abstracts. PLoS Comput. Biol. 2009;5(7) doi: 10.1371/journal.pcbi.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gramatica R., Di Matteo T., Giorgetti S., Barbiani M., Bevec D., Aste T. Graph theory enables drug repurposing–how a mathematical model can drive the discovery of hidden mechanisms of action. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0084912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jang G., Lee T., Lee B.M., Yoon Y. Literature-based prediction of novel drug indications considering relationships between entities. Mol. Biosyst. 2017;13(7):1399–1405. doi: 10.1039/c7mb00020k. [DOI] [PubMed] [Google Scholar]
- 117.Kuusisto F., Steill J., Kuang Z., Thomson J., Page D., Stewart R. AMIA Summits on Translational Science Proceedings. 2017. A simple text mining approach for ranking pairwise associations in biomedical applications; p. 166. 2017. [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang M., Schmitt-Ulms G., Sato C., Xi Z., Zhang Y., Zhou Y.,., Rogaeva E. Drug repositioning for Alzheimer's disease based on systematic ‘omics’ data mining. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0168812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fleuren W.W., Alkema W. Application of text mining in the biomedical domain. Methods. 2015;74:97–106. doi: 10.1016/j.ymeth.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 120.Krallinger M., Erhardt R.A.A., Valencia A. Text-mining approaches in molecular biology and biomedicine. Drug Discov. Today. 2005;10(6):439–445. doi: 10.1016/S1359-6446(05)03376-3. [DOI] [PubMed] [Google Scholar]
- 121.Papanikolaou N., Pavlopoulos G.A., Theodosiou T., Vizirianakis I.S., Iliopoulos I. DrugQuest-a text mining workflow for drug association discovery. BMC Bioinf. 2016;17(5):182. doi: 10.1186/s12859-016-1041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheng D., Knox C., Young N., Stothard P., Damaraju S., Wishart D.S. PolySearch: a web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. 2008;36(suppl_2):W399–W405. doi: 10.1093/nar/gkn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Perovšek M., Kranjc J., Erjavec T., Cestnik B., Lavrač N. TextFlows: a visual programming platform for text mining and natural language processing. Sci. Comput. Program. 2016;121:128–152. [Google Scholar]
- 124.Moreno I., Moreda P., Romá-Ferri M.T. International Conference on Applications of Natural Language to Information Systems. Springer; Cham: 2015. June). MaNER: a MedicAl named entity recogniser; pp. 418–423. [Google Scholar]
- 125.Lee S., Kim D., Lee K., Choi J., Kim S., Jeon M.,., Kang J. BEST: next-generation biomedical entity search tool for knowledge discovery from biomedical literature. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Plake C., Schiemann T., Pankalla M., Hakenberg J., Leser U. AliBaba: PubMed as a graph. Bioinformatics. 2006;22(19):2444–2445. doi: 10.1093/bioinformatics/btl408. [DOI] [PubMed] [Google Scholar]
- 127.Wu H., Gao L., Dong J., Yang X. Detecting overlapping protein complexes by rough-fuzzy clustering in protein-protein interaction networks. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0091856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu L., Huang J., Ma Z., Zhang J., Zou Y., Gao L. Inferring drug-disease associations based on known protein complexes. BMC Med. Genomics. 2015;8(2):S2. doi: 10.1186/1755-8794-8-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu C., Gudivada R.C., Aronow B.J., Jegga A.G. Computational drug repositioning through heterogeneous network clustering. BMC Syst. Biol. 2013;7(5):S6. doi: 10.1186/1752-0509-7-S5-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Šubelj L., Bajec M. Unfolding communities in large complex networks: combining defensive and offensive label propagation for core extraction. Phys. Rev. E. 2011;83(3) doi: 10.1103/PhysRevE.83.036103. [DOI] [PubMed] [Google Scholar]
- 131.Luo H., Wang J., Li M., Luo J., Peng X., Wu F.X., Pan Y. Drug repositioning based on comprehensive similarity measures and Bi-Random walk algorithm. Bioinformatics. 2016;32(17):2664–2671. doi: 10.1093/bioinformatics/btw228. [DOI] [PubMed] [Google Scholar]
- 132.Emig D., Ivliev A., Pustovalova O., Lancashire L., Bureeva S., Nikolsky Y., Bessarabova M. Drug target prediction and repositioning using an integrated network-based approach. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mei J.P., Kwoh C.K., Yang P., Li X.L., Zheng J. Drug–target interaction prediction by learning from local information and neighbors. Bioinformatics. 2012;29(2):238–245. doi: 10.1093/bioinformatics/bts670. [DOI] [PubMed] [Google Scholar]
- 134.Köhler S., Bauer S., Horn D., Robinson P.N. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82(4):949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Adie E.A., Adams R.R., Evans K.L., Porteous D.J., Pickard B.S. Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinf. 2005;6(1):55. doi: 10.1186/1471-2105-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vanunu O., Magger O., Ruppin E., Shlomi T., Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput. Biol. 2010;6(1) doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martínez V., Navarro C., Cano C., Fajardo W., Blanco A. DrugNet: network-based drug–disease prioritization by integrating heterogeneous data. Artif. Intell. Med. 2015;63(1):41–49. doi: 10.1016/j.artmed.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 138.Palma G., Vidal M.E., Raschid L. International Semantic Web Conference. Springer; Cham: 2014. Drug-target interaction prediction using semantic similarity and edge partitioning; pp. 131–146. [Google Scholar]
- 139.Mullen J., Cockell S.J., Woollard P., Wipat A. An integrated data driven approach to drug repositioning using gene-disease associations. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0155811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen B., Ding Y., Wild D.J. Assessing drug target association using semantic linked data. PLoS Comput. Biol. 2012;8(7) doi: 10.1371/journal.pcbi.1002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhu Q., Tao C., Shen F., Chute C.G. Exploring the pharmacogenomics knowledge base (PharmGKB) for repositioning breast cancer drugs by leveraging Web ontology language (OWL) and cheminformatics approaches. Biocomputing. 2014;2014:172–182. [PMC free article] [PubMed] [Google Scholar]
- 142.Hearst M.A. Proceedings of the 37th Annual Meeting of the Association for Computational Linguistics on Computational Linguistics. Association for Computational Linguistics; 1999. June). Untangling text data mining; pp. 3–10. [Google Scholar]
- 143.Agrawal R., Gehrke J., Gunopulos D., Raghavan P. vol. 27. ACM; 1998. pp. 94–105. (Automatic Subspace Clustering of High Dimensional Data for Data Mining Applications). No. 2. [Google Scholar]
- 144.Sander J., Ester M., Kriegel H.P., Xu X. Density-based clustering in spatial databases: the algorithm gdbscan and its applications. Data Min. Knowl. Discov. 1998;2(2):169–194. [Google Scholar]
- 145.Ankerst M., Breunig M.M., Kriegel H.P., Sander J. vol. 28. ACM; 1999. June). OPTICS: ordering points to identify the clustering structure; pp. 49–60. (ACM Sigmod Record). No. 2. [Google Scholar]
- 146.Wang W., Yang J., Muntz R. VLDB. Vol. 97. 1997. STING: a statistical information grid approach to spatial data mining; pp. 186–195. August. [Google Scholar]
- 147.Emig D., Ivliev A., Pustovalova O., Lancashire L., Bureeva S., Nikolsky Y., Bessarabova M. Drug target prediction and repositioning using an integrated network-based approach. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mei J.P., Kwoh C.K., Yang P., Li X.L., Zheng J. Drug–target interaction prediction by learning from local information and neighbors. Bioinformatics. 2012;29(2):238–245. doi: 10.1093/bioinformatics/bts670. [DOI] [PubMed] [Google Scholar]
- 149.Pacini C., Iorio F., Gonçalves E., Iskar M., Klabunde T., Bork P., Saez-Rodriguez J. DvD: an R/Cytoscape pipeline for drug repurposing using public repositories of gene expression data. Bioinformatics. 2012;29(1):132–134. doi: 10.1093/bioinformatics/bts656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu Y., Hu B., Fu C., Chen X. DCDB: drug combination database. Bioinformatics. 2009;26(4):587–588. doi: 10.1093/bioinformatics/btp697. [DOI] [PubMed] [Google Scholar]
- 151.Ekins S., Bunin B.A. Silico Models for Drug Discovery. Humana Press; Totowa, NJ: 2013. The collaborative drug discovery (CDD) database; pp. 139–154. [DOI] [PubMed] [Google Scholar]
- 152.Fu C., Jin G., Gao J., Zhu R., Ballesteros-Villagrana E., Wong S.T. DrugMap Central: an on-line query and visualization tool to facilitate drug repositioning studies. Bioinformatics. 2013;29(14):1834–1836. doi: 10.1093/bioinformatics/btt279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Barabási A.L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12(1):56. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Thorn C.F., Klein T.E., Altman R.B. Pharmacogenomics. Humana Press; Totowa, NJ: 2013. PharmGKB: the pharmacogenomics knowledge base; pp. 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A.,., Overington J.P. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011;40(D1):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen B., Dong X., Jiao D., Wang H., Zhu Q., Ding Y., Wild D.J. Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data. BMC Bioinf. 2010;11(1):255. doi: 10.1186/1471-2105-11-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen J., Swamidass S.J., Dou Y., Bruand J., Baldi P. ChemDB: a public database of small molecules and related chemoinformatics resources. Bioinformatics. 2005;21(22):4133–4139. doi: 10.1093/bioinformatics/bti683. [DOI] [PubMed] [Google Scholar]
- 158.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Bryant S.H. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37(suppl_2):W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bolton E.E., Wang Y., Thiessen P.A., Bryant S.H. vol. 4. Elsevier; 2008. PubChem: integrated platform of small molecules and biological activities; pp. 217–241. (Annual Reports in Computational Chemistry). [Google Scholar]
- 160.Bernstein F.C., Koetzle T.F., Williams G.J., Meyer E.F., Jr., Brice M.D., Rodgers J.R., et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 161.Goel R., Muthusamy B., Pandey A., Prasad T.K. Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol. Biotechnol. 2011;48(1):87–95. doi: 10.1007/s12033-010-9336-8. [DOI] [PubMed] [Google Scholar]
- 162.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33(suppl_1):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2010;39(suppl_1):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pujol A., Mosca R., Farrés J., Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol. Sci. 2010;31(3):115–123. doi: 10.1016/j.tips.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 165.Oprea T.I., Bauman J.E., Bologa C.G., Buranda T., Chigaev A., Edwards B.S., et al. Drug repurposing from an academic perspective. Drug Discov. Today Ther. Strat. 2011;8(3–4):61–69. doi: 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Roin B.N. Unpatentable drugs and the standards of patentability. Tex. Law Rev. 2008;87:503. [Google Scholar]
- 167.Swinney D.C., Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011;10(7):507. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 168.Fields J.D., Bhardwaj A. 2009. Non-peptide arginine-vasopressin antagonists (vaptans) for the treatment of hyponatremia in neurocritical care: a new alternative? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kuter D.J. New thrombopoietic growth factors. Blood. 2007;109:4607–4616. doi: 10.1182/blood-2006-10-019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Selby P., Buick R.N., Tannock I. A critical appraisal of the human tumor stem-cell assay. N. Engl. J. Med. 1983;308(3):129–134. doi: 10.1056/NEJM198301203080304. [DOI] [PubMed] [Google Scholar]
- 171.Shah E.T., Upadhyaya A., Philp L.K., Tang T., Skalamera D., Gunter J., et al. Repositioning “old” drugs for new causes: identifying new inhibitors of prostate cancer cell migration and invasion. Clin. Exp. Metastasis. 2016;33(4):385–399. doi: 10.1007/s10585-016-9785-y. [DOI] [PubMed] [Google Scholar]
- 172.Eriksson A., Österroos A., Hassan S., Gullbo J., Rickardson L., Jarvius M.,., Larsson R. Drug screen in patient cells suggests quinacrine to be repositioned for treatment of acute myeloid leukemia. Blood Canc. J. 2015;5(4):e307. doi: 10.1038/bcj.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sant'Anna R., Gallego P., Robinson L.Z., Pereira-Henriques A., Ferreira N., Pinheiro F.,., Reixach N. Repositioning tolcapone as a potent inhibitor of transthyretin amyloidogenesis and associated cellular toxicity. Nat. Commun. 2016;7:10787. doi: 10.1038/ncomms10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Camus S., Quevedo C., Menéndez S., Paramonov I., Stouten P.F.W., Janssen R.A.J., et al. Identification of phosphorylase kinase as a novel therapeutic target through high-throughput screening for anti-angiogenesis compounds in zebrafish. Oncogene. 2012;31(39):4333. doi: 10.1038/onc.2011.594. [DOI] [PubMed] [Google Scholar]
- 175.Ridges S., Heaton W.L., Joshi D., Choi H., Eiring A., Batchelor L., et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119(24):5621–5631. doi: 10.1182/blood-2011-12-398818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tomlinson M.L., Hendry A.E., Wheeler G.N. Xenopus Protocols. Humana Press; Totowa, NJ: 2012. Chemical genetics and drug discovery in Xenopus; pp. 155–166. [DOI] [PubMed] [Google Scholar]
- 177.Tat J., Liu M., Wen X.Y. Zebrafish cancer and metastasis models for in vivo drug discovery. Drug Discov. Today Technol. 2013;10(1):e83–e89. doi: 10.1016/j.ddtec.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 178.Ai N., Wood R.D., Welsh W.J. Identification of nitazoxanide as a group I metabotropic glutamate receptor negative modulator for the treatment of neuropathic pain: an in silico drug repositioning study. Pharm. Res. 2015;32(8):2798–2807. doi: 10.1007/s11095-015-1665-7. [DOI] [PubMed] [Google Scholar]
- 179.Amelio I., Gostev M., Knight R.A., Willis A.E., Melino G., Antonov A.V. DRUGSURV: a resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell Death Dis. 2014;5(2) doi: 10.1038/cddis.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee H., Kang S., Kim W. Drug repositioning for cancer therapy based on large-scale drug-induced transcriptional signatures. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0150460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Turanli B., Grøtli M., Boren J., Nielsen J., Uhlen M., Arga K.Y., Mardinoglu A. Drug repositioning for effective prostate cancer treatment. Front. Physiol. 2018;9 doi: 10.3389/fphys.2018.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gaspar H.A., Gerring Z., Hübel C., Middeldorp C.M., Derks E.M., Breen G. Using genetic drug-target networks to develop new drug hypotheses for major depressive disorder. Transl. Psychiatry. 2019;9(1):117. doi: 10.1038/s41398-019-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Uddin Md Sahab, Kabir M.T., Al Mamun A., Abdel-Daim M.M., Barreto G.E., Ashraf G.M. APOE and Alzheimer’s disease: evidence mounts that targeting APOE4 may combat Alzheimer’s pathogenesis. Mol. Neurobiol. 2019;56:2450–2465. doi: 10.1007/s12035-018-1237-z. [DOI] [PubMed] [Google Scholar]
- 184.Buee L., Bussiere T., Buee-Scherrer V., Delacourte A., Hof P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Rev. 2000;33(1):95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 185.Hossain M.F., Uddin M.S., Uddin G.M.S., Sumsuzzman D.M., Islam M.S., Barreto G.E., Mathew B., Ashraf G.M. Melatonin in Alzheimer’s disease: a latent endogenous regulator of neurogenesis to mitigate Alzheimer’s neuropathology. Mol. Neurobiol. 2019 doi: 10.1007/s12035-019-01660-3. In Press. [DOI] [PubMed] [Google Scholar]
- 186.Kim E.K., Choi E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010:396–405. doi: 10.1016/j.bbadis.2009.12.009. 1802. [DOI] [PubMed] [Google Scholar]
- 187.Munoz L., Ammit A.J. Targeting p38 MAPK pathway for the treatment of Alzheimer's disease. Neuropharmacology. 2010;58:561–568. doi: 10.1016/j.neuropharm.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 188.Wang J.Z., Grundke-Iqbal I., Iqbal K. Kinases and phosphatases and tau sites involved in Alzheimer neurofibrillary degeneration. Eur. J. Neurosci. 2007;25:59–68. doi: 10.1111/j.1460-9568.2006.05226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang Y., Li H.L., Wang D.L., Liu S.J., Wang J.Z. A transitory activation of protein kinase-A induces a sustained tau hyperphosphorylation at multiple sites in N2a cells-imply a new mechanism in Alzheimer pathology. J. Neural Transm. 2006;113:1487–1497. doi: 10.1007/s00702-005-0421-2. [DOI] [PubMed] [Google Scholar]
- 190.Nykanen N.P. 2016. Cellular Physiology and Cell-To-Cell Propagation of Tau in Neurodegeneration: the Impact of Late-Onset Alzheimer's Disease Susceptibility Genes. [Google Scholar]
- 191.Harilal S., Jose J., Parambi D.G.T., Kumar R., Mathew G.E., Uddin M.S., et al. Advancements in nanotherapeutics for Alzheimer's disease: current perspectives. J. Pharm. Pharmacol. 2019;71:1370–1383. doi: 10.1111/jphp.13132. [DOI] [PubMed] [Google Scholar]
- 192.Silva T., Mohamed T., Shakeri A., Rao P.P., da Silva P.S., Remião F., Borges F. Repurposing nitrocatechols: 5-Nitro-α-cyanocarboxamide derivatives of caffeic acid and caffeic acid phenethyl ester effectively inhibit aggregation of tau-derived hexapeptide AcPHF6. Eur. J. Med. Chem. 2019;167:146–152. doi: 10.1016/j.ejmech.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 193.Bourque M., Morissette M., Di Paolo T. Repurposing sex steroids and related drugs as potential treatment for Parkinson's disease. Neuropharmacology. 2019;147:37–54. doi: 10.1016/j.neuropharm.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 194.Johnston T.H., Lacoste A.M., Visanji N.P., Lang A.E., Fox S.H., Brotchie J.M. Repurposing drugs to treat l-DOPA-induced dyskinesia in Parkinson's disease. Neuropharmacology. 2019;147:11–27. doi: 10.1016/j.neuropharm.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 195.Zhu K., Sun J., Kang Z., Zou Z., Wu X., Wang Y., et al. Repurposing of omeprazole for oligodendrocyte differentiation and remyelination. Brain Res. 2019;1710:33–42. doi: 10.1016/j.brainres.2018.12.037. [DOI] [PubMed] [Google Scholar]
- 196.Yong V.W. Prospects for neuroprotection in multiple sclerosis. Front. Biosci. 2004;9(864):1. doi: 10.2741/1276. [DOI] [PubMed] [Google Scholar]
- 197.Tranfaglia M.R., Thibodeaux C., Mason D.J., Brown D., Roberts I., Smith R.,., Cogram P. Repurposing available drugs for neurodevelopmental disorders: the fragile X experience. Neuropharmacology. 2019;147:74–86. doi: 10.1016/j.neuropharm.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 198.Wong A.S., Lee R.H., Cheung A.Y., Yeung P.K., Chung S.K., Cheung Z.H., Ip N.Y. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson's disease. Nat. Cell Biol. 2011;13(5):568. doi: 10.1038/ncb2217. [DOI] [PubMed] [Google Scholar]
- 199.Pandey V., Ranjan N., Narne P., Babu P.P. Roscovitine effectively enhances antitumor activity of temozolomide in vitro and in vivo mediated by increased autophagy and Caspase-3 dependent apoptosis. Sci. Rep. 2019;9(1):5012. doi: 10.1038/s41598-019-41380-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bach S., Knockaert M., Reinhardt J., Lozach O., Schmitt S., Baratte B., et al. Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 2005;280(35):31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 201.Lewis D.B. Developmental immunology and role of host defenses in neonatal susceptibility to infection. Infect. Dis. Fetus N.born Infant. 1992:20–98. [Google Scholar]
- 202.Dvorak H.F., Nagy J.A., Feng D., Brown L.F., Dvorak A.M. In: Vascular Growth Factors and Angiogenesis (Pp. 97-132) HeidelbergFerrara N., editor. vol. 56. Springer; Berlin: 1999. Vascular permeability factor/vascular endothelial growth factor and the significance of microvascular hyperpermeability in angiogenesis; pp. 794–814. (1999) Kidney Int. [DOI] [PubMed] [Google Scholar]
- 203.Ferrara N. Role of vascular endothelial growth factor in regulation of physiological angiogenesis. Am. J. Physiol. Cell Physiol. 2001;280(6):C1358–C1366. doi: 10.1152/ajpcell.2001.280.6.C1358. [DOI] [PubMed] [Google Scholar]
- 204.Carmeliet P., Jain R.K. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 205.Chan A.S., Leung S.Y., Wong M.P., Yuen S.T., Cheung N., Fan Y.W., Chung L.P. Am. J. Surg. Pathol. 1998;22:816–826. doi: 10.1097/00000478-199807000-00004. [DOI] [PubMed] [Google Scholar]
- 206.Xu L., Fukumura D., Jain R.K. Acidic extracellular pH induces vascular endothelial growth factor (VEGF) in human glioblastoma cells via ERK1/2 MAPK signaling pathway Mechanism of low pH-induced VEGF. J. Biol. Chem. 2002;277(13):11368–11374. doi: 10.1074/jbc.M108347200. [DOI] [PubMed] [Google Scholar]
- 207.Heinrich M.C., Corless C.L., Demetri G.D., Blanke C.D., Von Mehren M., Joensuu H., et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 2003;21(23):4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 208.Wardelmann E., Merkelbach-Bruse S., Pauls K., Thomas N., Schildhaus H.U., Heinicke T., et al. Polyclonal evolution of multiple secondary KIT mutations in gastrointestinal stromal tumors under treatment with imatinib mesylate. Clin. Cancer Res. 2006;12(6):1743–1749. doi: 10.1158/1078-0432.CCR-05-1211. [DOI] [PubMed] [Google Scholar]
- 209.Guo T., Hajdu M., Agaram N.P., Shinoda H., Veach D., Clarkson B.D., et al. Mechanisms of sunitinib resistance in gastrointestinal stromal tumors harboring KITAY502-3ins mutation: an in vitro mutagenesis screen for drug resistance. Clin. Cancer Res. 2009;15(22):6862–6870. doi: 10.1158/1078-0432.CCR-09-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lu T., Chen C., Wang A., Jiang Z., Qi Z., Hu Z., et al. Repurposing cabozantinib to GISTs: overcoming multiple imatinib-resistant cKIT mutations including gatekeeper and activation loop mutants in GISTs preclinical models. Cancer Lett. 2019;447:105–114. doi: 10.1016/j.canlet.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 211.Chatziathanasiadou M.V., Stylos E.K., Giannopoulou E., Spyridaki M.H., Briasoulis E., Kalofonos H.P., et al. Development of a validated LC-MS/MS method for the in vitro and in vivo quantitation of sunitinib in glioblastoma cells and cancer patients. J. Pharm. Biomed. Anal. 2019;164:690–697. doi: 10.1016/j.jpba.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 212.Kuenzi B.M., Rix L.L.R., Kinose F., Kroeger J.L., Lancet J.E., Padron E., Rix U. Off-target based drug repurposing opportunities for tivantinib in acute myeloid leukemia. Sci. Rep. 2019;9(1):606. doi: 10.1038/s41598-018-37174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Banerji V., Frumm S.M., Ross K.N., Li L.S., Schinzel A.C., Hahn C.K., et al. The intersection of genetic and chemical genomic screens identifies GSK-3α as a target in human acute myeloid leukemia. J. Clin. Investig. 2012;122(3):935–947. doi: 10.1172/JCI46465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cohen, P. Frame S 2001 the renaissance of GSK3. Nat. Rev. Mol. Cell Biol., 2, 769-776. [DOI] [PubMed]
- 215.Mihai D.P., Nitulescu G.M., Smith J.L., Hirsch A.J., Stecoza C.E. Dengue virus replication inhibition by dibenzothiepin derivatives. Med. Chem. Res. 2019;28(3):320–328. [Google Scholar]
- 216.Lv B.M., Tong X.Y., Quan Y., Liu M.Y., Zhang Q.Y., Song Y.F., Zhang H.Y. Drug repurposing for Japanese encephalitis virus infection by systems biology methods. Molecules. 2018;23(12):3346. doi: 10.3390/molecules23123346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Field-Smith A., Morgan G.J., Davies F.E. Bortezomib (Velcade™) in the treatment of multiple myeloma. Ther. Clin. Risk Manag. 2006;2(3):271. doi: 10.2147/tcrm.2006.2.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang S., Liu H., Zu X., Liu Y., Chen L., Zhu X., et al. The ubiquitin-proteasome system is essential for the productive entry of Japanese encephalitis virus. Virology. 2016;498:116–127. doi: 10.1016/j.virol.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 219.Passi A., Rajput N.K., Wild D.J., Bhardwaj A. RepTB: a gene ontology based drug repurposing approach for tuberculosis. J. Cheminf. 2018;10(1):24. doi: 10.1186/s13321-018-0276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Alderwick L.J., Lloyd G.S., Ghadbane H., May J.W., Bhatt A., Eggeling L., et al. The C-terminal domain of the arabinosyltransferase Mycobacterium tuberculosis EmbC is a lectin-like carbohydrate binding module. PLoS Pathog. 2011;7(2) doi: 10.1371/journal.ppat.1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Singh A., Somvanshi P., Grover A. Drug repurposing against arabinosyl transferase (EmbC) of Mycobacterium tuberculosis: essential dynamics and free energy minima based binding mechanics analysis. Gene. 2019;693:114–126. doi: 10.1016/j.gene.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 222.Keighobadi M., Emami S., Fakhar M., Shokri A., Mirzaei H., Teshnizi S.H. Repurposing azole antifungals into antileishmanials: novel 3-triazolylflavanones with promising in vitro antileishmanial activity against Leishmania major. Parasitol. Int. 2019;69:103–109. doi: 10.1016/j.parint.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 223.Bravo-Santano N., Stölting H., Cooper F., Bileckaja N., Majstorovic A., Ihle N.,., Letek M. Host-directed kinase inhibitors act as novel therapies against intracellular Staphylococcus aureus. Sci. Rep. 2019;9(1):4876. doi: 10.1038/s41598-019-41260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Li C.C., Wang X.J., Wang H.C.R. Repurposing host-based therapeutics to control coronavirus and influenza virus. Drug Discov. Today. 2019;24(3):726–736. doi: 10.1016/j.drudis.2019.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Oliveira I.M., Borges A., Borges F., Simões M. Repurposing ibuprofen to control Staphylococcus aureus biofilms. Eur. J. Med. Chem. 2019;166:197–205. doi: 10.1016/j.ejmech.2019.01.046. [DOI] [PubMed] [Google Scholar]
- 226.Chowdhury N., Wood T.L., Martínez-Vázquez M., García-Contreras R., Wood T.K. DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol. Bioeng. 2016;113(9):1984–1992. doi: 10.1002/bit.25963. [DOI] [PubMed] [Google Scholar]
- 227.Yuan M., Chua S.L., Liu Y., Drautz-Moses D.I., Yam J.K.H., Aung T.T., et al. Repurposing the anticancer drug cisplatin with the aim of developing novel Pseudomonas aeruginosa infection control agents. Beilstein J. Org. Chem. 2018;14(1):3059–3069. doi: 10.3762/bjoc.14.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pic E., Burgain A., Sellam A. Repurposing the anthelminthic salicylanilide oxyclozanide against susceptible and clinical resistant Candida albicans strains. Med. Mycol. 2018;57(3):387–390. doi: 10.1093/mmy/myy027. [DOI] [PubMed] [Google Scholar]
- 229.Patrick M.T., Stuart P.E., Raja K., Gudjonsson J.E., Tejasvi T., Yang J., et al. Genetic signature to provide robust risk assessment of psoriatic arthritis development in psoriasis patients. Nat. Commun. 2018;9(1):4178. doi: 10.1038/s41467-018-06672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kurayoshi M., Yamamoto H., Izumi S., Kikuchi A. Post-translational palmitoylation and glycosylation of Wnt-5a are necessary for its signalling. Biochem. J. 2007;402(3):515–523. doi: 10.1042/BJ20061476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Aumiller V., Balsara N., Wilhelm J., Günther A., Königshoff M. WNT/β-catenin signaling induces IL-1β expression by alveolar epithelial cells in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2013;49(1):96–104. doi: 10.1165/rcmb.2012-0524OC. [DOI] [PubMed] [Google Scholar]
- 232.Chen X., Shi C., Meng X., Zhang K., Li X., Wang C., et al. Inhibition of Wnt/β-catenin signaling suppresses bleomycin-induced pulmonary fibrosis by attenuating the expression of TGF-β1 and FGF-2. Exp. Mol. Pathol. 2016;101(1):22–30. doi: 10.1016/j.yexmp.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Boyapally R., Pulivendala G., Bale S., Godugu C. Niclosamide alleviates pulmonary fibrosis in vitro and in vivo by attenuation of epithelial-to-mesenchymal transition, matrix proteins & Wnt/β-catenin signaling: a drug repurposing study. Life Sci. 2019;220:8–20. doi: 10.1016/j.lfs.2018.12.061. [DOI] [PubMed] [Google Scholar]
- 234.Fang J., Cai C., Chai Y., Zhou J., Huang Y., Gao L., et al. Quantitative and systems pharmacology 4. Network-based analysis of drug pleiotropy on coronary artery disease. Eur. J. Med. Chem. 2019;161:192–204. doi: 10.1016/j.ejmech.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]






