Abstract
Recently, RNA viral infections caused by respiratory viruses, such as influenza, parainfluenza, respiratory syncytial virus, coronavirus, and Middle East respiratory syndrome-coronavirus (MERS-CoV), and Zika virus, are a major public health threats in the world. Although myriads of diagnostic methods based on RNA amplification have been developed in the last decades, they continue to lack speed, sensitivity, and specificity for clinical use. A rapid and accurate diagnostic method is needed for appropriate control, including isolation and treatment of the patients. Here, we report an isothermal, label-free, one-step RNA amplification and detection system, termed as iROAD, for the diagnosis of respiratory diseases. It couples a one-step isothermal RNA amplification method and a bio-optical sensor for simultaneous viral RNA amplification/detection in a label-free and real-time manner. The iROAD assay offers a one-step viral RNA amplification/detection example to rapid analysis (<20 min). The detection limit of iROAD assay was found to be 10-times more sensitive than that of real-time reverse transcription-PCR method. We confirmed the clinical utility of the iROAD assay by detecting viral RNAs obtained from 63 human respiratory samples. We envision that the iROAD assay will be useful and potentially adaptable for better diagnosis of emerging infectious diseases including respiratory diseases.
Keywords: Respiratory disease, Emerging infectious disease, RNA amplification/detection, Label-free biosensor
Highlights
-
•
Employed isothermal amplification with bio-photonic sensor for detection of viral RNAs.
-
•
Rapid RNA amplification and detection system simultaneously within 20 min.
-
•
Validated the utility of the system in human respiratory viral samples.
-
•
Versatile technology can be readily applied to other RNA-based studies and diseases.
1. Introduction
Emerging respiratory infections caused by viruses, such as influenza (IFN) type-A and B, respiratory syncytial virus (RSV) type-A and B, human coronavirus types OC43 and 229E, parainfluenza virus (PIV) types 1–4, adenovirus, and rhinovirus, present a major public health hazard (Cox and Subbarao, 2000; van der Hoek et al., 2004; Guy et al., 2000; Loeffelholz and Chonmaitree, 2010). Especially, acute respiratory tract infection is the leading cause of hospitalization, and a major reason of death in infants and children in developing countries (Tregoning and Schwarze, 2010; Bourgeois et al., 2009). In particular, the failure to rapidly and accurately diagnose contagious respiratory viruses results in an increased number of patients and duration of infection. Hence, the testing methods for accurate pathogen identification have been explored to improve the patient's outcome, allow appropriate use of antibiotics, and facilitate cost-effective care. Recently, the Middle East respiratory syndrome-coronavirus (MERS-CoV) and ZIKA virus outbreaks in 2015 has also heightened our awareness regarding the need for improved diagnostic methods (Cho et al., 2016, Kim et al., 2016, Gourinat et al., 2015). Therefore, there is a great need for an affordable, robust, rapid, accurate, flexible, and simple point-of-care (POC) testing in order to control the unpredictable pandemics.
Although a myriad of techniques for rapid diagnosis of pathogens have been developed, the cell culture method remains the gold standard in many laboratories and hospitals owing to the lack of an alternative high sensitivity technique (Moesker et al., 2016; Ginocchio and McAdam 2011). Although cell culture is a time-consuming and laborious method, it can generally detect over 90% of the viruses within 48 h (Ginocchio and McAdam, 2011 ; Pozzetto et al., 2010). Rapid antigen direct tests (RADT) and direct fluorescent antibody testing (DFA) have been developed as alternative techniques, which are simple, cheap, with a rapid turnaround time of 15–30 min (Mahony et al., 2007; Leland and Ginocchio, 2007; Welch and Ginocchio, 2010; Landry, 2009). ). However, these methods are limited by the availability of antibodies against newly found viruses and sub-families of pathogens (Guy et al., 2000, Borg et al., 2003, Renois et al., 2010). In the absence of appropriate diagnostic methods, it is impossible to rapidly predict the type of pathogen from clinical signs and symptoms, due to an overlap in the clinical phenotype of various infections. Alternatively, real-time reverse transcriptase (RT) polymerase chain reaction (PCR) is the current gold standard because of its superior sensitivity, rapid turnaround time (2–4 h), and ability to identify multiple types of pathogens in a single test (Moesker et al., 2016, Olofsson et al., 2011, Pillet et al., 2013, Mentel et al., 2003). As a result, many techniques based on the real-time RT-PCR, such as RespPlex (Qiagen), Infiniti system (AutoGenomics), Jaguar system (BD), FilmArray system (BioFire Diagnostics), and PLEX-ID (Abbott Molecular), have been commercialized for the detection of respiratory viruses in clinical use (Caliendo, 2011; Hammond et al., 2012). However, these are not well implemented in all hospitals, as they require a molecular diagnostic laboratory with specialized personnel, equipment, and time to validate the products with large clinical samples.
Meanwhile, isothermal RNA amplification techniques have emerged as an alternative to RT-PCR in order to bypass thermal constraints, as RNA is easily degraded at high temperatures, and by materials and reagents (Agrawal et al., 2007; Compton, 1991; Notomi et al., 2000; Lizardi et al., 1998; Vincent et al., 2004). Many isothermal amplification methods have been developed to allow exponential amplification at both constant and low temperatures. Among the methods, recombinase polymerase amplification (RPA) does not require thermal cycling and operates at a single temperature (Piepenburg et al., 2006, Shin et al., 2013a, Shin et al., 2015a). RPA forms a complex of a primer and a recombinase enzyme to extend the DNA, which negates the need for a polymerase and cycle repetition. Recently, reverse transcription-based RPA technique (RPA-RT) for RNA has been developed and applied for the amplification of RNA from Bovine coronavirus (BCoV), human immunodeficiency virus (HIV), influenza (IFN), dengue virus (DENV), and ebola virus (EVD) (Yang et al., 2016, Lillis et al., 2014, El Wahed et al., 2013, Teoh et al., 2015; El Wahed et al., 2015; Amer et al., 2013). Despite the advantages of the isothermal methods, these methods still require improved sensitivity, additional steps including gel electrophoresis, and labeling with a fluorescent dye for detection. Together with this, biosensors that are label-free with real-time detection have been developed to overcome the limitations of isothermal methods, such as reduced reaction time and cost by eliminating the need for cycle completion, gel electrophoresis, and labeling (Liu et al., 2016, Baeumner et al., 2003, Yan et al., 2016). Recently, silicon-based photonic biosensors have been developed as a sensitive bio-molecules detection technology that does not require additional materials for signal enhancement, such as chemical amplification or labeling of the analyte (Iqbal et al., 2010, Bogaerts et al., 2012). Particularly, silicon microring resonators (SMRs) are refractive index-based optical sensors that provide highly sensitive, label-free, real-time multiplexed detection of biomolecules near the sensor surface. The silicon photonic sensors transduce the presence of target molecules based on binding-induced changes in the refractive index proximal to the waveguide surface (Iqbal et al., 2010, Bogaerts et al., 2012). Furthermore, the fabrication of the SMRs by complementary metal oxide-semiconductor (CMOS) technology ensures the ability to minimize costs while providing large scale-up capabilities and makes the microring resonator a good candidate for a disposable sensor in POC diagnostics (Park et al., 2013, Shin et al., 2015a, De Vos et al., 2007, De Vos et al., 2009).
In this study, we report an isothermal and rapid one-step RNA amplification/detection (iROAD) assay to simultaneously amplify and detect the viral RNA in a label-free and real-time manner. The assay is rapid, affordable, simple, and accurate. Moreover, to the best of our knowledge, this is the first proof-of concept study that combines isothermal reverse transcriptase RNA amplification with RPA-RT reagents as an asymmetric RNA amplification method and a SMR biophotonic sensor as a label-free and real-time detection in a single chamber. We demonstrated that the detection limit of iROAD assay was 10-times higher than that of real-time RT-PCR method. Furthermore, we demonstrated the clinical utility of the iROAD assay by detecting respiratory viral RNAs extracted from the 63 nasopharyngeal samples with either IFN-A/B or HCoV-OC43/229E or RSV-A/B. The iROAD assay combines amplification and detection mechanism of RNA, resulting in increased sensitivity and specificity of viral RNA detection. Using this strategy, respiratory virus nucleic acid from human specimens was simultaneously amplified and detected within 20 min by a grafted complementary primer on the SMR in a label-free and real-time manner. Therefore, it is potentially adaptable for better diagnosis across various clinical applications involving RNA.
2. Experimental
2.1. Development and operation of iROAD chip
To use the iROAD chip as a detection system, a previously described protocol was used with slight modifications for the detailed structure and fabrication of silicon microring resonators (SMRs) (Shin et al., 2013a, Shin et al., 2013b, Park et al., 2013). The SMR sensor device was provided from One BioMed Pte. Ltd. Briefly, the iROAD chip [2.5 cm x 1 cm x 0.3 cm] structures such as microring structures, waveguides, and gratings, were patterned on a commercially available 200 mm Silicon-On-Insulator (SOI) wafer with a 220 nm thick top silicon layer, and 2 µm thick buried oxide layer by 210 nm deep ultraviolet (UV) lithography. The structures were then etched into the buried oxide layer by a reactive ion-etching process, followed by the deposition of 1.5 mm high-density plasma (HDP) oxide as a top cladding layer (Park et al., 2013, Shin et al., 2015a). We checked the layer thickness after every lay by layer deposition as a process control for the sensor uniformity during the chip fabrication. An array of microrings was designed to consist of four rings that are connected to a common input waveguide (through). Each ring had a dedicated output waveguide (drop). One of the microrings, left under the SiO2 cladding, is used as a reference sensor to monitor temperature-induced drift. The output signals of the 3 remaining microrings are collected through a vertical grating coupler connected to a single-mode fiber optic probe (Fig. S1). The tunable laser emits light from wavelength 1510–1612 nm, which corresponds to frequency from 1.861×105 GHz to 1.987×105 GHz. The ring resonance structure has multiple resonant wavelengths (frequencies) within the above wavelength range and the resonant peak we used for monitoring the wavelength shift is at ~1550 nm (1.987×105 GHz). The insertion loss (IL) spectrum was measured using an EXFO IQS-2600B DWDM passive component test system (Park et al., 2013, Shin et al., 2015a).
For operation of the iROAD, the chip was prepared in three steps. First, the surface of the chip was functionalized with an amine group for immobilization of the primer, as an asymmetric technique. The SMR sensors were treated with oxygen plasma and immersed in a solution of 2% 3-aminopropyltriethoxysiane (APTES) in a mixture of ethanol–H2O (95:5, v/v) for 2 h, followed by thorough rinsing with ethanol and diethylpyrocarbonate (DEPC)-treated deionized (DI) water. The sensors were cured by drying under a nitrogen stream and heating to 120 °C for 15 min. The sensors were then incubated with 2.5% glutaraldehyde (GAD) in DEPC-DI water containing 5 mM sodium cyanoborohydride for 1 h, rinsed with DEPC-DI water, and dried under a nitrogen stream. Second, for the immobilization of the target primers (Table S1), the pre-treated sensor was prepared by incubation with the primers of IFN, HCoV, and RSV in PBS (1 mM) containing 5 mM sodium cyanoborohydride for 16 h at room temperature. After the incubation, unbound DNA probes were washed away with PBS and the sensors were dried using nitrogen. To streamline the assay procedure, an acrylic well [6 mm x 1.5 mm x 1 mm] was used to enclose the sensing area. At this time, the chips were considered ready for optical measurements. Lastly, we prepared the RPA-RT solution for amplification and detection of the target RNA using the iROAD. For an optimized reaction, 29.5 mL of rehydration buffer, 15 mL of RNase inhibitor and water, 2 μM DTT, 2.5 mL of primers (10 mM) were mixed. One dried enzyme pellet was added to each solution and vortexed. Then, 2.5 mL of magnesium acetate solution was dispensed into the cap of each tube. A unidirectional shake mode mixing protocol guaranteed a homogeneous distribution of the molecules that are necessary for the reaction in the buffer. After mixing, the total volume of 50 μL of reaction buffer was split into five 10 μL aliquots. The viral RNA samples obtained from the patient samples were used for the detection of viral RNA. To start the reactions, 5 μL of target RNA (wild-type) were added to each 10 μL reaction aliquot. Finally, we added the RPA-RT solutions containing the viral RNA targets or genomic RNA (negative control) to the acrylic well at room temperature. Before closing the acrylic well, we added mineral oil to protect the solution from evaporation during the amplification. The iROAD assay was operated at a constant temperature (43 °C). A thermo-electric cooler (TEC), connected to a proportional integral derivative controller (Alpha Omega Instruments, USA), was employed to maintain the constant temperature. The resonance spectrum of the device was immediately measured and used as a reference to obtain a baseline. The SMRs allow target molecules to selectively bind to the immobilized primers in the evanescent field of the resonator waveguide, subsequently causing an increase in the proportion of each wavelength. During the amplification process, the wavelength shift was collected every 5 min for up to 30 min to monitor the amplification of target RNA in a label-free and real-time manner.
2.2. Conventional assays
Conventional assays, such as end-point reverse transcription (RT)-PCR and real-time RT-PCR, were compared with the iROAD assay to test its utility. The forward and reverse primers were synthesized at the usual length of around 24 bp (Table S1). The target viral RNA as a template for the conventional assays was obtained from the clinical samples. The human genomic RNA was used as a non-target control (negative control). The end-point RT-PCR process consisted of an initial cDNA synthesis step of 30 min at 50 °C, followed by 15 min at 95 °C and 45 cycles of 30 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C, and a final elongation step at 72 °C 10 min. Viral RNA (5 μL) was amplified in a total volume of 25 μL, containing 5X one-step RT-PCT buffer (Qiagen One-step RT-PCR kit), 0.25 mM deoxynucleotide triphosphate, 25 pmol of each primer, and 1 unit of one-step RT-PCR Enzyme Mix (Qiagen, Germany). Gel electrophoresis was used to separate PCR products on a 2% agarose gel containing ethidium bromide (EtBr). The gel was visualized using a Gel Doc System (Clinx Science Instruments). For real-time RT-PCR, the following procedure was modified in the AriaMx (Aligent) Instrument protocol. Briefly, 5 μL of RNA was amplified in a total volume of 20 μL, containing 2x brilliant SYBR green RT-qPCR master mix, 25 pmol of each primer, and 5 μL of RNA template. An initial cDNA synthesis step of 20 min at 50 °C, followed by 15 min at 95 °C, fifty cycles of 15 s at 95 °C, 20 s at 55 °C, and 20 s at 72 °C, and by cooling step of 40 °C for 30 s. The amplified products with SYBR Green signals were obtained using an AriaMx Real-Time PCR System (Agilent).
2.3. T7 in vitro transcribed RNA
To check the detection limit of the iROAD assay, T7 in vitro transcribed RNA was generated with either IFN-B or HCoV-OC43 (MEGAscript T7 kit, Ambion, Austin, TX, USA) (Moll et al., 2004; Bustin, 2000). First, we performed end-point RT-PCR to obtain the PCR product of IFN-B or HCoV-OC43. Then, a mixture including 2 μL of 10X reaction buffer, 75 mM NTP (ATP, CTP, GTP, UTP), enzyme mix, and 0.2 μg of PCR product in a volume of 20 μL was incubated at 37 °C overnight. To get rid of cDNA in the mixture, 1 μL of DNase was added to the mixture and incubated at 37 °C for 15 min. After the T7 in vitro transcription, we performed the purification of T7 in vitro transcribed RNA (MEGA clear kit, Ambion, Austin, TX, USA). Finally, we obtained the purified T7 in vitro transcribed RNA with either IFN-B or HCoV-OC43. The purified T7 RNA of IFN-B was stored at −80 °C until use.
2.4. RNA extraction from clinical specimens
The nasopharyngeal samples from the patients infected with influenza (IFN)-A/B, human coronavirus (HCoV)-OC43/229E, or respiratory syncytial virus (RSV)-A/B were obtained using protocols approved by the institutional review board of Asan Medical Center (AMC), Republic of Korea. Institutional approval and written informed consent from the patients were obtained. The viral RNA samples were extracted from the patient samples using QIAamp viral RNA mini kit (Qiagen, Germany) according to the manufacturer's instructions (Guy et al., 2000; Leland and Ginocchio, 2007; Welch and Ginocchio, 2010). We used samples at a starting volume of 200 μL each, and eluted around 60 μL using viral elution buffer. The extracted RNA was then aliquoted, and stored at −80 °C until use.
3. Results and discussion
3.1. iROAD assay for viral RNA detection
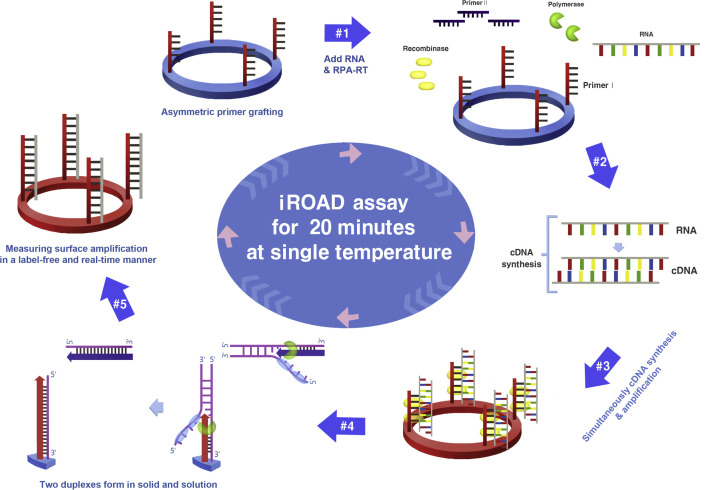
Fig. 1 illustrates the iROAD assay that was designed for the clinical detection of viral RNAs, which were extracted from the nasopharyngeal swab samples with either Influenza (IFN)-A/B or human coronavirus (HCoV)-OC43/229E or respiratory syncytial virus (RSV)-A/B, by the QIAamp viral RNA mini kit. Following the extraction of viral RNA, the target region was amplified by an isothermal-based asymmetric RNA amplification method through recombinase polymerase amplification-reverse transcription (RPA-RT) reagents. For the reaction of iROAD assay, one primer was grafted covalently to an optical sensor surface and the other was in solution, while the temperature was kept constant at 43 °C. The amplified viral RNA targets were simultaneously detected with the grafted primer on the sensor surface in a label-free and real-time manner (Fig. 1). First, selection of primer pairs used for amplification and detection in both solid- and solution-phases was critical for the specific amplification of viral RNA. The primer pair sequences from each viral infection were complementary to the target sequences (Table S1). In order to avoid the formation of primer-dimers, one primer was immobilized and possessed a 5′- amine group as an asymmetric assay on the silicon microring resonators (SMR), which could be used as unique refractive index sensitive sensors that allow target molecules to selectively bind to the immobilized receptors in the evanescent field of the resonator waveguide, subsequently causing an increase in the proportion of each wavelength during the reaction. Second, during the amplification process, complementary DNA (cDNA) was obtained from the viral RNA template after reverse transcription (RT) reaction. As a result, the cDNA was hybridized with the immobilized primer on the amine-modified SMR surface, and then the target amplification commenced from the RPA-RT mixture. This resulted in binding of the recombinase-primer complex to double-stranded cDNA by components of the proteins (namely gp32, uvsX, and uvsY) to facilitate strand exchange, leading to strand elongation and an exponential increase in the number of target cDNA copies. This assay was placed on an in-house thermal pad with an optical instrument to enable the simultaneous amplification and detection of viral RNA. Finally, the exponentially amplified targets by repetition of the reaction on the sensor surface are monitored by the wavelength shift of the microring resonator without any labeling and in a real-time manner. Subsequently, the iROAD assay detected the respiratory viral targets from the clinical sample by measuring the resonance wavelength shift within 15–20 min (Fig. 1).
Fig. 1.
Schematic representation of the principle of an isothermal, rapid and label-free one-step RNA amplification/detection (iROAD) assay. First, preparation of the iROAD chip through the primers (forward) grafting on the optical sensor would be needed for a ready-to-use viral RNA detection assay (#1). Then, the mixture containing recombinase polymerase amplification-reverse transcription (RPA-RT) reagents, reverse primers, and extracted RNA is added into the reaction chip (#2). During the isothermal reaction, complementary DNA (cDNA) is synthesized from the RNA template via RPA-RT kit (#3). Thereafter, recombinase/primer complexes bind to double-stranded target cDNA and facilitate strand exchange at a constant temperature. After the displaced strand forms a D-loop by gp32 (sky blue), the immobilized primers are extended by polymerase (light green) on the surface of the silicon microring resonator (#4). The formation of two duplexes is caused by the amplification of the solid and the solution. The exponential RNA amplification after the reverse transcription based on the asymmetric assay is achieved by the repetition of the process (#5). The amplification and detection of the target is simultaneously monitored by measuring the wavelength shift on an optical sensor for 20 min. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. iROAD assay optimization
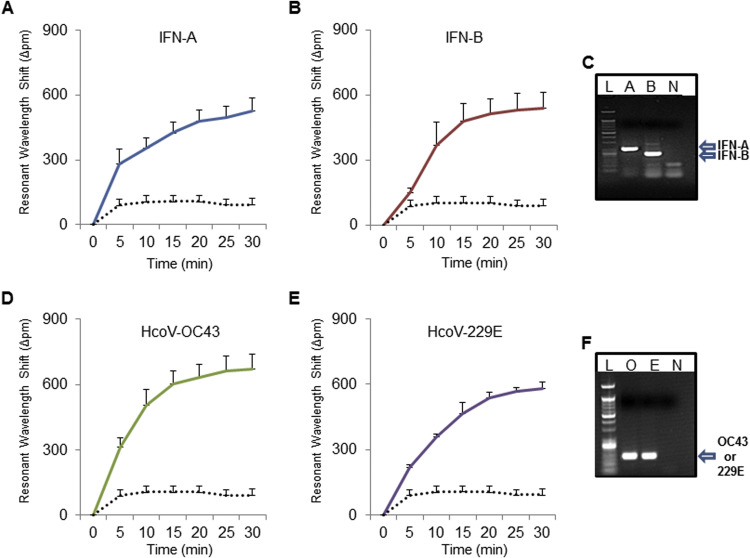
We first optimized the assay protocol to simultaneous amplify and detect the viral RNAs extracted from the samples. The viral RNAs were extracted from several types of viral infections, such as seasonal IFN-A/B, HCoV-OC43/229E, and RSV-A/B. In order to determine whether the iROAD assay could be useful for detection of viral RNAs for clinical use in a label-free and real-time manner, we examined the utility of the iROAD assay compared to the one-step end-point reverse transcription (RT)-PCR method. As shown in Fig. 2 and Fig. S1, the resonance wavelength shift in 30 min using the iROAD assay was 527.64 pm±56.79 for IFN-A, 540.59 pm±75.07 for IFN-B, 672.27 pm±65.68 for HCoV-OC43, 579.92 pm±30.66 for HCoV-229E, 616.10 pm±33.27 for RSV-A, and 769.51 pm±42.47 for RSV-B in the presence of the target viral RNAs. We also confirmed that the same target viral RNA strands were strongly amplified using the one-step end-point RT-PCR (Fig. 2C, F and Fig. S2C). The human genomic RNA from HCT116 cells was used as a negative control to monitor non-specific interaction with the non-target RNA. The wavelength shift in the absence of viral RNAs was 93.08 pm±45.94 owing to the background caused by the binding of nonspecific components such as polymerase and other chemical compounds to the sensor surface (Fig. 2A-B, D-E and Fig. S2A-B). The difference between the presence and absence of target RNA in the wavelength shift is easily distinguishable after 5 min. As a result, the data clearly indicate that the iROAD assay is able to amplify and detect the target RNA simultaneously on the optical sensor in a label-free and real-time manner, as compared to the non-target RNA.
Fig. 2.
iROAD assay optimization. Resonance wavelength shift using the iROAD assay shows the results of viral RNA amplification/detection in a label-free and real-time manner. (A, B) Resonance wavelength shift shows the result of the amplification/detection of influenza (IFN)-A (blue), B (dark red), and negative control (black dot). Error bars indicate standard deviation from the mean, based on at least 3 independent experiments. (C) Gel electrophoresis data for end-point reverse transcription (RT)-PCR product from IFN-A and B. (D, E) Resonance wavelength shift shows the amplification of human coronavirus (HCoV)-OC43 (green), HCoV-229E (purple), and negative control (black dot). (F) Gel electrophoresis data for end-point reverse transcription (RT)-PCR product from HCoV-OC43 and HCoV-229E. Error bars indicate standard deviation from the mean, based on at least 3 independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Detection limit of iROAD assay
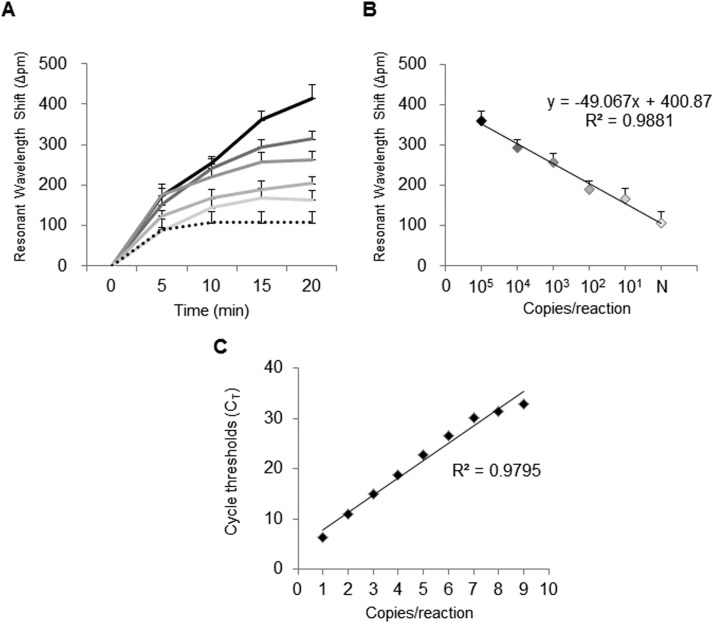
The detection limit of the iROAD assay was comprehensively characterized using the purified viral RNA samples with either T7 in vitro transcribed IFN-B or HCoV-OC43. The serially diluted samples ranging from 2.5×101 to 109 copies/reaction were used as RNA templates for determination of the absolute detection limit. We determined the relative detection limit of the iROAD assay, as compared to the conventional methods (end-point RT-PCR and real-time RT-PCR), using the serially diluted samples ( Fig. 3). In case of the iROAD assay with label-free and real-time capabilities, the wavelength shift was sequentially increased based on the concentrations (2.5×101 to 105 copies/reaction) of IFN-B within 20 min, as compared to the negative control. The resonant wavelength shift from 2.5×101 copies/reaction of IFN-B sample was clearly distinguishable from that of human genomic RNA (Fig. 3A). Moreover, the difference between the target (IFN-B) and non-target (human genomic RNA) was observed as early as 15 min after simultaneous amplification and detection. Fig. 3B shows good linearity (=0.9881) for different concentrations of the target after 20 min of amplification. In the case of the real-time RT-PCR assay, the fluorescent SYBR Green signal was observed in RNA samples diluted up to 2.5×102 copies/reaction and showed good linearity (=0.9795) for different concentrations of the target (Fig. 3C). Fig. 3 shows that the limit of detection in the iROAD assay was 10 times more sensitive than that of the real-time RT-PCR method. In addition, we also determined the relative detection limit of the iROAD assay with the serially diluted samples of HCoV-OC43 (Fig. S3). In the iROAD assay, the resonant wavelength shift from 2.5×101 copies/reaction of HCoV-OC43 sample is clearly distinguishable from that of human gRNA within 20 min (Fig. S3A). Fig. S2B shows good linearity (=0.9601) for different concentrations of the target in 20 min of amplification. The fluorescent SYBR Green signal in the real-time RT-PCR method was observed in RNA samples diluted up to 2.5×102 copies/reaction and showed good linearity (=0.9486) for the different concentration of the target (Fig. S3C). In the HCoV-OC43 sample, the detection limit of the iROAD assay was superior to that of real-time RT-PCR method. Therefore, this device would be useful as a rapid and highly sensitive method for respiratory viral RNA detection, based on a molecular diagnostic method.
Fig. 3.
Comparison of limit of detection with iROAD assay and conventional method using T7-in vitro transcribed IFN-B RNA. (A) Resonance wavelength shift in iROAD assay. The colors represent the amount of the target: black (2.5×105 copies/reaction), gray with darker 50% (2.5×104 copies/reaction), gray with darker 35% (2.5×103 copies/mL), gray with darker 25% (2.5×102 copies/mL), gray with darker 15% (2.5×101 copies/reaction), and black dot (negative). (B) Linear relationship between wavelength by iROAD assay and the concentration of target in 20 min. Error bars indicate standard deviation from the mean, based on at least 3 independent experiments. (C) Linear relationship between the concentration of target and Ct value of fluorescence signal by real-time RT-PCR. 1: 2.5×109 copies/reaction, 2: 2.5×108 copies/reaction, 3: 2.5×107 copies/reaction, 4: 2.5×106 copies/reaction, 5: 2.5×105 copies/reaction, 6: 2.5×104 copies/reaction, 7: 2.5×103 copies/reaction, 8: 2.5×102 copies/reaction and 9: negative control..
3.4. Sensitivity and specificity of iROAD assay in clinical samples
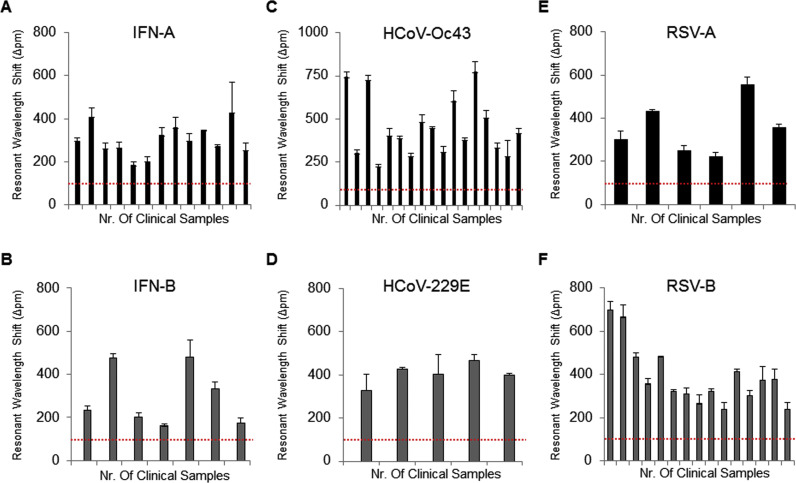
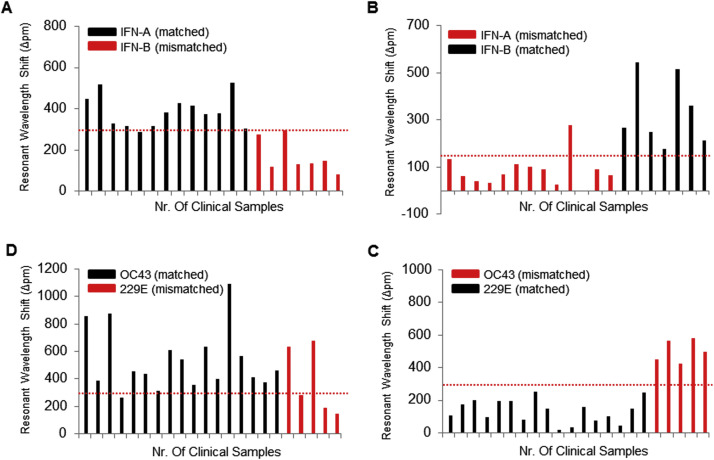
To validate the clinical utility of the iROAD assay, we analyzed 63 clinical nasopharyngeal samples from 13 IFN-A patients, 7 IFN-B patients, 17 HCoV-OC43 patients, 5 HCoV-229E patients, 6 RSV-A patients, and 15 RSV-B patients ( Fig. 4 and Fig. S4). We also compared efficiency of the iROAD assay with that of real-time RT-PCR assay using the same samples. The primers used for the detection of IFN-A/B, HCoV-OC43/229E, and RSV-A/B are shown in Table S1. Fig. 4 shows that the resonant wavelength shifts using the iROAD assay were above 150 pm within 20 min when the target samples were amplified with the matched target primers (Fig. 4A-B for IFN-A/B, Fig. 4C-D for HCoV-OC43/229E, and Fig. 4E-F for RSV-A/B). In order to further verify whether the target primer was amplified specifically, we used human genomic RNA obtained from HCT116 cell line as a non-target sample. The wavelength shift was below 100 pm by non-specific targeting. As a result, the viral RNAs from the variety samples were amplified and detected as positive samples when the human genomic RNA samples were used as negative controls (Fig. 4 and Fig. S4). Furthermore, the respiratory viruses subtypes may cause inaccurate detection due to cross-reactivity that should be distinguishable. Hence, we performed the cross-reactivity testing of the iROAD assay using the clinical samples ( Fig. 5 and Fig. S5). For example, when we analyzed the 13 IFN-A samples with the IFN-A primer for the sensitivity of the assay, the 7 IFN-B samples with the IFN-A primer were used as negative controls for the specificity of the assay or vice versa (Fig. 5A). Using the IFN-A primer, 12 out of 13 IFN-A samples were detected as true positives. One sample was observed as a false negative sample. On the other hand, 6 out of 7 IFN-B were detected as true negatives and one was observed as a false positive. In addition, when we analyzed the 7 IFN-B samples with the IFN-B primer, 13 IFN-A samples were used as negative controls (Fig. 5B). Using the IFN-B primer, all 7 IFN-B samples were detected as true positives. On the other hand, 12 out of 13 IFN-A samples were detected as true negatives. One sample was observed as a false positive sample (Fig. 5B). In case of HCoV, when we analyzed the 17 HCoV-OC43 samples with the HCoV-OC43 primer, 5 HCoV-229E samples were used as negative controls or vice versa (Fig. 5C-D). Using the HCoV-OC43 primer, 16 out of 17 HCoV-OC43 samples were detected as true positives. One sample was observed as a false negative. On the other hand, 2 out of 5 HCoV-229E samples were detected as false positives. Three samples were observed as true negatives. In addition, when we analyzed the 5 HCoV-229E samples with the HCoV-229E primer, 17 HCoV-OC43 samples were used as negative controls (Fig. 5C). Using the HCoV-229E primer, all 5 HCoV-229E samples were detected as true positives. All 17 HCoV-OC43 samples were detected as true negatives (Fig. 5D). As shown in Fig. S5 and Table S2, the iROAD assay with IFN-A from 15 min showed a value of 84–92% and 85.7% for sensitivity and specificity, respectively. The iROAD assay with IFN-B from 15 min showed a value of 100% and 92.3% for sensitivity and specificity, respectively. Furthermore, the iROAD assay with HCoV-OC43 from 15 min showed a value of 94.1% and 40–60% for sensitivity and specificity, respectively. The iROAD assay with HCoV-229E from 15 min showed a value of 100% for both sensitivity and specificity. Although the specificity of iROAD for the HCoV-OC43 was very low (40–60%) due to the lack of the samples, the iROAD was found to be a rapid (<20 min), highly sensitive, and specific assay for viral RNA detection. On the other hand, the sensitivity and specificity of the real-time RT-PCR assay were found to be insufficient using the same samples (Table S2). Therefore, we showed that the sensitivity and specificity of the iROAD assay was superior to that of real-time RT-PCR method.
Fig. 4.
Clinical utility of the iROAD assay. Analysis of 63 clinical nasopharyngeal samples from 13 IFN-A patients (A), 7 IFN-B patients (B), 17 HCoV-OC43 patients (C), 5 HCoV-229E patients (D), 6 RSV-A patients (E), and 15 RSV-B patients (F) with the matched primers in 20 min. The red dots represent as a negative control by non-specific binding. Error bars indicate standard deviation from the mean, based on at least 3 independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Cross-reactivity testing of the iROAD assay in clinical samples. (A) Analysis of 20 clinical nasopharyngeal samples from 13 IFN-A patients (as targets, black) and 7 IFN-B patients (as non-targets, red) with IFN-A primer in 30 min (B) Analysis of 20 clinical nasopharyngeal samples from 13 IFN-A patients (as non-targets, red) and 7 IFN-B patients (as targets, black) with IFN-B primer in 30 min (C) Analysis of 22 clinical nasopharyngeal samples from 17 HCoV-OC43 patients (as targets, black) and 5 HCoV-229E patients (as non-targets, red) with HCoV-OC43 primer in 30 min (D) Analysis of 22 clinical nasopharyngeal samples from 17 HCoV-OC43 patients (as non-targets, red) and 5 HCoV-229E patients (as targets, black) with HCoV-229E primer in 30 min. The red dots represent the cut off (criterion) for reporting a sample as virus (positive/negative) detected. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Conclusions
We have developed a new isothermal RNA amplification and detection assay system for rapid, simple, and label-free detection of viral RNAs. The combination of isothermal amplification and optical sensor-based detection not only simplifies the assay protocol considerably, but also enhances the sensitivity of detection. The iROAD assay has many innovative features, presenting a new multidisciplinary approach to diagnosis of respiratory viral infection. First, the iROAD assay enhances RNA amplification and detection speed through real-time detection using an optical sensor. This is a significant progress from our previous approach that utilized DNA oligonucleotides to target DNA; a method limited to DNA only. In contrast to DNA, RNA is easily degraded due to instability of the samples. Hence, the system should be compatible with RNA to avoid degradation. In addition, the cDNA synthesis step from RNA is a prerequisite for the amplification and detection of RNA. The iROAD assay is performed by the reverse transcription to cDNA synthesis in a single chip, followed by simultaneous amplification and detection in a real-time manner. Second, the iROAD assay is adapting the RPA-RT isothermal method (43 °C), which is widely used to avoid RNA degradation at a high temperature. Third, this assay is based on a label-free SMR sensor system and it exhibits the following properties: simplicity, scalability, multiplexing capability, affordability, and rapid analysis time. Although only single viral RNA detection has been reported in this study, this assay can be used to simultaneously target multiple RNA molecules. Finally, the iROAD is a versatile technology that could be readily applied to other RNA-based studies and diseases. By changing the primer sequences, it could be used to detect emerging pathogens in hospitals. This prototype is being improved for more robust operation by using an array of microrings (Fig. S1) for multiple detection in clinical use. Moreover, we are developing a sample-processing device to construct a fully integrated device with iROAD assay. Based on low-cost thin film and non-chaotropic reagents for nucleic acid extraction (Shin et al., 2015b), the platform enables robust nucleic acid extraction from a variety of sample sources such as blood, urine, and sputum. Such a system will further optimize the protocol with a large clinical cohort for the improvement of the sensitivity and specificity in clinical applications. We envision the ultimate integration of sample processing and detection into a single device to enable technology for point-of-care testing.
Acknowledgement
This work was supported by the grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI15C-2774-020015), and also supported by a grant (W16-701) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Republic of Korea.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bios.2016.11.051.
Contributor Information
Sung-Han Kim, Email: kimsunghanmd@hotmail.com.
Yong Shin, Email: shinyongno1@gmail.com.
Appendix A. Supplementary material
Supplementary material
.
References
- Agrawal N., Hassan Y.A., Ugaz V.M. Angew. Chem. Int. Ed. 2007;46:4316–4319. doi: 10.1002/anie.200700306. [DOI] [PubMed] [Google Scholar]
- Amer H.M., El Wahed A.A., Shalaby M.A., Almajhdi F.N., Hufert F.T., Weidmann M. J. Virol. Methods. 2013;193:337–340. doi: 10.1016/j.jviromet.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeumner A.J., Cohen R.N., Miksic V., Min J. Biosens. Bioelectron. 2003;18:405–413. doi: 10.1016/s0956-5663(02)00162-8. [DOI] [PubMed] [Google Scholar]
- Bogaerts W., De Heyn P., Van Vaerenbergh T., De Vos K., Selvaraja S.K., Claes T., Dumon P., Bienstman P., Thourhout D.V., Baets R. Laser Photonics Rev. 2012;6:47–73. [Google Scholar]
- Borg I., Rohde G., Loseke S., Bittscheidt J., Schultze-Werninghaus G., Stephan V., Bufe A. Eur. Respir. J. 2003;21:944–951. doi: 10.1183/09031936.03.00088102. [DOI] [PubMed] [Google Scholar]
- Bourgeois F.T., Valim C., McAdam A.J., Mandl K.D. Pediatrics. 2009;124:e1072–e1080. doi: 10.1542/peds.2008-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Caliendo A.M. Clin. Infect. Dis. 2011;52:S326–S330. doi: 10.1093/cid/cir047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.Y., Kang J.M., Ha Y.E., Park G.E., Lee J.Y., Ko J.H., Lee J.Y., Kim J.M., Kang C.I., Jo I.J., Ryu J.G., Choi J.R., Kim S. Lancet. 2016;16:30623–30627. [Google Scholar]
- Compton J. Nature. 1991;1991(350):91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Cox N.J., Subbarao K. Annu. Rev. Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- De Vos K., Bartolozzi I., Schacht E., Bienstman P., Baets R. Opt. Express. 2007;15:7610–7615. doi: 10.1364/oe.15.007610. [DOI] [PubMed] [Google Scholar]
- De Vos K., Girones J., Popelka S., Schacht E., Baets R., Bienstman P. Biosens. Bioelectron. 2009;24:2528–2533. doi: 10.1016/j.bios.2009.01.009. [DOI] [PubMed] [Google Scholar]
- El Wahed A.A., El-Deeb A., El-Tholoth M., El Kader H.A., Ahmed A., Hassan S., Hoffmann B., Haas B., Shalaby M.A., Hufert F.T., Weidmann M. PLOS One. 2013;8:e71642. doi: 10.1371/journal.pone.0071642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Wahed A.A., Weidmann M., Hufert F.T. J. Clin. Virol. 2015;69:16–21. doi: 10.1016/j.jcv.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginocchio C.C., McAdam A.J. J. Clin. Microbiol. 2011;49:S44–S48. [Google Scholar]
- Gourinat A.C., O’Connor O., Calvez E., Goarant C., Dupont-Rouzeyrol M. Emerg. Infect. Dis. 2015;21:84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J.S., Breslin J.J., Breuhaus B., Vivrette S., Smith L.G. J. Clin. Microbiol. 2000;38:4523–4526. doi: 10.1128/jcm.38.12.4523-4526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.P., Gagne L.S., Stock S.R., Marty F.M., Gelman R.S., Marasco W.A., Poritz M.A., Baden L.R. J. Clin. Microbiol. 2012;50:3216–3221. doi: 10.1128/JCM.00538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M., Gleeson M.A., Spaugh B., Tybor F., Gunn W.G., Hochberg M., Baehr-Jones M.T., Bailey R.C., Gunn L.C. IEEE J. Sel. Top. Quantum Electron. 2010;16:654–661. [Google Scholar]
- Kim S.H., Chang S.Y., Sung M., Park J.H., Kim B.H., Lee H., Choi J.P., Choi W.S., Min J.Y. Clin. Infect. Dis. 2016;63:363–369. doi: 10.1093/cid/ciw239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry M.L. Clin. Lab. Med. 2009;29:635–647. doi: 10.1016/j.cll.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Leland D.S., Ginocchio C.C. Clin. Microbiol. Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillis L., Lehman D., Singhal M.C., Cantera J., Singleton J., Labarre P., Toyama A., Piepenburg O., Parker M., Wood R., Overbaugh J., Boyle D.S. PLOS One. 2014;9:e108189. doi: 10.1371/journal.pone.0108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Nam J., Kim S., Lim C.T., Park M.K., Shin Y. Biosens. Bioelectron. 2016;82:1–8. doi: 10.1016/j.bios.2016.03.050. [DOI] [PubMed] [Google Scholar]
- Lizardi P.M., Huang X., Zhu T., Bray-Ward P., Thomas D.C., Ward D.C. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Loeffelholz M., Chonmaitree T. Int. J. Microbiol. 2010 doi: 10.1155/2010/126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J., Chong S., Merante F., Yaghoubian S., Sinha T., Lisle C., Janeczko R. J. Clin. Microbiol. 2007;45:2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentel R., Wegner U., Bruns R., Gurtler L. J. Med. Microbiol. 2003;52:893–896. doi: 10.1099/jmm.0.05290-0. [DOI] [PubMed] [Google Scholar]
- Moesker F.M., van Kampen J.J.A., Aron G., Schutten M., van der Vijver D.A.M.C., Koopmans M.P.G., Osterhaus A.D.M.E., Fraij P.L.A. J. Clin. Virol. 2016;79:12–17. doi: 10.1016/j.jcv.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll P.R., Duschl J., Richter K. Anal. Chem. 2004;334:164–174. doi: 10.1016/j.ab.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson S., Brittain-Long R., Andersson L.M., Westin J., Lindh M. Expert. Rev. Anti. Infect. Ther. 2011;9:615–626. doi: 10.1586/eri.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.K., Kee J.S., Quah J.Y., Netto V., Song J. Sens. Actuators B. 2013;176:552–559. [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet S., Lardeux M., Dina J., Grattard F., Verhoeven P., Le Goff J., Vabret A., Pozzetto B. PLoS One. 2013;8:e72174. doi: 10.1371/journal.pone.0072174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzetto B., Grattard F., Pillet S. Expert. Rev. Anti. Infect. Ther. 2010;8:251–253. doi: 10.1586/eri.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renois F., Talmud D., Huguenin A., Moutte L., Strady C., Cousson J., Leveque N., Andreoletti L. J. Clin. Microbiol. 2010;48:3836–3842. doi: 10.1128/JCM.00733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Perera A.P., Kim K.W., Park M.K. Lab Chip. 2013;13:2106–2114. doi: 10.1039/c3lc50129a. [DOI] [PubMed] [Google Scholar]
- Shin Y., Perera A.P., Kee J.S., Song J., Lo G.Q., Park M.K. Sens. Actuators B. 2013;177:404–411. [Google Scholar]
- Shin Y., Perera A.P., Tang W.Y., Fu D.L., Liu Q., Sheng J.K., Gu Z., Lee T.Y., Barkham T., Park M.K. Biosens. Bioelectron. 2015;68:390–396. doi: 10.1016/j.bios.2015.01.030. [DOI] [PubMed] [Google Scholar]
- Shin Y., Lim S.Y., Lee T.Y., Park M.K. Sci. Rep. 2015;5:14127. doi: 10.1038/srep14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh B.T., Sam S.S., Tan K.K., Danlami M.B., Shu M.H., Johari J., Hooi P.S., Brooks D., Piepenburg O., Nentwich O., Wilder-Smith A., Franco L., Tenorio A., Abubakar S. J. Clin. Microbiol. 2015;53:830–837. doi: 10.1128/JCM.02648-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregoning J.S., Schwarze J. Clin. Microbiol. Rev. 2010;23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim- van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Xu Y., Kong H. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch D.F., Ginocchio C.C. J. Clin. Microbiol. 2010;48:22–25. doi: 10.1128/JCM.02268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Ding S., Yuan R., Zhang Y., Cheng W. Sci. Rep. 2016;6:18810. doi: 10.1038/srep18810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Ke Y., Wang X., Ren H., Liu W., Lu H., Zhang W., Liu S., Chang G., Tian S., Wang L., Huang L., Liu C., Yang R., Chen Z. Sci. Rep. 2016;6:26943. doi: 10.1038/srep26943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material