Abstract
Cyclic GMP–AMP synthase (cGAS) is a signaling enzyme in human cells that controls immune-sensing of cytosolic DNA. The recent discoveries of diverse structural homologs of cGAS in animals and bacteria reveal that cGAS-like signaling is surprisingly ancient and widespread in biology. Together with the Vibrio cholerae protein dinucleotide cyclase in Vibrio (DncV), cGAS and DncV homologs comprise a family of cGAS/DncV-like nucleotidyltransferase (CD-NTase) enzymes that synthesize noncanonical RNA signals including cyclic dinucleotides, cyclic trinucleotides, and linear oligonucleotides. Structural and biochemical breakthroughs provide a framework to understand how CD-NTase signaling allows cells to respond to changing environmental conditions. The CD-NTase family also includes uncharacterized human genes like MB21D2 and Mab21L1, highlighting emerging functions of cGAS-like signaling beyond innate immunity.
Current Opinion in Structural Biology 2019, 59:178–187
This review comes from a themed issue on Protein nucleic acid interactions
Edited by Frédéric H-T Allain and Martin Jinek
For a complete overview see the Issue and the Editorial
Available online 6th October 2019
https://doi.org/10.1016/j.sbi.2019.08.003
0959-440X/© 2019 The Author. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
Cyclic GMP–AMP synthase (cGAS) is a vertebrate enzyme that catalyzes production of the nucleotide second messenger 2′–5′/3′–5′ cyclic GMP–AMP (2′3′-cGAMP) in response to cytosolic double-stranded DNA [1,2,3••,4••,5]. Since the discovery of cGAS in late 2012 [4••], cGAS-dependent sensing of mislocalized DNA has emerged as a central event in the cellular responses to pathogen replication, nuclear and mitochondrial stress, and DNA damage [6]. cGAS signaling is also an important component of antitumor immunity [7,8], and analogs of 2′3′-cGAMP are currently in development for cancer immunotherapy. The diverse roles for cGAS in cell biology and the potent therapeutic potential of 2′3′-cGAMP illustrate the importance of discovering and mechanistically understanding the enzymes that synthesize nucleotide signals in human cells.
Recent findings overturn the idea that cGAS signaling is a specialized pathway unique to the innate immune response to cytosolic DNA and instead reveal a striking diversity of cGAS-like enzymes in nature. Bioinformatic and structural findings identified homology between cGAS and the innate immunity enzyme oligo adenylate synthase 1 (OAS1) that senses cytosolic double-stranded RNA [3••,4••,9••,10,11••,12, 13, 14,15•]. Likewise, structural analysis of the Vibrio cholerae enzyme dinucleotide cyclase in Vibrio (DncV), a 3′–5′/3′–5′ cGAMP (3′3′-cGAMP) synthase, revealed that DncV is a structural and functional homolog of human cGAS and that cGAS-like enzymes exist in the bacterial kingdom [16••,17•,18•]. Thousands of functional cGAS homologs have now been identified in bacteria [19••,20•], revealing that cGAS and DncV are founding members of a large family of cGAS/DncV-like nucleotidyltransferase (CD-NTase) enzymes. Bacterial CD-NTases comprise at least seven distinct protein clades including newly identified enzymes like Escherichia coli CdnE that produces 3′3′ cUMP–AMP and Enterobacter cloacae CdnD that produces the cyclic trinucleotide 3′3′3′ cAMP–AMP–GMP [19••]. Eukaryotic CD-NTases include cGAS and OAS1, hundreds of metazoan enzymes, and nine proteins in the human genome of unknown function.
The remarkable abundance of CD-NTases encoded in animal and bacterial genomes provides a unique opportunity to understand the cellular roles of specialized nucleotide signals. Recent breakthroughs in biochemistry and structural biology establish a model of cGAS and CD-NTase function, and demonstrate that CD-NTase enzymes synthesize diverse nucleotide signals including cyclic dinucleotides, cyclic trinucleotides, and oligonucleotides with an assortment of base and phosphodiester-linkage specificities. This review outlines the results that define CD-NTases as a new family of signaling enzymes, overviews conserved structural features that control CD-NTase function, and provides a framework for classification of CD-NTase signaling pathways. Although individual CD-NTase enzymes synthesize distinct nucleotide signals and control independent downstream effector functions, conserved biochemical mechanisms unite activation and regulation of each signaling pathway. Advances in understanding of CD-NTase biology provide an opportunity to explain the role of uncharacterized CD-NTase family members and potentially unveil new nucleotide signals in human cells.
cGAS and CD-NTase enzymes as chemical sensors
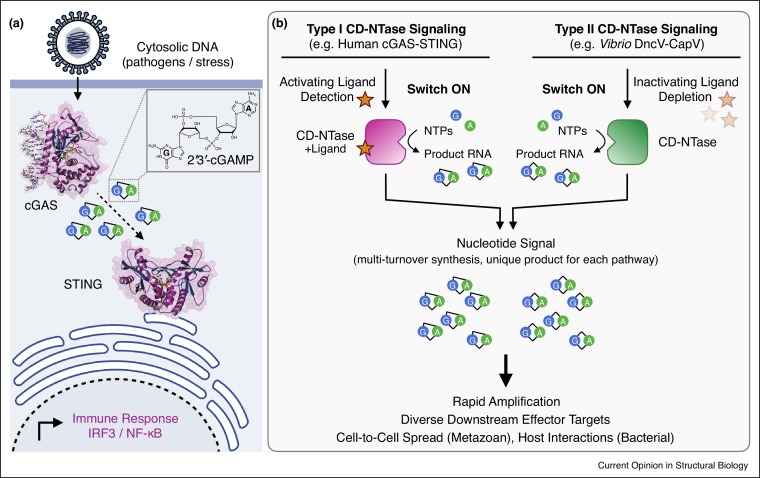
CD-NTase enzymes function as nucleic acid and chemical sensors, allowing animal and bacterial cells to respond to changing environmental conditions and rapidly initiate signaling responses. In the active state, CD-NTases catalyze multi-turnover synthesis of cyclic and oligonucleotide signals built from common nucleotide triphosphate precursors [3••,4••,11••,17•,19••]. This iterative process allows relatively few copies of an active CD-NTase enzyme to produce many nucleotide second messenger molecules and significantly amplify a signaling response (Figure 1 a). For example, activation of a few molecules of cGAS bound to DNA results in multi-turnover production, with >750 molecules of 2′3′-cGAMP produced for each active enzyme [3••]. 2′3′-cGAMP diffuses throughout the cell, and is recognized by the receptor Stimulator of Interferon Genes (STING) to initiate downstream IRF3/NF-κB immune responses and autophagy [4••,6,21,22]. In addition to STING-dependent responses in the primary activated cell, 2′3′-cGAMP is highly stable [23] and can be transferred to neighboring cells in gap-junctions, released from tumors to activate immune cells, and packaged into viral particles [8,24, 25, 26]. The dramatic signal amplification following activation of cGAS and CD-NTase enzymes is a key feature of how these pathways propagate responses.
Figure 1.
CD-NTase signaling in animals and bacteria.
(a) Cartoon schematic of the human cGAS-STING signaling pathway. cGAS binds to cytosolic double-stranded DNA and is activated to produce the second messenger 2′3′-cGAMP. 2′3′-cGAMP is recognized by STING, and initiates a downstream IRF3/NF-κB-dependent immune response. (b) Overview of Type I and Type II CD-NTase Signaling. Type I CD-NTase pathways like human cGAS-STING rely on detection of an activating ligand (e.g. cytosolic DNA) depicted here as a star. In contrast, Type II CD-NTase pathways like Vibrio DncV-CapV are active in the absence of ligand, and may rely on depletion of a ligand (e.g. folate-like metabolites) that represses enzyme activation. In either case, activated CD-NTases catalyze multi-turnover synthesis of a nucleotide second messenger (e.g. 2′3′-cGAMP or 3′3′-cGAMP) to amplify signaling and initiate downstream effector responses.
Human cGAS and Vibrio DncV use distinct mechanisms to couple enzyme activation and ligand detection, illustrating the existence of at least two separate types of CD-NTase pathways (Table 1 ). Designated here as ‘Type I’ CD-NTase signaling, cGAS and OAS1 are inactive and unable to catalyze nucleotide signal synthesis until ligand binding induces a conformational change that remodels the active site into an enzymatically competent state (Figure 1b) [3••,9••,10,11••,12,13,27,28,29•]. In contrast, ‘Type II’ CD-NTases like Vibrio DncV and many bacterial CD-NTases are catalytically active in the absence of ligand and constitutively synthesize nucleotide signals in vitro [16••,17•,18•,19••]. Vibrio DncV is inhibited upon binding to folate-like compounds including 5-methyltetrahydrofolic acid [18•], a metabolite required for nucleotide biosynthesis, suggesting that regulation of Type II enzymes can occur through ligand binding events that repress enzyme function (Figure 1b). Bacterial CD-NTase operons are frequently encoded within defense islands and have been hypothesized to be involved in phage resistance [19••,20•]. Although the exact function of Type II CD-NTase pathways in bacteria remains unknown [16••,18•,19••,30], one current model is that these CD-NTases respond to conditions like phage infection by sensing depletion of intracellular metabolites or nutrient starvation.
Table 1.
CD-NTase signaling pathways
| CD-NTase enzyme | Classification | Ligand | Nucleotide signal | Effector | Downstream response | Signal degradation | ||
|---|---|---|---|---|---|---|---|---|
| Mammalian | cGAS | Type I | dsDNA | 2′3′-cGAMP | 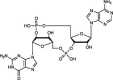 |
STING | Type I interferon, NF-κB signaling | ENPP1, poxin, CdnP |
| OAS1/2/3 | Type I | dsRNA | 2′–5′ Oligoadenylate | 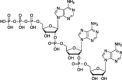 |
RNase L | Translational arrest | PDE12, AKAP7, coronavirus NS2, rotavirus VP3 | |
| MB21D2 | Type I? | ? | ? | ? | Cancer resistance? | ? | ||
| Mab21L1/2/3/4 | Type I? | ? | - - /? | ? | Transcription control | ? | ||
| MID49/51 | ? | - - | - - | DRP1 | Mitochondrial fission | - - /? | ||
| Bacterial | V. cholerae DncV | Type II | Folate metabolites | 3′3′-cGAMP |  |
CapV | Nutrient sensing | cGAP1/2/3 |
| E. coli CdnE | Type II | ? | 3′3′-cUMP–AMP | 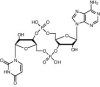 |
CapE | ? | ? | |
| E. cloacae CdnD | Type II | ? | 3′3′-cAMP–AMP–GMP | 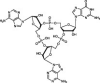 |
? | ? | Ring nucleases? | |
Summary of known CD-NTase signaling pathways including recognized ligand, product nucleotide signal, downstream receptor and effector response, and dedicated machinery for signal degradation. Undefined pathway components are designated as unknown (?) or likely to be not involved (--).
Structural anatomy of CD-NTase enzymes
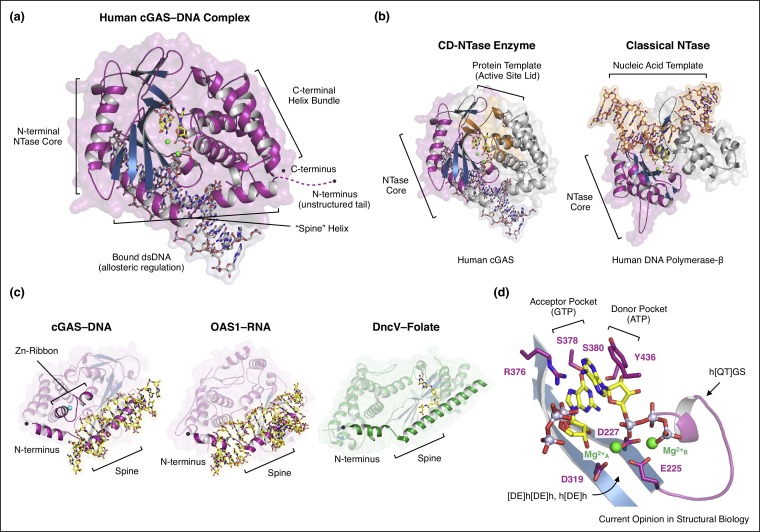
All CD-NTases share a common structural architecture. The enzymatic domain is typically 250–400 amino acids (∼35–50 kDa) and adopts a caged tertiary structure formed by a long α-helix ‘spine’ that bridges an N-terminal nucleotidyltransferase (NTase) core and a C-terminal helix bundle (Figure 2 a) [3••,9••,10,11••,17•,19••]. The extreme N-terminal and C-terminal residues pack against each other in the helix bundle, and the overall CD-NTase cage-like architecture creates a recessed pocket to catalyze nucleotide signal formation. The N-terminal NTase core is a common fold found in many diverse enzymes [14]. There is no one structural feature that uniquely identifies CD-NTases from classical NTase proteins, and the commonality of the core NTase fold is in part why bioinformatic identification of CD-NTase homologs from primary sequence alone remains a challenge [20•]. Whereas classical NTases like DNA polymerase-β adopt a flat, open conformation for recruitment of a nucleic acid template [31], the deep catalytic pocket of CD-NTase enzymes allows amino-acid side chains extending from the active-site lid to control nucleotide base specificity and direct product formation (Figure 2b). Structures of the cGAS–DNA, OAS1–RNA and DncV–folate complexes reveal that both Type I and Type II CD-NTases utilize a shared ligand binding surface located behind the enzyme active site, along the α-helix spine (Figure 2c) [3••,9••,12,18•,27,28,29•]. In cGAS, a zinc-ribbon insertion within the ligand binding surface in part explains how this enzyme evolved to bind double-stranded DNA [9••,10]. The shared location of CD-NTase–ligand interactions indicates that diversification of this surface likely allows enzyme activity to be coupled to recognition of a wide variety of chemical signals beyond nucleic acids and folate-like compounds.
Figure 2.
Structural anatomy of cGAS and CD-NTase enzymes.
(a) Structural overview of the human cGAS–DNA complex. CD-NTase enzymes adopt a conserved cage-like structure where a long α-helix ‘spine’ braces an N-terminal NTase core and C-terminal helix bundle. Note, a 1:1 unit of cGAS–DNA is shown for clarity, but the minimally active complex is a 2:2 unit. (b) The cage-like architecture creates a deep cavity for nucleotide substrate coordination and enzymatic catalysis. Unlike classical nucleotidyltransferases (NTases) like DNA polymerase-β that require a nucleic acid template (orange, right) to dictate nucleotide substrate specificity, amino-acid side chains in the CD-NTase active-site lid (orange, left) directly control nucleotide interactions. (c) Structural overview of the ligand binding surfaces in the cGAS–DNA, OAS–RNA, and DncV–folate complexes. Structures are shown rotated 180° from the orientation in (a). Activating and inactivating CD-NTase ligands (yellow) bind on the back of the enzyme along the α-helix spine. Insertion of a zinc-ribbon (Zn-Ribbon) along the ligand-binding surface in part explains evolution of cGAS specificity for DNA interactions. (d) CD-NTase active sites contain a highly conserved h[QT]GS, DE[h[DE]h, h[DE]h catalytic triad and distinct ‘acceptor’ and ‘donor’ nucleotide pockets. Each CD-NTase catalyzes a multi-step reaction that proceeds through a defined reaction path (e.g. the cGAS intermediate product is pppG[2′–5′]pA).
The active site of CD-NTase enzymes comprises a catalytic triad at the base and additional amino acids lining the lid that are required for nucleotide substrate coordination. All catalytically active CD-NTases encode a highly conserved h[QT]GS [X8–20] [DE]h[DE]h [X50–90] h[DE]h sequence (where h is a hydrophobic amino acid) that is located on a short α-helix (h[QT]GS) and two adjacent beta strands ([DE]h[DE]h, h[DE]h) of the nucleotidyltransferase core (Figure 2d) [3••,9••,10,11••,14,17•,19••]. The active site of CD-NTases contains discrete ‘donor’ and ‘acceptor’ nucleotide binding pockets. Once activated, CD-NTases bind nucleotide substrates and catalyze an SN2 nucleophilic substitution reaction that results in the transfer of the α-phosphate and nucleobase from the donor nucleotide onto the ribose 2′ or 3′ OH of the acceptor nucleotide. The reaction is divalent metal-dependent (Mg2+ or Mn2+), requiring two metals in the enzyme–substrate complex and likely a third metal transiently coordinated in the transition or product-bound states [3••,9••,17•,32]. Metal ion A is coordinated by all three Asp/Glu side chains of the active-site triad ([DE]h[DE]h, h[DE]h; e.g. E225, D227, D319 in human cGAS) and participates in deprotonation and positioning of the 2′ or 3′ OH of the accepting nucleotide to attack the donor nucleotide α-phosphate. Metal ion B is coordinated by carboxyl groups from the first and second Asp/Glu side chains of the active-site triad ([DE]h[DE]h; e.g. E225, D227 in human cGAS) and the triphosphate of the donor nucleotide, and functions to position the donor nucleotide and help stabilize the reaction intermediate (Figure 2d) [3••,9••,17•,18•,19••,27,28,29•]. The donor pocket contains a highly conserved hydrophobic side chain (typically Tyr/Phe/Ile; e.g. Y436 in human cGAS) in the C-terminal helix bundle that stacks against the nucleobase, and sequence-specific hydrogen-bond interactions with the side chains in the active-site lid control donor and acceptor pocket nucleotide specificity (Figure 2d). Importantly, recent studies with cGAS have demonstrated that a space formed above the nucleotide donor pocket in CD-NTase enzymes is amenable to targeting by small-molecule therapeutics [33•,34•].
A unique feature of the CD-NTase family is the use of a single site to catalyze nucleotide signal production through a sequential, multi-step reaction. The CD-NTase reaction is distinct from other nucleotide second messenger synthase families including GGDEF-like and DisA-like enzymes that require protein dimerization to produce 3′3′ c-di-GMP and 3′3′ c-di-AMP [35,36]. In the case of cGAS, in the first reaction state ATP and GTP occupy the donor and acceptor nucleotide positions respectively, and AMP is transferred onto the 2′ OH of GTP to form pppG[2′–5′]pA [3••,9••]. In the second reaction state, the intermediate molecule is reoriented such that the 3′ OH of AMP attacks the guanine α-phosphate to produce a final cyclized c(G[2′–5′]pA[3′–5′]p) product 2′3′-cGAMP. Each CD-NTase catalyzes signal production in a specific reaction order. For example, Vibrio DncV catalyzes the opposite reaction path where GTP first occupies the donor pocket to produce a pppA[3′–5′]pG intermediate during 3′3′-cGAMP synthesis [17•,18•], while the E. coli CdnE enzyme that produces 3′3′ cUMP–AMP uses a reaction order more similar to cGAS that proceeds through a pppU[3′–5′]pA intermediate [19••]. For CD-NTase enzymes like OAS1 that produce linear oligonucleotide signals, sequential transfer and repositioning of the donor nucleotide to the acceptor pocket allows extension of the oligonucleotide chain [13]. Open mechanistic questions include how repositioning of CD-NTase reaction intermediates is controlled and how product chain-length is specified in the active site [15•,37].
Although CD-NTase activity requires a single monomeric active site, an emerging regulatory feature is the role of enzyme oligomerization. The minimally active cGAS–DNA complex is a 2:2 unit where two molecules of cGAS embrace two molecules of double-stranded DNA [27,28,29•]. Recent results highlight the role of further higher-order complex formation in cGAS regulation where DNA ‘laddering’ and a liquid–liquid phase separation process are required for robust enzyme activation [38•,39•]. Other complex oligomerization events include an enigmatic star-shaped pentameric complex formed by the human protein Male abnormal 21-Like 1 (Mab21L1) [40]. Eukaryotic CD-NTases often contain >100 amino acid residues of charged, unstructured sequence within N-terminal or C-terminal tails appended to the core enzymatic module (Figure 1a). The cGAS N-terminal tail has been implicated in DNA interactions, protein stability, localization, and higher-order complex formation, suggesting that these extensions are important to regulate enzyme activation [10,39•,41, 42, 43]. In addition to oligomerization, CD-NTase function is further regulated by intrinsic species-specific substitutions that fine-tune enzyme activation [29•,44], additional domains like tandem arrays of the CD-NTase module or fusion to ubiquitin-like proteins in some OAS homologs [45], and post-translational modifications [46, 47, 48].
Nucleotide signals and downstream effector functions
CD-NTase enzymes synthesize a remarkable array of nucleotide products to control downstream effector functions. Although the overall protein structure is conserved between distantly related animal and bacterial CD-NTases, a high degree of amino-acid sequence variability exists in the side chains extending from the active-site lid and in the ligand binding surface formed on the back of the enzyme behind the active site (Figure 2c,d). This variability affords CD-NTases the plasticity necessary to recognize diverse chemical ligands and to synthesize distinct nucleotide products with alternative base and phosphodiester-linkage specificities. Currently, known CD-NTase products include di-purine, di-pyrimidine, and hybrid purine–pyrimidine cyclic dinucleotides [2,3••,4••,9••,16••,19••]. In addition to cyclic dinucleotides, CD-NTase enzymes also produce linear oligonucleotides and larger cyclic RNA species including cyclic trinucleotides (Table 1). Thus far, the products of bacterial CD-NTases contain canonical 3′–5′ phosphodiester linkages [16••,17•,18•,19••], while both known metazoan CD-NTase products contain noncanonical 2′–5′ linkages [1,2,3••,5]. Given the simple reaction chemistry and enormous diversity of CD-NTase enzymes in bacteria and metazoan systems, it is likely that many more CD-NTase products remain to be discovered.
Once synthesized, CD-NTase products function as nucleotide signals to control downstream effector functions. In mammalian cells, 2′3′-cGAMP and 2′–5′ oligoadenylate are recognized by the protein receptors STING and RNase L to activate innate immune responses [21,49]. In bacteria, known receptors include cGAMP-activated phospholipase in Vibrio (CapV) and cUMP–AMP-activated phospholipase in E. coli (CapE), phospholipases that are selectively activated by 3′3′-cGAMP and 3′3′-cUMP–AMP [19••,30]. Additionally, candidate receptors in bacteria include riboswitches [50,51], and a diverse array of nuclease, protease, and pore-forming effector proteins in CD-NTase-containing operons [20•]. Interestingly, animal receptors including STING and the mouse oxidoreductase RECON recognize diverse bacterial CD-NTase products, indicating that cross-kingdom CD-NTase signaling may be an important component of host–microbe interactions [19••,21,52]. Specific nuclease and phosphodiesterase enzymes have been identified that degrade CD-NTase product signals [23,53, 54, 55, 56, 57, 58], and it is likely that many CD-NTase pathways require dedicated mechanisms for signal inactivation in animal and bacterial cells (Table 1).
CD-NTase family members sometimes lack the conserved catalytic triad and instead perform non-enzymatic functions. MID49 and MID51 are CD-NTases crucial for recruitment of dynamin-related protein 1 to the mitochondria outer-membrane, and crystal structures of MID51 bound to an adenosine diphosphate molecule demonstrate that the CD-NTase domain may have been coopted for nucleotide recognition [59,60]. Catalytically inactive OAS homologs also function as co-factors to modulate cellular RNA and DNA sensing [61], regulate mRNA translation [62], and even appear to have been co-opted as viral factors for host immune suppression [63]. Other CD-NTases, like homologs of the developmental regulatory protein Mab21L1 [40], exhibit partial loss of conserved active-site residues and it remains unclear if these factors are capable of synthesizing nucleotide signals.
Evolutionary origin and diversification of CD-NTase signaling
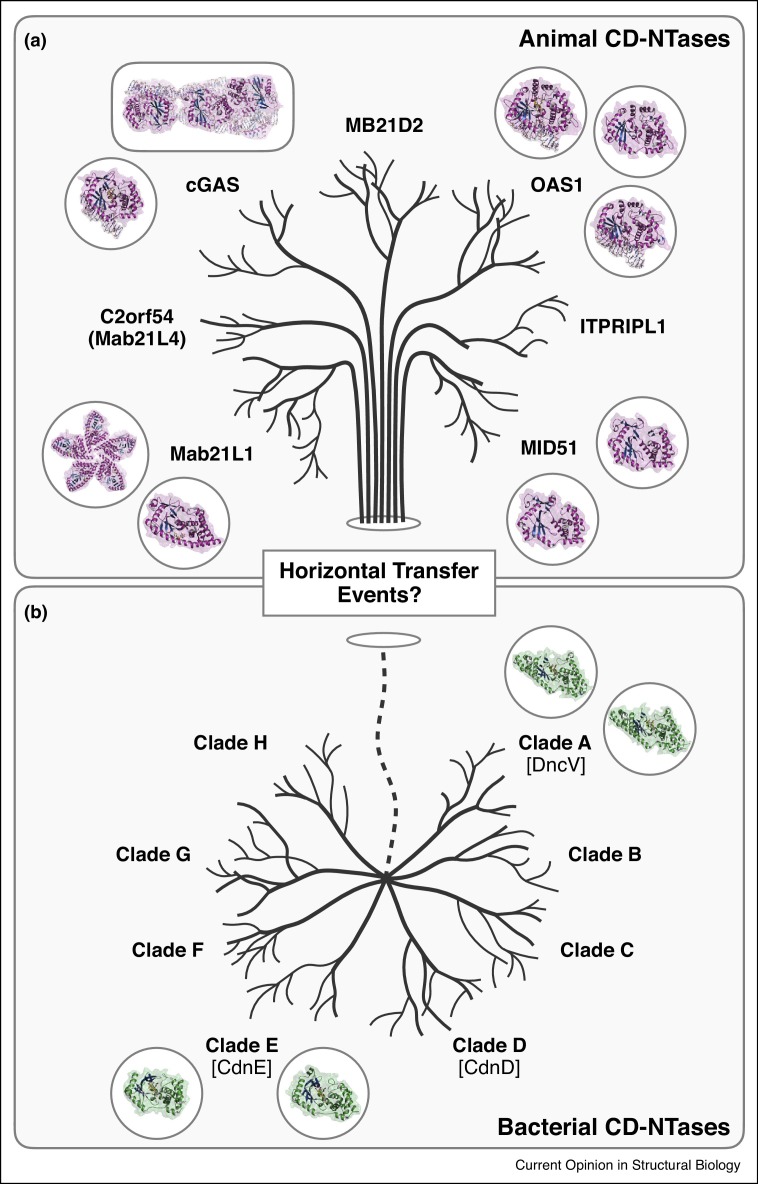
The conservation of CD-NTase enzymes in both the animal and bacterial kingdoms provides a framework to understand pathway regulation and evolution. Structural and biochemical comparisons of human cGAS signaling with ancestrally related bacterial and primitive metazoan CD-NTase pathways have revealed key details in cGAS activation and downstream STING signaling [17•,22,64, 65, 66, 67]. No CD-NTase enzymes have been identified in plants or single-cell eukaryotes, supporting an evolutionary model where animal CD-NTases may have originated through horizontal transfer (Figure 3 ). Interestingly, the active-site lid of OAS proteins contains an extended α-helix and shares structural homology with non-templated nucleotidyltransferases like CCA-adding enzyme and poly-A polymerase [45]. This extended α-helix is not conserved in the structures of cGAS, DncV, or CdnE but is predicted to exist in the bacterial CD-NTase clade C01 [19••], indicating that metazoan OAS1 and cGAS potentially represent separate evolutionary lineages of CD-NTase enzymes. Bacterial CD-NTases are ubiquitously encoded on mobile genetic elements found in nearly every bacterial phylum, and thus may represent an ancestral reservoir of antiviral defense genes from which multiple immune-factors like human cGAS and OAS1 separately originated.
Figure 3.
Conservation and evolution of the CD-NTase enzyme family.
(a, b) Overview of shared conservation of animal (top, magenta) and bacterial (bottom, green) CD-NTases. Distinct branches of individual animal CD-NTase proteins may have arisen from ancestral enzymes rooted in bacteria. Known structures are shown as schematics, and the hypothesized horizontal transfer events are indicated as a dashed line.
While the best understood roles for CD-NTase signaling are in innate immunity, the function of uncharacterized CD-NTases in the human genome represents an important opportunity for future discovery. Mab-21 domain-containing protein 2 (MB21D2), the human gene most closely related to cGAS, is predicted to have a complete active site and appears capable of catalytic function. MB21D2 is frequently mutated in lung cancers [68], supporting a possible role in tumor suppression and cancer resistance. Additionally, the Male abnormal 21 genes are CD-NTase family proteins conserved in nearly all metazoan animals. Human mutations in Mab21L1 and Mab21L2 cause severe birth defects including skeletal dysplasia, eye malformations, and intellectual disability [69]. This suggests possible new non-immune roles of CD-NTase signaling pathways in early embryonic development.
Conclusion
Although only recently discovered, cGAS-dependent signaling has emerged as a key feature of many aspects of human biology and disease [6]. The recognition of cGAS as part of a diverse family of CD-NTase enzymes in animals and bacteria now provides a foundation to define unified mechanisms that control nucleotide signaling and to address open questions in the field. What are the functions of diverse metazoan CD-NTases including uncharacterized enzymes like MB21D2 and Mab21L1 in the human genome? How can bioinformatic classification of CD-NTase homologs be improved to predict nucleotide product signals from primary enzyme sequence? Which ligands are recognized by CD-NTases other than nucleic acid and folate-like metabolites? Are bacterial CD-NTase operons involved in defense against phage infection, and is there a direct evolutionary link between animal and bacterial antiviral immunity? Combined genetic, biochemical, and structural studies of CD-NTases in both animal and bacterial cells provide a path to answer these questions and expand our understanding of the role of nucleotide signaling in human cells.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
I thank A. Lee, A. Whiteley, J. Eaglesham, B. Morehouse, A. Govande, C. de Oliveira Mann, and all members of the lab for helpful discussion; and credit A. Whiteley for the idea of “Type I” and “Type II” CD-NTase designations. I acknowledge funding through the Richard and Susan Smith Family Foundation and the Parker Institute for Cancer Immunotherapy, and apologize to the many scientists in the field who were not cited due to the space constraints of this short review.
Glossary
- cGAS
Cyclic GMP–AMP synthase
- DncV
Dinucleotide cyclase in Vibrio
- CD-NTase
cGAS/DncV-like nucleotidyltransferase
- 2′3′-cGAMP
2′–5′/3′–5′ cyclic GMP–AMP, c(G[2′–5′]pA[3′–5′]p)
References
- 1.Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Rohl I., Hopfner K.P., Ludwig J., Hornung V. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diner E.J., Burdette D.L., Wilson S.C., Monroe K.M., Kellenberger C.A., Hyodo M., Hayakawa Y., Hammond M.C., Vance R.E. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3••.Gao P., Ascano M., Wu Y., Barchet W., Gaffney B.L., Zillinger T., Serganov A.A., Liu Y., Jones R.A., Hartmann G., et al. Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of two papers (see also Civril et al. [9]) reporting the first structure of the cGAS–DNA complex and the mechanism of 2'3' cGAMP product formation.
- 4••.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]; Discovery of cGAS as a cytosolic DNA sensor and first reported connection between cGAS and OAS1.
- 5.Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C., Chen Z.J. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ablasser A., Chen Z.J. cGAS in action: expanding roles in immunity and inflammation. Science. 2019;363 doi: 10.1126/science.aat8657. [DOI] [PubMed] [Google Scholar]
- 7.Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y., Duggan R., Wang Y., Barber G.N., Fitzgerald K.A., et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcus A., Mao A.J., Lensink-Vasan M., Wang L., Vance R.E., Raulet D.H. Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity. 2018;49:754–763. doi: 10.1016/j.immuni.2018.09.016. e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Civril F., Deimling T., de Oliveira Mann C.C., Ablasser A., Moldt M., Witte G., Hornung V., Hopfner K.P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of two papers (see also Gao et al. [3]) reporting the first structure of the cGAS–DNA complex and the mechanism of 2'3' cGAMP product formation.
- 10.Kranzusch P.J., Lee A.S., Berger J.M., Doudna J.A. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Hartmann R., Justesen J., Sarkar S.N., Sen G.C., Yee V.C. Crystal structure of the 2’-specific and double-stranded RNA-activated interferon-induced antiviral protein 2’-5’-oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]; Structural analysis of OAS1 and mechanism of 2'–5' oligoadenylate synthesis.
- 12.Donovan J., Dufner M., Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohofener J., Steinke N., Kay-Fedorov P., Baruch P., Nikulin A., Tishchenko S., Manstein D.J., Fedorov R. The activation mechanism of 2’-5’-oligoadenylate synthetase gives new insights into OAS/cGAS triggers of innate immunity. Structure. 2015;23:851–862. doi: 10.1016/j.str.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Kuchta K., Knizewski L., Wyrwicz L.S., Rychlewski L., Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37:7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Hornung V., Hartmann R., Ablasser A., Hopfner K.P. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review outlining the emerging connection between cGAS and OAS1 and the detailed mechanism of nucleotide product formation.
- 16••.Davies B.W., Bogard R.W., Young T.S., Mekalanos J.J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; Discovery of Vibrio DncV and identification of 3′3′ cGAMP.
- 17•.Kranzusch P.J., Lee A.S.Y., Wilson S.C., Solovykh M.S., Vance R.E., Berger J.M., Doudna J.A. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell. 2014;158:1011–1021. doi: 10.1016/j.cell.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of two papers (see also Zhu et al. [18]) reporting the structure of Vibrio DncV and the first connection between cGAS and bacterial enzymes.
- 18•.Zhu D., Wang L., Shang G., Liu X., Zhu J., Lu D., Wang L., Kan B., Zhang J.R., Xiang Y. Structural biochemistry of a Vibrio cholerae dinucleotide cyclase reveals cyclase activity regulation by folates. Mol Cell. 2014;55:931–937. doi: 10.1016/j.molcel.2014.08.001. [DOI] [PubMed] [Google Scholar]; One of two papers (see also Kranzusch et al. [17]) reporting the structure of Vibrio DncV and the first connection between cGAS and bacterial enzymes.
- 19••.Whiteley A.T., Eaglesham J.B., de Oliveira Mann C.C., Morehouse B.R., Lowey B., Nieminen E.A., Danilchanka O., King D.S., Lee A.S.Y., Mekalanos J.J., et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature. 2019 doi: 10.1038/s41586-019-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of diverse bacterial cGAS-like enzymes and establishment of CD-NTase enzyme family.
- 20•.Burroughs A.M., Zhang D., Schaffer D.E., Iyer L.M., Aravind L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 2015;43:10633–10654. doi: 10.1093/nar/gkv1267. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bioinformatic analysis identifying diverse cGAS and DncV homologs in bacteria.
- 21.Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M., Hayakawa Y., Vance R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gui X., Yang H., Li T., Tan X., Shi P., Li M., Du F., Chen Z.J. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eaglesham J.B., Pan Y., Kupper T.S., Kranzusch P.J. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature. 2019;566:259–263. doi: 10.1038/s41586-019-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E., Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentili M., Kowal J., Tkach M., Satoh T., Lahaye X., Conrad C., Boyron M., Lombard B., Durand S., Kroemer G., et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349:1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 26.Bridgeman A., Maelfait J., Davenne T., Partridge T., Peng Y., Mayer A., Dong T., Kaever V., Borrow P., Rehwinkel J. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349:1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Shu C., Yi G., Chaton C.T., Shelton C.L., Diao J., Zuo X., Kao C.C., Herr A.B., Li P. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Wu J., Du F., Xu H., Sun L., Chen Z., Brautigam C.A., Zhang X., Chen Z.J. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Zhou W., Whiteley A.T., de Oliveira Mann C.C., Morehouse B.R., Nowak R.P., Fischer E.S., Gray N.S., Mekalanos J.J., Kranzusch P.J. Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell. 2018;174:300–311. doi: 10.1016/j.cell.2018.06.026. e311. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structure of the human cGAS–DNA complex, and mechanism of human-specific DNA-length specificity.
- 30.Severin G.B., Ramliden M.S., Hawver L.A., Wang K., Pell M.E., Kieninger A.K., Khataokar A., O’Hara B.J., Behrmann L.V., Neiditch M.B., et al. Direct activation of a phospholipase by cyclic GMP-AMP in El Tor Vibrio cholerae. Proc Natl Acad Sci U S A. 2018;115:E6048–E6055. doi: 10.1073/pnas.1801233115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelletier H., Sawaya M.R., Kumar A., Wilson S.H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 32.Freudenthal B.D., Beard W.A., Shock D.D., Wilson S.H. Observing a DNA polymerase choose right from wrong. Cell. 2013;154:157–168. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Hall J., Brault A., Vincent F., Weng S., Wang H., Dumlao D., Aulabaugh A., Aivazian D., Castro D., Chen M., et al. Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184843. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of two papers (see also Vincent et al. [34]) reporting a small molecule inhibitor of cGAS.
- 34•.Vincent J., Adura C., Gao P., Luz A., Lama L., Asano Y., Okamoto R., Imaeda T., Aida J., Rothamel K., et al. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of two papers (see also Hall et al. [33•]) reporting a small molecule inhibitor of cGAS.
- 35.Corrigan R.M., Grundling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 36.Jenal U., Reinders A., Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 2017;15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 37.Hall J., Ralph E.C., Shanker S., Wang H., Byrnes L.J., Horst R., Wong J., Brault A., Dumlao D., Smith J.F., et al. The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition. Protein Sci. 2017;26:2367–2380. doi: 10.1002/pro.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Andreeva L., Hiller B., Kostrewa D., Lassig C., de Oliveira Mann C.C., Jan Drexler D., Maiser A., Gaidt M., Leonhardt H., Hornung V., et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature. 2017;549:394–398. doi: 10.1038/nature23890. [DOI] [PubMed] [Google Scholar]; One of two papers (see also Du and Chen [39•]) reporting higher-order complex formation crucial for cGAS activation. Here the authors report a highly structured mouse cGAS–DNA ‘ladder’ complex that enables detection of long DNA.
- 39•.Du M., Chen Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of two papers (see also Andreeva et al. [38]) reporting higher-order complex formation crucial for cGAS activation. Here the authors identify a liquid–liquid phase separation process that enhances cGAS activity.
- 40.de Oliveira Mann C.C., Kiefersauer R., Witte G., Hopfner K.P. Structural and biochemical characterization of the cell fate determining nucleotidyltransferase fold protein MAB21L1. Sci Rep. 2016;6 doi: 10.1038/srep27498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hooy R.M., Sohn J. The allosteric activation of cGAS underpins its dynamic signaling landscape. eLife. 2018;7 doi: 10.7554/eLife.39984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnett K.C., Coronas-Serna J.M., Zhou W., Ernandes M.J., Cao A., Kranzusch P.J., Kagan J.C. Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell. 2019;176:1432–1446. doi: 10.1016/j.cell.2019.01.049. e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentili M., Lahaye X., Nadalin F., Nader G.F.P., Puig Lombardi E., Herve S., De Silva N.S., Rookhuizen D.C., Zueva E., Goudot C., et al. The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 2019;26:2377–2393. doi: 10.1016/j.celrep.2019.01.105. e2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carey C.M., Govande A.A., Cooper J.M., Hartley M.K., Kranzusch P.J., Elde N.C. Recurrent loss-of-function mutations reveal costs to OAS1 antiviral activity in primates. Cell Host Microbe. 2019;25:336–343. doi: 10.1016/j.chom.2019.01.001. e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kristiansen H., Gad H.H., Eskildsen-Larsen S., Despres P., Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 46.Seo G.J., Kim C., Shin W.J., Sklan E.H., Eoh H., Jung J.U. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat Commun. 2018;9 doi: 10.1038/s41467-018-02936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia P., Ye B., Wang S., Zhu X., Du Y., Xiong Z., Tian Y., Fan Z. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol. 2016;17:369–378. doi: 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- 48.Dai J., Huang Y.J., He X., Zhao M., Wang X., Liu Z.S., Xue W., Cai H., Zhan X.Y., Huang S.Y., et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell. 2019;176:1447–1460. doi: 10.1016/j.cell.2019.01.016. e1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong B., Silverman R.H. A bipartite model of 2-5A-dependent RNase L. J Biol Chem. 1997;272:22236–22242. doi: 10.1074/jbc.272.35.22236. [DOI] [PubMed] [Google Scholar]
- 50.Kellenberger C.A., Wilson S.C., Hickey S.F., Gonzalez T.L., Su Y., Hallberg Z.F., Brewer T.F., Iavarone A.T., Carlson H.K., Hsieh Y.F., et al. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc Natl Acad Sci U S A. 2015;112:5383–5388. doi: 10.1073/pnas.1419328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson J.W., Sudarsan N., Phillips G.E., Stav S., Lunse C.E., McCown P.J., Breaker R.R. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc Natl Acad Sci U S A. 2015;112:5389–5394. doi: 10.1073/pnas.1419264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McFarland A.P., Luo S., Ahmed-Qadri F., Zuck M., Thayer E.F., Goo Y.A., Hybiske K., Tong L., Woodward J.J. Sensing of bacterial cyclic dinucleotides by the oxidoreductase RECON promotes NF-kappaB activation and shapes a proinflammatory antibacterial state. Immunity. 2017;46:433–445. doi: 10.1016/j.immuni.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L., Yin Q., Kuss P., Maliga Z., Millan J.L., Wu H., Mitchison T.J. Hydrolysis of 2’3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat Chem Biol. 2014;10:1043–1048. doi: 10.1038/nchembio.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J., Tao J., Liang W., Zhao M., Du X., Cui S., Duan H., Kan B., Su X., Jiang Z. Identification and characterization of phosphodiesterases that specifically degrade 3’3’-cyclic GMP-AMP. Cell Res. 2015;25:539–550. doi: 10.1038/cr.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kubota K., Nakahara K., Ohtsuka T., Yoshida S., Kawaguchi J., Fujita Y., Ozeki Y., Hara A., Yoshimura C., Furukawa H., et al. Identification of 2’-phosphodiesterase, which plays a role in the 2-5A system regulated by interferon. J Biol Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 56.Gusho E., Zhang R., Jha B.K., Thornbrough J.M., Dong B., Gaughan C., Elliott R., Weiss S.R., Silverman R.H. Murine AKAP7 has a 2',5'-phosphodiesterase domain that can complement an inactive murine coronavirus ns2 gene. MBio. 2014;5:e01312–e01314. doi: 10.1128/mBio.01312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dey R.J., Dey B., Zheng Y., Cheung L.S., Zhou J., Sayre D., Kumar P., Guo H., Lamichhane G., Sintim H.O., et al. Inhibition of innate immune cytosolic surveillance by an M. tuberculosis phosphodiesterase. Nat Chem Biol. 2017;13:210–217. doi: 10.1038/nchembio.2254. [DOI] [PubMed] [Google Scholar]
- 58.Athukoralage J.S., Rouillon C., Graham S., Gruschow S., White M.F. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature. 2018;562:277–280. doi: 10.1038/s41586-018-0557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samangouei P., Crespo-Avilan G.E., Cabrera-Fuentes H., Hernandez-Resendiz S., Ismail N.I., Katwadi K.B., Boisvert W.A., Hausenloy D.J. MiD49 and MiD51: new mediators of mitochondrial fission and novel targets for cardioprotection. Cond Med. 2018;1:239–246. [PMC free article] [PubMed] [Google Scholar]
- 60.Loson O.C., Liu R., Rome M.E., Meng S., Kaiser J.T., Shan S.O., Chan D.C. The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure. 2014;22:367–377. doi: 10.1016/j.str.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghosh A., Shao L., Sampath P., Zhao B., Patel N.V., Zhu J., Behl B., Parise R.A., Beumer J.H., O’Sullivan R.J., et al. Oligoadenylate-synthetase-family protein OASL inhibits activity of the DNA sensor cGAS during DNA virus infection to limit interferon production. Immunity. 2019;50:51–63. doi: 10.1016/j.immuni.2018.12.013. e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee M.S., Kim B., Oh G.T., Kim Y.J. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol. 2013;14:346–355. doi: 10.1038/ni.2535. [DOI] [PubMed] [Google Scholar]
- 63.Darby A.C., McInnes C.J., Kjaer K.H., Wood A.R., Hughes M., Martensen P.M., Radford A.D., Hall N., Chantrey J. Novel host-related virulence factors are encoded by squirrelpox virus, the main causative agent of epidemic disease in red squirrels in the UK. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kranzusch P.J., Wilson S.C., Lee A.S., Berger J.M., Doudna J.A., Vance R.E. Ancient origin of cGAS-STING reveals mechanism of universal 2',3' cGAMP signaling. Mol Cell. 2015;59:891–903. doi: 10.1016/j.molcel.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin M., Hiroyasu A., Guzman R.M., Roberts S.A., Goodman A.G. Analysis of Drosophila STING reveals an evolutionarily conserved antimicrobial function. Cell Rep. 2018;23:3537–3550. doi: 10.1016/j.celrep.2018.05.029. e3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y., Gordesky-Gold B., Leney-Greene M., Weinbren N.L., Tudor M., Cherry S. Inflammation-induced, STING-dependent autophagy restricts Zika virus infection in the Drosophila brain. Cell Host Microbe. 2018;24:57–68.e3. doi: 10.1016/j.chom.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goto A., Okado K., Martins N., Cai H., Barbier V., Lamiable O., Troxler L., Santiago E., Kuhn L., Paik D., et al. The kinase IKKbeta regulates a STING- and NF-kappaB-dependent antiviral response pathway in Drosophila. Immunity. 2018;49:225–234. doi: 10.1016/j.immuni.2018.07.013. e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campbell J.D., Alexandrov A., Kim J., Wala J., Berger A.H., Pedamallu C.S., Shukla S.A., Guo G., Brooks A.N., Murray B.A., et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horn D., Prescott T., Houge G., Braekke K., Rosendahl K., Nishimura G., FitzPatrick D.R., Spranger J. A novel Oculo-Skeletal syndrome with intellectual disability caused by a particular MAB21L2 mutation. Eur J Med Genet. 2015;58:387–391. doi: 10.1016/j.ejmg.2015.06.003. [DOI] [PubMed] [Google Scholar]