Dear Editor,
We read with interest the study that antibodies induced by receptor binding domain in spike protein of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) do not cross-neutralize Middle East respiratory syndrome coronavirus (MERS-CoV) reported in this journal recently.1
To date, six CoVs, including human CoV-229E, -NL63, -OC43, -HKU1, SARS-CoV, and MERS-CoV, are known to infect humans. The number of MERS-CoV infection cases in the world has sharply increased since mid-March 2014 and the infections have spread from the Middle East to Europe, North Africa, Asia, and America. The World Health Organization (WHO) has encouraged all countries to monitor MRES-CoV and to carefully review any unusual patterns. However, for mild or unusual symptomatic infection, it is not always possible to identify the infection with MERS-CoV using PCR assay. Hence, it is important to perform seroepidemiological studies in natural populations to analyze HCoVs' epidemiologic spectrum.
The CoV nuleocapide (N) protein is abundantly produced during infection and exhibits strong immunogenicity, which can act as an ideal antigen for viral antibody detection.2, 3, 4 However, the antigenic and serologic relationship between N proteins within subgroups of the six HCoVs, such as NL63 and 229E (both subgroup 1b), OC43 and HKU1 (both subgroup 2a), has not been fully understood, which can affect seropositive data of HCoVs. A recent study showed that BtCoV HKU5 and BtCoV HKU4 N proteins within the 2c subgroups share cross-reactive epitopes with MERS-CoV. In addition, BtCoV HKU3 and BtCoV 279 N proteins within the 2b subgroups share cross-reactive epitopes with SARS-CoV.5
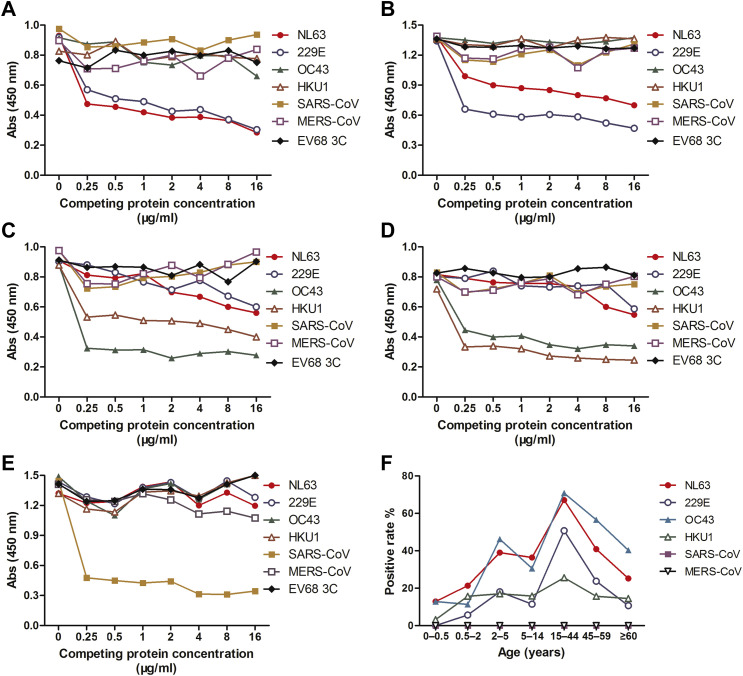
To evaluate the cross-reactivities among HCoVs, we developed a competitive ELISA (cELISA) as previously described for detecting anti-N IgG antibodies of six HCoVs.6 To this end, we first identify HCoV-positive (including HCoV-229E, -OC43 -NL63, -HKU1, and SARS-CoV) and -negative human serum samples by Western blot analysis. Using these positive controls, we found that 1.0 μg/ml of competing N protein was sufficient for the competition assays in cELISA (Fig. 1 A–E).
Figure 1.
Cross-reactivity among human coronaviruses (HCoVs) and seropositive rates of IgG antibodies against HCoVs in different age groups. Human positive sera against HCoV-NL63 (A), -229E (B), -OC43 (C), -HKU1 (D) and SARS-CoV (E) were tested for reactivity to N proteins of HCoV-NL63, -229E, OC43, -HKU1, SARS-CoV and MERS-CoV, respectively, using a competive ELISA assay. EV68 3C protein was used as the control. The absorbance values (Abs) at 450 nm for each concentration of coating antigens are shown on the y-axis; the competing protein concentrations in ELISA assay are shown on the x-axis. (F) IgG antibodies against HCoV-NL63, -229E, -OC43, -HKU1, SARS-CoV and MERS-CoV were detected by competition ELISA at a dilution of 1:200. All serum samples were grouped based on age, as indicated by the x-axis labels.
We then evaluated the possible cross-reactivity among N proteins using cELISA with the positive control sera. Our results suggest that HCoV-229E and -NL63 (subgroup 1b), and HCoV-OC43 and -HKU1 (subgroup 2a) share subgroup cross-reactive epitopes among their N proteins (Fig. 1A–E). However, no cross-reactivity was observed between the N proteins across groups or subgroups. Therefore, cELISA assays were performed with N proteins between HCoV-NL63 and -229E, and HCoV-OC43 and -HKU1 to minimize the cross-reactivity for seroprevalence determination. Since we did not have access to the positive human serum against MERS-CoV, the cross-reactivity of MERS-CoV antibodies with other HCoV N proteins could not be determined. We performed cELISA of MERS-CoV competing with the other five N proteins together.
To evaluate the seroprevalence of HCoVs in China, we determined the cut-off values of the cELISA for six HCoVs as previously described.7, 8 We obtained the cut-off values of HCoV-NL63, -229E, -OC43, -HKU1, SARS-CoV, and MERS-CoV as 0.25, 0.25, 0.23, 0.25, 0.27, and 0.25, respectively. A tested sample was considered positive if its A450 was above the cut-off value.
Then we tested anti-N IgG in sera from 695 healthy adults ≥15 yr by cELISA (Fig. 1F). We obtained seroprevalences of 50.8% for HCoV-229E, 67.1% for -NL63, 70.8% for -OC43, and 25.6.7% for -HKU1 for the age group of 15–44 year olds. Seroprevalences decreased with age, with 10.7% for HCoV-229E, 25.2% for -NL63, 40.3% for -OC43, and 14.5% for -HKU1 for old adults of ≥60 yr. No SARS-CoV and MERS-CoV IgG were detected in these serum samples. The seropositive rates of HCoV-NL63 and -OC43 were higher than those of HCoV-229E and -HKU1 (χ2 tests. χ2 = 130.8, P = 3.9 × 10−29 for HCoV-NL63 vs -229E and -HKU1; χ2 = 239, P = 1.2 × 10−52 for HCoV-OC43 vs -229E and -HKU1) (Fig. 1F). Our data suggest that there is an age-related waning of HCoV-229E, -NL63, -OC43, and -HKU1 specific antibodies.
The seroprevalence of 492 healthy children ≤14 yr were 12.4% for HCoV-229E, 33.7% for -NL63, 32.5% for -OC43, 15.4% for -HKU1, 0% for SARS-CoV, and 0% for MERS-CoV IgG (Fig. 1F). Anti-N-IgG antibodies of HCoV-229E, -NL63, -OC43, and -HKU1 were detected in children between 0.5 and 2 yr in this study population, suggesting that exposure to HCoV-229E, -NL63, -OC43, and -HKU1 may occur early in childhood.
To assess the relationship between anti-N-IgG and HCoV infection, we measured the IgG antibody in 361 serum samples from children with lower respiratory infections (Table 1 ). Of 246 samples from HCoV-negative patients, 124 (50.4%) had serologic evidence for past exposure of HCoV-229E, 142 (57.7%) for -NL63, 123 (50%) for -OC43, and 133 (54.1%) for -HKU1. However, among the 30 children who were HCoV-NL63-positive, only 7 (23.3%) were seropositive for anti-N-IgG. Similar results were found in serum samples from those who were positive for HCoV-NL63 (23.8%), -OC43 (18.4%), and -HKU1 (23.1%). Further analysis showed that IgG seropositive rates of HCoV-negative patients were significantly higher than those from HCoV-positive patients (χ2 tests. χ2 = 7.68, P = 0.0056 for HCoV-NL63; χ2 = 6.81, P = 0.00906 for HCoV-229E; χ2 = 11.98, P = 0.00054 for HCoV-OC43; χ2 = 7.84, P = 0.00511 for HCoV-HKU1), suggesting that the low anti-N IgG level may be used as a predictive index for susceptibility to HCoV in a population.
Table 1.
Comparison of serum IgG antibody levels against N proteins for ARTI patients positive and negative for HCoVs.
| HCoVs | Positive |
Negative |
χ2, P value | ||||
|---|---|---|---|---|---|---|---|
| All | IgG+ (%) | IgG− (%) | All | IgG+ (%) | IgG− (%) | ||
| NL63 | 21 | 5a(23.8)b | 16 (76.2) | 246 | 142 (57.7) | 104 (42.3) | χ2 = 7.68, P = 0.0056 |
| 229E | 30 | 7 (23.3) | 23 (76.7) | 246 | 124 (50.4) | 122 (49.6) | χ2 = 6.81, P = 0.00906 |
| OC43 | 38 | 7 (18.4) | 31 (81.6) | 246 | 123 (50.0) | 123 (50.0) | χ2 = 11.98, P = 0.00054 |
| HKU1 | 26 | 6 (23.1) | 20 (76.9) | 246 | 133 (54.1) | 113 (45.9) | χ2 = 7.84, P = 0.00511 |
Number of positive samples.
Percentage of positive samples.
In summary, our results suggest that the development of specific serologic diagnosis for HCoVs infection based on N proteins needs to take into consideration the cross-reactivities existing in the same subgroup. However, a common diagnostic platform for HCoVs should include a panel of phylogenetically distinct N proteins. Further, the anti-N IgG may serve as an indication of susceptibility to HCoV infections. Our study is informative for developing HCoV immunoassays and provides insights into the prevalence and pathologic roles of HCoVs.
Potentials conflicts of interest
No reported conflicts.
Acknowledgments
We thank the Beijing Children's Hospital, Beijing Blood Center, and Shandong Medicinal Biotechnology Center for providing blood samples. This study was supported in part by the Chinese National Major S & T Project (2012ZX10004-206, 2014ZX10004-001), the National Foundation for Distinguished Young Scientists (81225014), the Program for Changjiang Scholars and Innovative Research Team in University (IRT13007) and the Fondation Mérieux.
Contributor Information
Li Guo, Email: gnyny0803@163.com.
Jianwei Wang, Email: wangjw28@163.com.
References
- 1.Du L., Ma C., Jiang S. Antibodies induced by receptor-binding domain in spike protein of SARS-CoV do not cross-neutralize the novel human coronavirus hCoV-EMC. J Infect. 2013;67:348–350. doi: 10.1016/j.jinf.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai M.M.C., Perlman S., Anderson L.J. Coronaviridae. In: Knipe D.M., Howley P.M., editors. 5th ed. vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1306–1336. (Fields virology). [Google Scholar]
- 3.Timani K.A., Ye L., Ye L., Zhu Y., Wu Z., Gong Z. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J Clin Virol. 2004;30:309–312. doi: 10.1016/j.jcv.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sastre P., Dijkman R., Camuñas A., Ruiz T., Jebbink M.F., van der Hoek L. Differentiation between human coronaviruses NL63 and 229E using a novel double-antibody sandwich enzyme-linked immunosorbent assay based on specific monoclonal antibodies. Clin Vaccin Immunol. 2011;18:113–118. doi: 10.1128/CVI.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agnihothram S., Gopal R., Yount B.L., Jr., Donaldson E.F., Menachery V.D., Graham R.L. Evaluation of serologic and antigenic relationships between middle eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J Infect Dis. 2014;209:995–1006. doi: 10.1093/infdis/jit609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo L., Wang Y., Zhou H., Wu C., Song J., Li J. Differential seroprevalence of human bocavirus species 1-4 in Beijing, China. PLoS One. 2012;7:e39644. doi: 10.1371/journal.pone.0039644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kean J.M., Rao S., Wang M., Garcea R.L. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hustedt J.W., Christie C., Hustedt M.M., Esposito D., Vazquez M. Seroepidemiology of human bocavirus infection in Jamaica. PLoS One. 2012;7:e38206. doi: 10.1371/journal.pone.0038206. [DOI] [PMC free article] [PubMed] [Google Scholar]