Abstract
Ticks are medically-important arthropods that maintain and transmit numerous emerging viruses. China suffers severely from tick-borne viral diseases such as tick-borne encephalitis and severe fever with thrombocytopenia syndrome (SFTS), but the background of tick-borne viruses is very limited. Here we report the virome profiling of ticks and goat sera from SFTS-epidemic areas, and serological investigation of SFTS virus (SFTSV) and Nairobi sheep disease virus (NSDV). Results revealed divergent viruses in ticks and goat sera, including SFTSV and NSDV. Sequence and phylogenetic analyses showed that the SFTSV identified here was most closely related to human SFTSV in sampling and surrounding areas, and the NSDV to the previously identified NSDV from northeast China. Serological investigation of SFTSV infection in goats revealed intensive activity in those areas. Surprisingly, two different methods of NSDV serological investigation showed no sera positive for this virus.
Keywords: Virome, SFTSV, NSDV, Genomes, Seroprevalence
1. Introduction
Ticks are a large group of ectoparasites that are exclusively specialized obligate hematophages. They are recognized, along with mosquitoes, as the main arthropod vectors of disease agents with hosts ranging from wildlife to humans and domestic animals globally (Dantas-Torres et al., 2012). Due to their propensity for feeding on a wide range of hosts, ticks readily transmit pathogens between different host species, thereby providing huge obstacles to the control and prevention of tick-borne diseases (TBD). Diverse microorganisms such as rickettsia, anaplasma, and ehrlichia are among the important pathogens transmitted by ticks, and tick-borne viruses (TBV) are increasingly being considered a major threat to humans and other animals (Dantas-Torres et al., 2012).
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging hemorrhagic fever first diagnosed in Xinyang, Henan province, China in 2009 (Yu et al., 2011). Currently, almost 10,000 cases of the disease have been reported in 23 provinces and etiologically confirmed in 18 provinces, with numbers increasing yearly ( Fig. 1) (Liu et al., 2014b, Zhan et al., 2017). Due to a lack of any prophylactic vaccine or effective antivirals, the mortality rate of SFTS cases can reach as high as 30%. SFTS has therefore become a significant public health problem (Liu et al., 2014b, Zhan et al., 2017). Beyond China, SFTS has also been reported in Japan and South Korea in 2013, with a similar case reported in the USA in 2012 (Kim et al., 2013, McMullan et al., 2012, Takahashi et al., 2014), indicative of a potentially global situation requiring control and prevention. The causative agent of SFTS is a recently described new phlebovirus (SFTSV) within the family Phenuiviridae, order Bunyavirales. The tick Haemaphysalis longicornis (H. longgicornis) is considered the most likely transmission vector (Zhang et al., 2012), but the natural animal hosts of SFTSV remain uncertain, even though many domestic animals in endemic areas, such as goats, cattle, dogs, pigs and chickens, and even rodents, have been found to exhibit a high seroprevalence of this virus without presenting any notable symptoms. It cannot be ruled out that those infections were caused by SFTSV transmitted to animals by SFTSV-infected ticks (Liu et al., 2013a, Niu et al., 2013, Zhan et al., 2017).
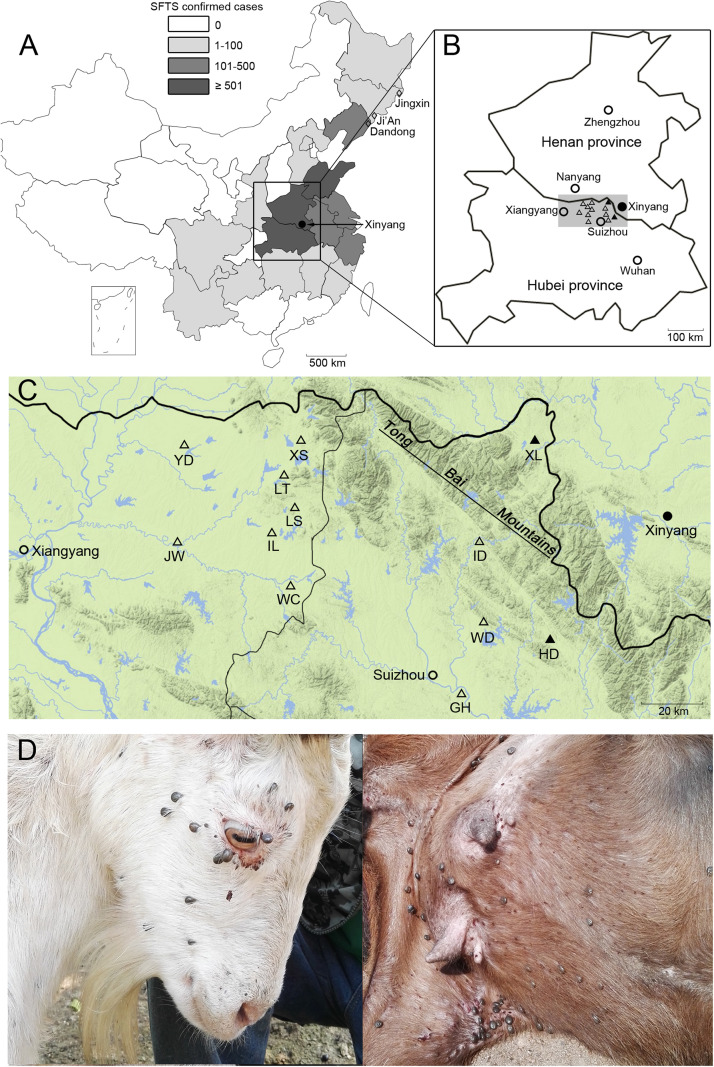
Fig. 1.
Sampling information in this study. The first location (Xinyang) of reported SFTS is identified by a filled circle; the prior detection sites of NSDV are shown as open diamonds. (B) Detailed locations (triangles) of sampling in this study with SFTSV- and NSDV-positive sites by RT-PCR highlighted by filled triangles. (C) Magnified landscape illustration (generated from QQ Map) of shadowed area in panel B. (D) Goats, showing heavy infestation by ticks. Site abbreviations are as in Table 1.
(A) SFTS confirmed cases in China from 2010 to 2016 [Adapted from Reference (Zhan et al., 2017)].
Nairobi sheep disease virus (NSDV) is another important tick-borne virus belonging to the genus Orthonairovirus, family Nairoviridae, order Bunyavirales (International Committee on Taxonomy of Viruses ICTV, 2018). NSDV can cause Nairobi sheep disease (NSD), an acute hemorrhagic gastroenteritis, in sheep and goats with mortalities of up to 70%, especially in exotic and crossbred animals, and has been responsible for several outbreaks of disease in East Africa (Baron and Holzer, 2015). The virus has also been identified in South Asia (India and Sri Lanka), where it is known as Ganjam virus (Marczinke and Nichol, 2002, Perera et al., 1996). NSDV infection of humans can cause a mild disease with symptoms such as fever, headache, nausea and vomiting, and therefore classifies as a zoonotic pathogen (Dandawate et al., 1969b). Extensive serological studies carried out in different parts of India have showed a certain seroprevalence of this virus in humans, sheep, goats and cattle, indicating a wide transmission of this virus within the subcontinent (Banergee, 1996, Dandawate et al., 1969b, Joshi et al., 1998, Sudeep et al., 2009). The virus spreads only through the feeding of competent infected ticks, and therefore its transmission is highly dependent on the geographic distribution of its hosts: Rhipicephlus appendiculata in Africa and H. intermedia in India (Baron and Holzer, 2015). Such a virus and associated disease remained unreported in east Asia until the first identification of a new NSDV variant from H. longicornis ticks collected in northeast China in 2015 (Gong et al., 2015).
To further understand the circulation of SFTSV and NSDV in China, we performed viral metagenomic analyses of ticks and goat sera sampled from SFTSV-endemic areas of Hubei province, China, with discovery of variants of SFTSV and NSDV in these samples, and then conducted genomic characterization as well as serological surveys of SFTSV and NSDV. Results obtained here contribute to understanding of the viral flora of ticks and goats, and genetic diversity and seroprevalence of SFTSV and NSDV in China.
2. Results
2.1. Sample collection
During 26th May and 5th June 2016, a total of 1079 goat sera (Boer and local crossbred) were collected at 5 locations in Suizhou (n = 697) and 7 locations in Xiangyang (n = 382), with sampling at each location from 1 to 3 adjacent farms. At the same time 4595 adult ticks were plucked from goats at 3 locations in Suizhou (n = 3206) and 2 locations in Xiangyang (n = 1389). Sample information is in Fig. 1 and Table 1. All goats freely grazed in the field daily with some heavily infested by ticks (Fig. 1D). All ticks were identified as H. longicornis.
Table 1.
Sample information, nucleic acids and serological detection of SFTSV and NSDV * .
| RT-PCR | S.I. | N.E. | N.I. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| City | Site | Ticks # | Pool # | SFTSV P | NSDV P | Sera # | 1:200 | 1:400 | 1:1600 | 1:6400 | 1:25,600 | 1:200 | 1:200 |
| SZ | HD | 1084 | 103 | 1(1.0) | 2(1.9) | 49 | 29(59.2) | 16(32.7) | 9(18.4) | 2(4.1) | 0(0) | 0(0) | 0(0) |
| XL | 1537 | 169 | 2(1.2) | 0 | 211 | 160(75.8) | 113(53.6) | 57(27.0) | 5(2.4) | 0(0) | 0(0) | 0(0) | |
| WD | 0 | – | – | – | 155 | 51(32.9) | 24(15.5) | 9(5.8) | 0(0) | 0(0) | 0(0) | 0(0) | |
| GH | 0 | – | – | – | 51 | 29(56.7) | 19(37.3) | 10(19.6) | 0(0) | 0(0) | 0(0) | 0(0) | |
| ID | 585 | 31 | 0(0) | 0(0) | 231 | 173(74.9) | 154(66.7) | 100(43.3) | 19(8.2) | 0(0) | 0(0) | 0(0) | |
| S-total | 3206 | 303 | 3(1.0) | 2(0.7) | 697 | 442(63.4) | 326(46.8) | 185(26.5) | 26(3.7) | 0(0) | 0(0) | 0(0) | |
| XY | XS | 1246 | 104 | 0(0) | 0(0) | 25 | 6(24.0) | 6(24.0) | 5(20.0) | 0(0) | 0(0) | 0(0) | 0(0) |
| IL | 0 | – | – | – | 38 | 8(21.1) | 8(21.1) | 5(13.2) | 0(0) | 0(0) | 0(0) | 0(0) | |
| JW | 0 | – | – | – | 14 | 3(21.4) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | |
| WC | 0 | – | – | – | 67 | 13(19.4) | 3(4.5) | 1(1.5) | 0(0) | 0(0) | 0(0) | 0(0) | |
| LS | 143 | 9 | 0(0) | 0(0) | 136 | 24(17.7) | 22(16.2) | 10(7.4) | 3(2.2) | 1(0.7) | 0(0) | 0(0) | |
| YD | 0 | – | – | – | 37 | 5(13.5) | 5(13.5) | 2(5.4) | 0(0) | 0(0) | 0(0) | 0(0) | |
| LT | 0 | – | – | – | 65 | 20(30.8) | 11(16.9) | 4(6.2) | 1(1.5) | 0(0) | 0(0) | 0(0) | |
| S-total | 1389 | 113 | 0(0) | 0(0) | 382 | 79(20.7) | 55(14.4) | 27(7.1) | 4(1.1) | 1(0.3) | 0(0) | 0(0) | |
| Total | 4595 | 416 | 3(0.7) | 2(0.5) | 1079 | 521(48.3) | 381(35.3) | 212(19.7) | 30(2.8) | 1(0.1) | 0(0) | 0(0) |
*Abbreviations: SZ, Suizhou; XY, Xiangyang; HD, Haodian; XL, Xiaolin; WD, Wandian; GH, Guanghua; ID, Yingdian; S-total, Sub-total; XS, Xinshi; IL, Xinglong; JW, Juwan; WC, Wangcheng; LS, Liushen; YD, Yangdang; LT, Lutou; Tick #, tick numbers; Pool #, tick pool numbers; SFTSV P, tick pools positive for SFTSV by RT-PCR tests with percentages in brackets; NSDV P, tick pools positive for NSDV by RT-PCR test with percentages in brackets; Sera #, numbers of goat sera; S.I., SFTSV positive numbers (percentages) of goat sera by IIFA test with 5 dilutions; N.E., NSDV positive numbers (percentage) of goat sera by ELISA at 1:200 dilution; N.I., NSDV positive numbers (percentages) of goat sera by IIFA at 1:200 dilution. Individual or sub-total percentages larger than total levels are highlighted in bold italics.
2.2. Virome profiling of ticks and goat sera
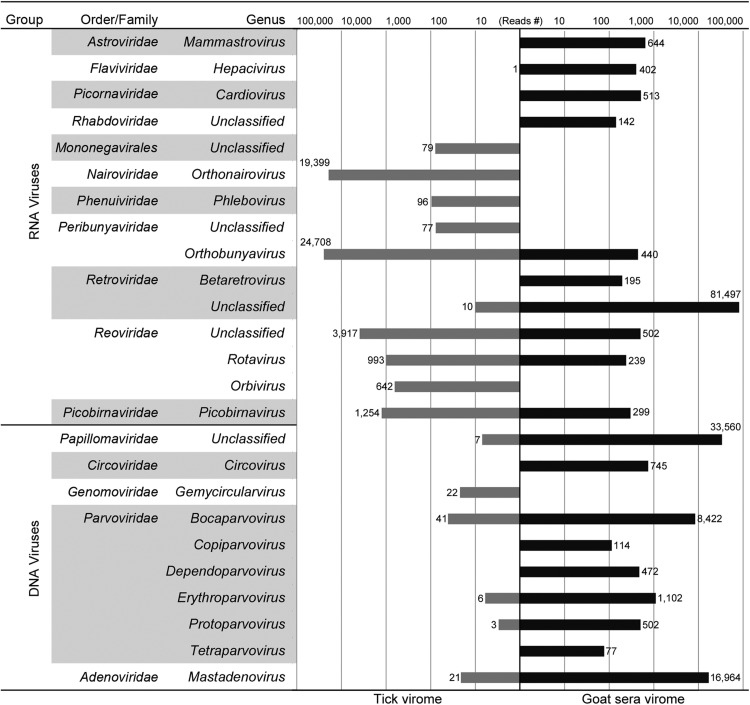
Ticks and goat sera were subjected to VIrome-RESearch (VIRES) analyses (see Materials and Methods), with 1.57 and 2.28 Gb Illumina sequencing reads for ticks and goats generated, respectively. After a quality check and removal of host genome data, 66.8% and 67.5% clean reads remained, which contained 3269,576 and 5738,180 reads of average length 182 bp. The primary annotation and validation showed that 51,255 (1.6%) and 12,790 (2.3%) reads were identifiable clearly as virus-like reads (VLR).
In ticks, the VLRs were cataloged into 17 genera within 12 families or orders, including many mammal-infecting viruses such as hepacivirus, orthonairovirus, phlebovirus, orthobunyavirus, retrovirus, rotavirus, orbivirus, picobirnavirus, and bocaparvovirus ( Fig. 2). VLRs annotated to genera Phlebovirus and Orthonairovirus were related to SFTSV and NSDV. In goat sera, VLRs could be cataloged into 19 genera and 12 families with the most being unclassified retrovirus-like reads (62.8%, 81,497/129,790) (Fig. 2). All VLRs in goat sera were annotated to the viruses that infect or are harbored by mammals, except for one unclassified rhabdovirus of unknown host.
Fig. 2.
Numbers of reads annotated to viruses classified by viral genus, family and nucleic acid group.
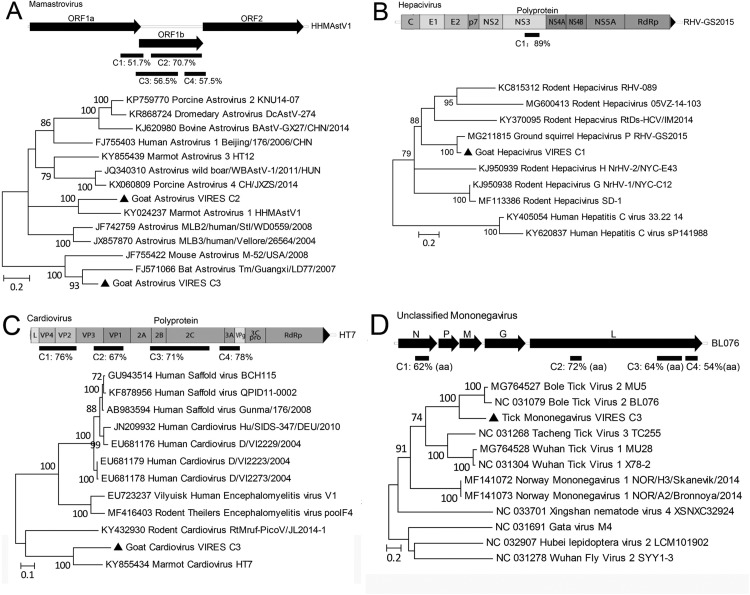
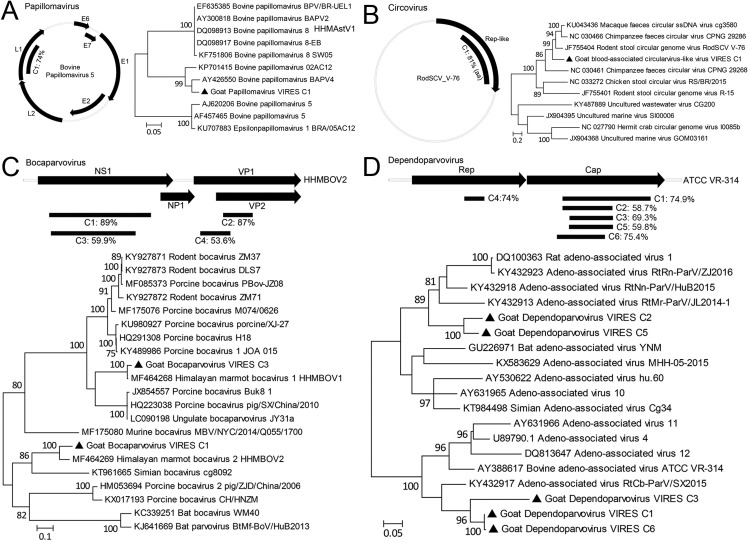
Contigs of 8 groups were first phylogenetically compared with Genbank reference sequences, detailed results were summarized in Fig. 3, Fig. 4. Generally contigs of astrovirus, cardiovirus, mononegavirus, papillomavirus, circovirus, bocaparvovirus (C3–4) and dependoparvovirus showed < 78% nt or < 81% aa identities with known viruses, but contigs of hepacivirus and bocaparvovirus (C1–2) had > 87% identities with corresponding references (Fig. 3, Fig. 4). Phylogenetic constructions showed that those contigs neighbored with viruses of different animal origins, such as marmot, squirrel, bovine, rodent (Fig. 3, Fig. 4).
Fig. 3.
Distribution and identities of contigs of goat mamstrovirus (A), hepacivirus (B), cardiovirus (C), and tick mononeavirus (D) against reference sequences, and their preliminary phylogenetic relationship (filled circles). Scale bars indicate substitutions per site.
Fig. 4.
Distribution and identities of contigs of goat papillomavirus (A), circovirus (B), bocaparvovirus (C) and dependoparvovirus (D) against reference sequences and their preliminary phylogenetic relationship (filled circles). Scale bars indicate substitutions per site.
2.3. Complete genome and phylogeny of SFTSV and NSDV
VIRES analysis of ticks resulted in detection of SFTSV and NSDV. Follow-up RT-PCR screening showed that 1 and 2 pools of ticks out of 103 and 169 from HD (0.97%) and XL (1.18%) respectively were SFTSV positive, with only 2 pools from HD NSDV positive (1.94%, 2/103) (Table 1). Three SFTSV and two NSDV amplicons, all from Suizhou ticks, showed 100% identities within the viruses (Fig. 1B). However, all goat sera were negative for both viruses in RT-PCR screening. Positive tick samples of NSDV were tested for the presence of virus in BHK-21 and Vero cell lines and in suckling mice by intracerebral (i.c.) injection. Five blind passages in the cells revealed no cytopathogenic effects (CPE) and none of the injected animals showed any symptoms. RT-PCR analyses of all passaged cultures as well as tissues of the injected animals were negative: i.e., no evidence of NSDV infection was obtained. Using degenerate primer pairs, the complete genome of an SFTSV isolate (named SZHL9) and the almost complete genome of an NSDV isolate (named SZHL16) were successfully obtained. The S, M and L segments of SFTSV SZHL9 had exactly the same lengths (1744, 3378 and 6368 nt, respectively) as those of previously identified viruses in China. The 3 nearly complete genomic segments of NSDV SZHL16 were 1602 (S), 5090 (M), and 12,054 (L) nt in length and covered the entire open-reading frames (ORF) of the genome, showing the same lengths as those previously identified in China, India and Africa, except for a 12 nt insertion immediately after the 3′ terminal of the SZHL16 S gene ORF.
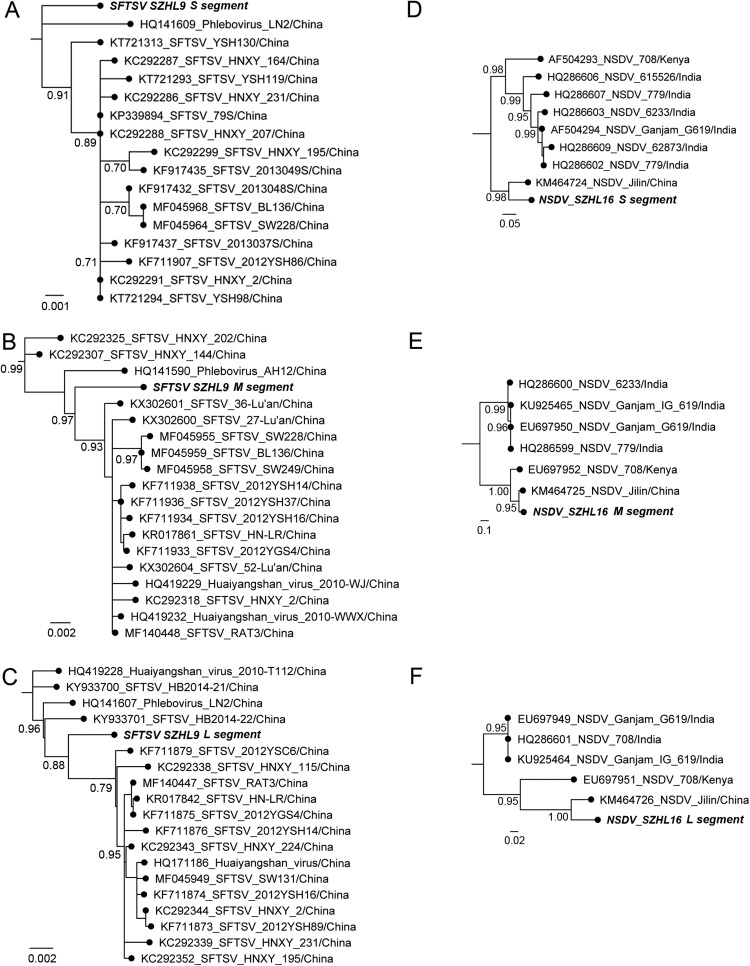
The full genomic sequences obtained above were aligned with all representative SFTSV and NSDV sequences retrieved from Genbank and subjected to phylogenetic analyses. The best-fit substitution models, under AIC1, for the S, M, and L segments of both viruses were selected as K80 + I (I= 0.86), TVM+I (I= 0.89), TVM+I (I= 0.88), GTR+I+G (I= 0.22, a= 0.91), GTR+I+G (I= 0.15, a= 1.14), and GTR + I + G (I= 0.30, a= 1.13). Phylogenetic trees are shown in Fig. 5. In the SFTSV tree, S, M and L segments of SZHL9 were closely similar to those of other SFTSVs with the S segment sharing the highest identity (99.4%) with SFTSV HNXY_27 (Liu et al., 2013b), and the M and L segments (99.4% and 99.2%) with SFTSV 2012YSH37 and 2012YSH14 respectively (Fig. 5A-C) (Huang et al., 2014). In three gene segment NSDV trees, NSDV SZHL16 clustered most closely with NSDV Jilin/China (93.2%, 94.9% and 95.4% for the S, M and L segments respectively) (Fig. 5D-F). Interestingly, in the S gene phylogeny, Chinese NSDVs shared nearly the same identity (~88.0%) with viruses from both Africa and India, but in M and L phylogenies Chinese NSDVs grouped more closely with African viruses (88.1% identity in M and 89.0% in L) than with Indian viruses (72.0–76% identity in M and 88.9% in L) (Fig. 5D-F).
Fig. 5.
Phylogenetic relationships of SFTSV (A: S segment; B: M segment; C: L segment) and NSDV (D: S segment; E: M segment; F: L segment). New viruses reported in the present study are identified in bold italics. Scale bars indicate substitutions per site.
2.4. Serological investigation of SFTSV
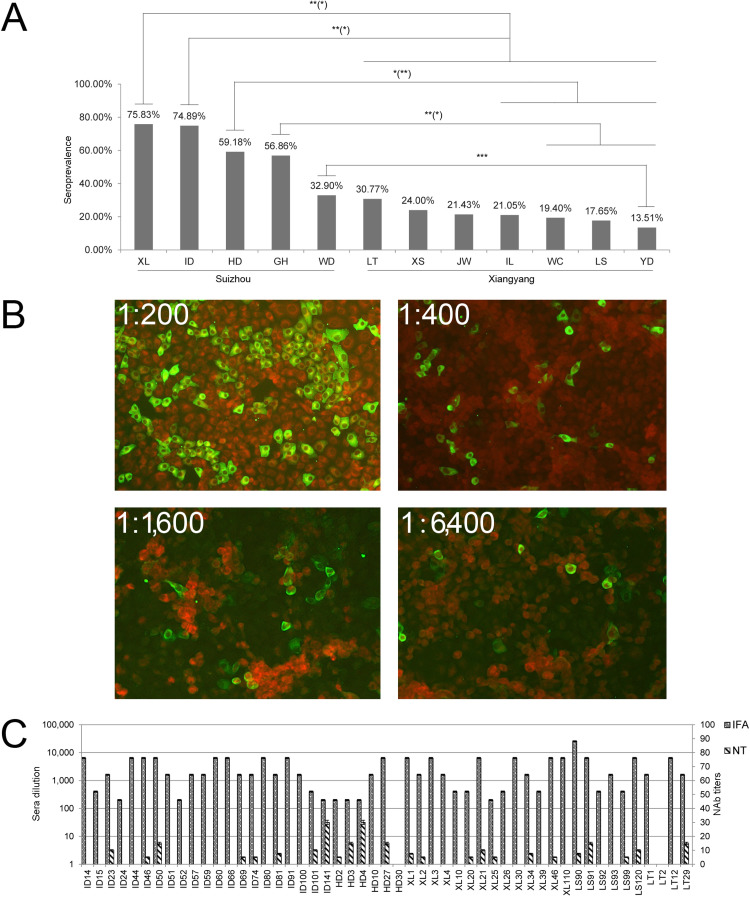
To investigate SFTSV infection of goats, the indirect immunofluorescence assay (IIFA) was established using SFTSV strain WF66. At dilution of 1:200, 521 of 1079 sera (48.3%) were antibody (Ab) positive: 442 of 697 sera (63.4%) from Suizhou and 79 of 382 (20.7%) from Xiangyang. By sampling site, the highest seroprevalence was in samples from Xiaolin (75.8%, 160/211) followed by Yindian (74.9%, 173/231), with Yangdan and Wangcheng goats showing the lowest (13.5%, 5/37% and 19.4%, 13/67 respectively). Of note is that Xiaolin is the site from which the collected ticks were SFTSV-positive, while another tick SFTSV-positive site, Haodian, showed 59.2% (29/49) seroprevalence in goats. IIFA results summarized in Table 1 show that goat SFTSV seroprevalence in Suizhou was much higher than in Xiangyang ( Fig. 6A). IIFA tested using 2-fold dilutions showed that 35.3% (46.8% of Suizhou, 14.4% of Xiangyang) had titers of 1:400, 19.7% of 1:1600 (26.5% in Suizhou, 7.1% in Xiangyang), and 2.8% of 1:6400 (3.7% in Suizhou, 1.1% in Xiangyang) (Fig. 6B). One serum from Liushen in Xiangyang even had a titer of 1:25,600 (Table 1). Fifty one sera (49 positive and 2 negative) were further tested for neutralizing antibody (NAb) levels against SFTSV WF66 by neutralization assay (NA) with results showing that 49.0% (24/49) of the IIFA-positive sera had NAb titers of between 5 and 30, whereas the 2 IIFA negative sera were also negative by NA (Fig. 6C).
Fig. 6.
Serological investigation of SFTSV. (A) Seroprevalence comparison between Suizhou and Xiangyang; * *(*), significant difference with p value < 0.05 or < 0.001; *(**), with p value < 0.01 or < 0.001; ***, and with p value < 0.001. (B) A serum sample from Xiaolin (sample code XL21) was applied at dilutions of 1:200–1:6400 for IIFA analyses. (C) Endpoint titers (left titer scale) and neutralizing titers (right index scale) of SFTSV-specific Abs in goat sera tested by IIFA and NA respectively. Site abbreviations are as in Table 1.
2.5. Serological investigation of NSDV
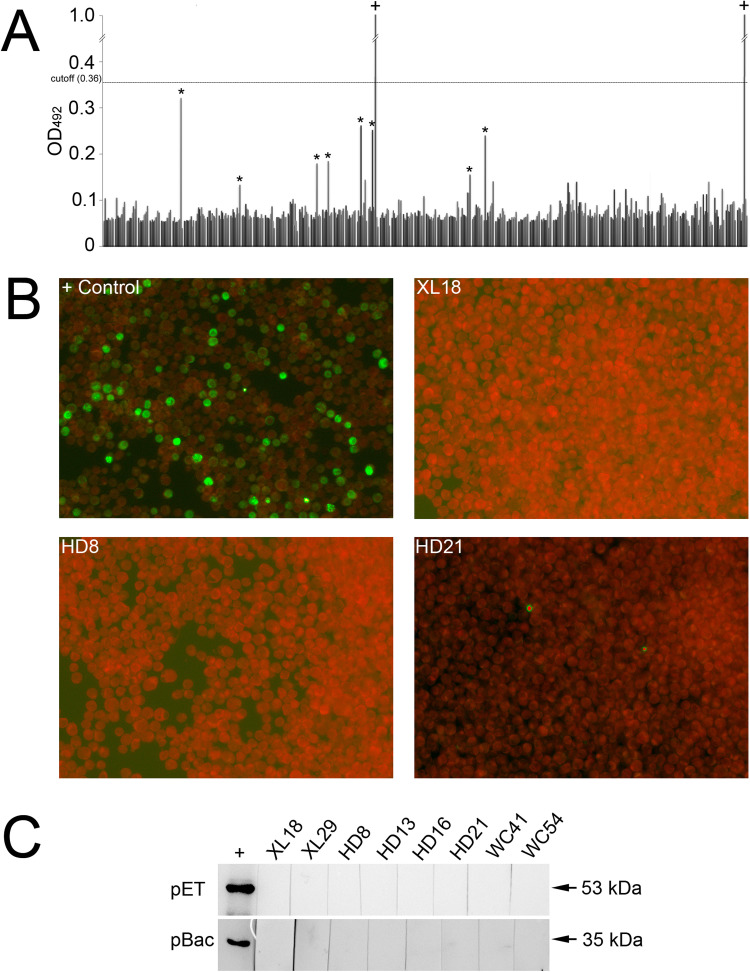
To investigate NSDV infection in goats, an indirect ELISA based on a prokaryotic-expressed NP was established. All goat sera were tested at 1:10 with the highest OD492 reading of 0.32 (less than cutoff value 0.36), and none being considered seropositive ( Fig. 7A). Additionally, to test for possible non-specific interference, 300 sera were retested at dilutions of 1:100 and 1:300 with no change in results. An IIFA method using truncated NP-recombinant baculovirus was established to confirm the ELISA results. Again, all sera tested seronegative (Fig. 7B). As a further check, 14 sera including 8 with the highest OD492 values (marked with an asterisk in Fig. 7A) were analyzed by Western blot (WB) using two types of NPs, expressed by E. coli and a recombinant baculovirus, as antigens. Results showed no significant reaction of goat sera with the NPs.
Fig. 7.
Serological investigation of NSDV. (A) ELISA OD492 reading profiling of 504 goat sera including 2 positive controls (noted by +); 8 sera with the OD492 readings are noted by asterisks. (B) Three (XL8, HD8 and HD21) of goat sera noted by asterisks in panel A were analyzed by IIFA with mouse anti-NP hyperimmune serum as a positive control. (C) Eight sera noted by asterisks in panel A were analyzed by WB using E. coli expressed (pET) and recombinant baculovirus system (pBac) expressed NPs as antigens. Lanes were taken from different gels with sample codes illustrated above; + , mouse anti-NP hyperimmune serum.
3. Discussion
3.1. Virome of goat sera and ticks
Domestic animals harbor a variety of pathogens with many capable of infecting humans and causing zoonoses, such as Brucella spp and Mycobacterium tuberculosis in ruminants, and MERS coronavirus in camels (Azhar et al., 2014, Ganter, 2015, Wolfe et al., 2007). Investigation into pathogens harbored by domestic animals does not therefore benefit only animal health but is also crucial to maintain human public health. Goats, as a major meat source, are widely farmed in China, but the pathogens they harbor have been poorly investigated compared to those of pigs and chickens. Here, our pioneer virome profiling of goat sera has revealed a variety of viruses that have not been previously reported in goats, including parvovirus, picobirnavirus, cardiovirus, hepacivirus and papillomavirus. Many viruses identified here in goats, such as astrovirus, cardiovirus, papillomavirus and circovirus, showed significant genetic divergences and are therefore likely newly discovered and suggestive of an independent evolution of these viruses in goats. In contrast, some other viruses, such as hepacivirus and bocaparvovirus, had up to 89% nt identities with corresponding viruses of other animal species, indicating a broad host tropism. Results here also indicated multiple infections of variants of mamastrovirus, bocaparvovirus and dependoparvovirus, because several alleles of genes of those viruses showed distinct divergences, but contigs of cardiovirus and mononegavirus could be from different viruses or from the same virus (recombinational or not), since their dispersed contigs showed different but close identities with references. Only astroviruses have been identified to be associated with a disease, encephalitis and ganglionitis in sheep (Toffan et al., 2009), and the pathogenicity of the other viruses remains unknown. Of note is the identification here of hepaciviruses, a species that includes hepatitis C virus causing hepatocellular carcinoma and lymphomas in humans (Hajarizadeh et al., 2013). Hepaciviruses have recently been identified in other mammals, including dogs, horses, rodents and bats, indicating its complicated evolutionary history (Scheel et al., 2015). The hepacivirus contig obtained here showed close relationship with rodent hepaciviruses and clustered together with these (Fig. 3B), indicating either a common ancestor between rodent and goat or possible interspecies transmission. Although major pathogenic viruses were not detected in serum samples of the healthy-looking goat flocks tested in the present study, virome profiling has revealed that goat sera can harbor a variety of viruses without evident manifestation. Regarding this, virome profiling of respiratory and digestive systems may provide insight into the biological functions of the viral flora in healthy animals and may open a new scientific area to investigate the symbiosis of viruses with their hosts. These viral reads revealed by VIRES most likely originate from viruses that infect or are harbored by goats, such as astrovirus, hepacivirus and parvovirus, but the possibility cannot be ruled out that some of these sequences, especially retrovirus-like contigs, derive from endogenized viral sequences (EVEs) since contigs containing intact ORF have not been obtained, even though host genomes and free nucleic acids were largely removed by digestion of nuclease.
The VIRES analyses of ticks have revealed some TBVs, such as phleboviruses and nairoviruses, and have also revealed viruses that are known to be non-vector-borne pathogens, such as hepaciviruses, erythroparvoviruses, bocaparvoviruses, and papillomaviruses (Fig. 2). These non-vector-borne viruses identified here in ticks presumably originated from goats by blood-taking, and show the ecological relationship of pathogen circulation between vectors and hosts. In China, a metagenomic profile of viral communities in Rhipicephalus ticks from Yunnan province has also been conducted and has revealed a broad range of viral families (Xia et al., 2015). Apart from phages, viruses from a variety of families including Herpesviridae, Poxviridae, Geminiviridae, Circoviridae and Virgaviridae, were identified in the Yunnan ticks, but not in this study, while viruses of the Reoviridae, Papillomaviridae, Picobirnaviridae, Flaviviridae, and Adenoviridae were identified in this study but not in Yunnan ticks (Xia et al., 2015). Such differences might be ascribed to: a) different methodologies of viral metagenomics; b) differences in the ways of sampling of ticks, since Yunnan ticks were collected by flag-dragging fields (Xia et al., 2015), whereas ticks here were directly plucked from animals upon which they have fed; and/or c) different tick species within the two widely separated locations. However, bunyaviruses were present in both studies, suggesting a wide distribution of these viruses in different tick species and locations.
3.2. Seroprevalence of SFTSV
SFTS is a severe disease in China, with Henan and Hubei provinces being the main epidemic regions (Fig. 1A) (Zhan et al., 2017). This disease is difficult to control and prevent, because of its complicated transmission chain and unknown natural hosts (Liu et al., 2014b). SFTSV has a broad genetic diversity with high mutation rates (Lam et al., 2013), and recombination and reassortment of SFTSVs frequently occur (Shi et al., 2017). In this study, SFTSV was identified from two sites next to Xinyang, Henan province (Fig. 1C), and were largely identical (up to 99.4% nt identities) to human viruses (HNXY_27 and 2012YSH37) identified in Xinyang (Huang et al., 2014, Liu et al., 2013b), indicating that infections within the SFTS epidemic area covering Suizhou and Xingyang were caused by the same viruses. Here we also screened goat sera for SFTSV by RT-PCR, but no viremia was detected. IIFA testing of these sera showed high IgG levels against this virus but low neutralizing titers, indicating a rapid virus clearance in goats following SFTSV infection. Experimental infection of macaques with SFTSV has also showed an short viremic phase and undetectable viral RNA levels by day 7 following injection (Jin et al., 2015). Considering the high seroprevalence of SFTSV in goats but low rate of SFTSV detection in ticks, the exact mechanism of virus circulation between vectors and vertebrate hosts remains unclear.
Several serological investigations of SFTSV have previously been conducted in animals and humans in China, but none has investigated the seroprevalence of SFTSV within different regions (Niu et al., 2013, Zhang et al., 2014). Here goat sera were collected from 12 sites, of which 5 within Suizhou were located in or close to the Tong Bai mountains (TBM). (Fig. 1C). Of note is that the seroprevalence of SFTSV in Suizhou was significantly higher than in Xiangyang (Fig. 1, Fig. 6). Along with the detection of SFTSV RNAs in ticks from Suizhou, all data indicate that circulation of SFTSV between animal hosts and vectors is wider in Suizhou, particularly in forest areas. Previous epidemiologic and environmental risk factor analyses of human cases in Xinyang, Henan province, and in Hubei province also proposed that shrub and forest regions showed strong associations with SFTSV infection (Liu et al., 2014a, Wang et al., 2017). Such an association is undoubtedly related to the vector, H. longicornis, which is predominantly found in shrub and forest areas (Xu and Sun, 2003).
3.3. NSDV in China
NSDV is a lethal pathogen to sheep and goats, and is mainly found in Africa and south Asia (Baron and Holzer, 2015). The virus was first reported in H. longicornis ticks in the northeast of China in 2015 by RT-PCR detection (Gong et al., 2015), but infection or disease outbreaks in goats or sheep have never been reported in China. Here we identified NSDV in the same tick species in central China, thereby demonstrating a wider distribution of this virus in ticks in China. Viruses identified here and in the previous study are of two distinct strains, indicating that NSDV in China has a genetic diversity. Of note is that Chinese NSDVs showed a closer relationship with African strains than with Indian ones in the M and L segments (Fig. 5E-F), suggesting an intriguing evolutionary route of this virus. Current data suggest that the most recent common ancestor of Chinese and African NSDVs existed within Indian viruses. Such a suggestion cannot be explained by geographic distribution, and it will be necessary to investigate NSDV epidemiologically over a wider area in China in order to determine if it is a true domestic virus or an imported one.
To further investigate the potential infection of goats with NSDV, two serological methods, ELISA and IIFA, using expressed NSDV rNPs, were developed to test goat sera, then further confirmed by WB. Although NSDV was identified in ticks collected from goat flocks, no NSDV-positive goat sera were found. Previous investigations have shown that this virus was not only isolated from ticks, humans, sheep, and mosquitoes in India and Africa, but that NP antibodies in human, sheep, goat and cattle sera were also reported (Dandawate et al., 1969a, Dandawate et al., 1969b, Joshi et al., 1998, Joshi et al., 2005, Sudeep et al., 2009). A possible reason for the negative sera in China is that the currently identified Chinese NSDVs are incapable of infecting goats to cause disease. This might explain why, although the virus does exist in China, no cases of NSD have been reported. Unfortunately, we were unable to isolate this virus during both the present study and a previous one (31) either by cell culture or animal injection.
TBDs are very difficult to control and prevent, and keep emerging or re-emerging (Fang et al., 2015), largely because of limited knowledge of their background and complicated methods of circulation in nature. Many ecological factors involved in the life cycle of TBVs remain unknown (Fuente et al., 2016). Considering that TBDs are associated with many important aspects of modern life, including animal and human health, ecological balance and climate change, One Health strategy for their control must be emphasized.
4. Materials and methods
4.1. Ethics statement
Experimental infection of mice in this study was reviewed and approved by the Administrative Committee on Animal Welfare of the Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, China (Laboratory Animal Care and Use Committee Authorization, permit number: JSY-DW-2016-02).
4.2. Sample collection and preparation
All ticks were directly plucked from infested goats in Suizhou and Xiangyang, Hubei province. Not all goats in a flock infested ticks. These two regions are neighbors of Xinyang of Henan province where SFTS was first identified and now a SFTS-endemic area. All collected ticks were adults and maintained alive in cases at room temperature before study. Tick species were identified morphologically by a field trained expert and confirmed by mitochondrial 16 S rDNA sequencing (Black and Piesman, 1994). Approximately 2 ml blood samples of all goats in a flock were collected from goat jugular veins and sera were obtained after blood clotting, then stored in dry ice and transported to the laboratory along with the ticks where all were stored at −80 °C until further processing. Sample details are listed in Fig. 1 and Table 1.
4.3. VIRES: sample-preprocessing and high-throughput sequencing
All samples were subjected to viral metagenomic analyses following our VIRES protocol which has integrated previously published but updated sample-preprocessing (He et al., 2013) and bioinformatics analyses. Ticks were divided into pools with each having 5–15 individuals according to their body size (Table 1). Tick sex was not determined when pooling. One half of the tick pools of each sample location were subjected to VIRES analyses following washing with sterile phosphate-buffered saline (PBS), homogenization in a Tissuelyzer-48 (Jingxin) at 4 °C and centrifugation at 10,000×g at 4 °C for 10 min. Fifty µl aliquots of each supernatant were transferred and pooled together in fresh tubes for further VIRES processing. The remaining supernatants with sediments were stored at −80 °C for later virus detection. Twenty µl of each goat serum were pooled. All supernatants and pooled sera were sterilized by filtration through 0.22-µm syringe filters, digested with nuclease, and immediately subjected to total nucleic acid automatic extraction in a QIAcube (Qiagen) using the RNeasy mini kit (Qiagen) with β-mercaptoethanol (1:100, v/v) added to the lysis buffer. Total nucleic acids were reverse transcribed with the Reverse-Transcription Kit (TaKaRa) using B17N8 primers (5′-CTTGAGCTCTGCAGTACNNNNNNNN-3′). The resulting cDNA products were digested by RNase H (TaKaRa), purified using cold isopropanol, then subjected to double-stranded cDNA synthesis using Klenow fragment (TaKaRa), followed by single-primer random PCR amplification using B17 primers (5′-CTTGAGCTCTGCAGTAC-3′). The PCR products were purified using a PCR purification kit (Axygen) and then fragmented to ~200 bp size by ultrasonication. The DNA fragments were subjected to Illumina pair-end (125 bp) sequencing in a HiSeq. 2500 sequencer. The resulting sequencing reads were quality trimmed and used for bioinformatics analyses as clean data.
4.4. VIRES: bioinformatic analyses
Host genomes were removed using SOAP with default settings. Tick genomes were referred to the Ixodes scapularis tick genome (Genbank accessions: ABJB010000000) (Gulia-Nuss et al., 2016), and goat genomes to the Capra hicus goat genome (Genbank accession: AJPT00000000) (Dong et al., 2012). The remaining sequence pairs were overlapped using FLASH and subjected to gene prediction using MetaGeneMark. Predicted reads were translated into protein sequences and then subjected to local BLASTp analyses against the non-phage viral reference database of Genbank (version: 20151121). Sequences with BLAST E value ≤ 10−5 were further compared with the nonredundant database of Genbank (version: 20151121) using BLASTn. Sequences matching to non-viral organisms were eliminated and the remaining sequences were defined as VLRs and used for further statistical analyses and assembly. All VLRs were cataloged at the level of genus according to their annotations, and then de-duplicated using CD-HIT at 99.9% similarity. Simplified VLRs were de novo assembled using MEGAHIT or SeqMan v7.0 (DNASTAR). The resulting contigs of ≥ 200 bp were mapped against their reference sequences to identify their corresponding genomic locations. The longest contigs or contigs of the same locations were aligned with representative sequences using ClustalW in MEGA 6.0, and preliminary phylogenetic analyses were conducted by the maximum-likelihood method with the GTR+G+I model and with 1000 bootstrap replicates.
4.5. SFTSV and NSDV detection and isolation
The remaining halves of the tick pool samples after VIRES analyses were washed, homogenized and centrifuged as described above. The obtained supernatants along with those used for VIRES analyses (150 µl for each) were subjected to viral RNA automatic extraction on an epMotion 5075 workshop (Eppendorf) using the QIAamp Virus BioRobotMDx Kit (Qiagen). Viral RNAs in goat sera were also extracted in the same way. The viral RNAs were eluted with 30 µl eluent and reverse transcribed into cDNA using the Reverse Transcription kit (TaKaRa). PCR detection of SFTSV and NSDV was conducted using our previously published methods (Gong et al., 2015, Liu et al., 2016) with double-distilled water as a negative control. No positive control was used in this study. Positive PCR amplicons were directly sequenced by the Sanger method on an ABI 3730 sequencer (ComateBio).
The supernatants of NSDV positive samples were sterilized by filtration through 0.22 µm syringe filters, then added to 24-well plates seeded with BHK-21 or Vero cells. After incubation for 2 h at 37 °C, the cells were gently washed with warm sterile PBS and incubated in DMEM with 2% FBS for 3 days. The cultures were observed microscopically daily and blind passaged 5×, following which they were tested for NSDV by RT-PCR. NSDV-positive supernatants were also 1:5 (v/v) diluted in PBS and injected i.c. into 8 3-day-old suckling mice (30 µl each). Two mice injected with PBS served as a negative control. Animals were inspected twice daily for up to 14 days. Groups were euthanized at 7 and 14 days post-injection and organs (brain, spleen, liver, kidney, lung and intestinal tissues) were collected and, following homogenization, etc., were subjected to RT-PCR analysis.
4.6. Complete genome sequencing, genomic analyses and evolution inspection
To obtain the full genomes of SFTSV and NSDV, previously developed primer pairs were used in the PCR systems with the Fast HiFidelity PCR kit (Tiangen) (Gong et al., 2015, Liu et al., 2016). The amplicons were blunt ligated into the pZeroBack vector (Tiangen), and used to transfect competent E. coli DH5α (Tiangen). Three clones of each amplicon were randomly picked for sequencing. Overlapping amplicons were assembled with SeqMan v7.0 (DNAstar).
Genomic structure predictions and phylogenetic characterizations were conducted using our previously published methods (Yang et al., 2018). Briefly, genomic structures were predicted by Vector NTI v.11.5, and then validated by comparison with genomes of other SFTSV and NSDV. Nucleotide reference sequences were retrieved from Genbank and aligned with counterparts of viruses identified here using MAFFT v7.394 (Katoh and Standley, 2013). Evolutionary models were determined using ModelGenerator v0.85 with Akaike Information Criterion 1 (AIC1) (Keane et al., 2006). Phylogenetic analyses were conducted using PhyML v3.3 by the maximum likelihood and best-fit substitution models with evaluation of 100 bootstraps, and visualized with FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree). Nucleotide and amino acids identities were calculated using MegAlign v7.1 (DNASTAR).
4.7. IIFA and NA of SFTSV
The IIFA procedure was modified from our previously published method (He et al., 2017). Mouse hyperimmune serum was first generated by intramuscular injection of adult SPF mice (purchased from the Breeding Laboratory of Jilin University) with SFTSV strain WF66. Vero cells were cultured in 96-well plates (Nunc) and incubated with 100 TCID50 of SFTSV strain WF66 for 72 h before washing 3x with sterile PBS. Following fixation with 5% paraformaldehyde, cells were incubated at 37 °C for 1 h with collected goat sera diluted 1:200, with mouse hyperimmune serum as a positive control, followed by three washes with PBS-Tween (PBST). Fluorescein isothiocyanate (FITC)-labeled rabbit anti-goat IgG (Bioss; 1:2000) was used to detect the primary Ab and visualized by immunofluorescence microscopy (LSM780, Zeiss). Ab titers of goat sera positive at 1:200 dilution were determined by endpoint titrations using 4-fold dilutions (1:400–1:25,600).
To determine their NAb titers, 51 goat sera, in duplicate, were 2-fold serially diluted from 1:5–1:640 and incubated with 100 TCID50 of SFTSV at 37 °C for 1 h. Mixtures were then transferred onto cultures of Vero cells and incubated for 72 h. Following fixation, the cells were incubated sequentially with mouse hyperimmune sera (1:4000) and FITC-labeled rabbit anti-mouse IgG (Bioss; 1:1000) Endpoints were taken as the highest dilutions still showing immunofluorescence and neutralization titers were calculated using the Kärber formula.
4.8. rNP of NSDV expression in E. coli and mouse anti-rNP specific hyperimmune serum preparation
The complete ORF of NSDV NP gene was amplified and subcloned into a prokaryotic expression vector pET-28a(+) with a His-Tag at the C terminus. Following transfection of E. coli BL21(DE3) (Tiangen), the resulting rNPs were expressed after 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) induction and confirmed by SDS-PAGE and WB. The rNPs were purified and quantified by Ni-NTA His Bind Resins (Novagen) and the BCA Protein Assay Kit (CWBio). To prepare specific hyperimmune serum, the purified rNPs were injected intramuscularly into adult SPF mice (50 μg protein in complete Freund's adjuvant (Sigma), 1:1 v/v). The same booster doses were administered at 14, 28 and 35 days following. Blood was collected from the orbital sinus for serum preparation and antibody titers were determined by rNP-based ELISA as described below.
4.9. ELISA investigation of NSDV
ELISAs using rNPs as the coating antigen were developed to detect NSDV antibodies in goat sera. Ninety six-well micro-titration plates (Corning) were coated with purified rNP (100 ng/well in NaHCO3-Na2CO3 buffer, pH9.6) at 4 °C overnight and blocked with 5% skimmed milk (Promega) at 37 °C for 1 h. After washing with PBST, PBS-diluted sera samples were added to wells in duplicate and incubated at 37 °C for 1 h. Mouse anti-rNP specific hyperimmune serum and SPF mouse serum were used as positive and negative controls. After additional washing, 100 µl HRP-conjugated donkey anti-goat IgG (Bioss) at 1:2000 dilution was added and incubation continued at 37 °C for 1 h. Freshly-prepared o-phenylenediamine (OPD) substrate (Sigma) was then added to each well for 5 min of color reaction and stopped by addition of 2 M sulfuric acid. OD492 values were immediately read and blanked by the OD630 value using a Multimode Microplate Reader (Infinite 200 PRO, Tecan). Samples with a mean OD492 value 3-fold or higher (OD492 value ≥0.36) than that of a negative control (OD492 value: 0.12) were considered positive.
Fourteen sera with different ELISA readings were further analyzed by WB, in which rNP was separated by SDS-PAGE, then transferred onto nitrocellulose membranes (Millipore) followed by blocking with 5% skimmed milk (Promega). The membranes were then incubated with selected goat sera (1:200) or mouse anti-rNP specific hyperimmune serum (1:1000; positive control) for 2 h at room temperature. After washing 3x with PBST, HRP-conjugated donkey anti-goat IgG (Bioss; 1:1000) or HRP-conjugated goat anti-mouse IgG (Bioss) was added and incubation continued for 50 min at room temperature. The protein bands were recorded using an automatic chemiluminescence instrument (Tanon).
4.10. Preparation of NSDV NP recombinant baculovirus
A 5′- and 3′-terminal-truncated NP of NSDV was amplified as described above and subcloned into pFastBac (Invitrogen) and then used to transfect E. coli DH10Bac (Invitrogen). The resulting recombinant bacmids were purified with the PureLinkHiPure Plasmid Miniprep kit (Invitrogen) and used to transfect sf9 insect cells (Invitrogen) with Cellfectin II reagent (Invitrogen). Following incubation for 4d, recombinant baculovirus particles were harvested and incubated with sf9 cells (Invitrogen) in 96-well plates for 3 days. The IIFA was performed in a similar manner to that of SFTSV described above.
4.11. Statistics
To compare the difference of positive rates among the sampling sites, Fisher's exact test was conducted with adjusted significance level by the Bonferroni method. All statistics were programed and calculated using the Statistical Analysis System (SAS) version 9.2.
4.12. Dataset illustration and accession numbers
Sequences of all assembled contigs have been deposited in GenBank under accession numbers MH835405-MH835448 and the complete genomes of SFTSV SZHL9 and NSDV SZHL16 under accession numbers MH790412-MH790414 and MH791449-MH791451. The raw data of Illumina sequencing of ticks and goat sera have been deposited in Short Reads Archives (SRA) under accession numbers SRR7985010 and SRR7985011.
Acknowledgements
We thank Dr. Chang Li, Institute of Military Veterinary Medicine, for the SFTSV strain WF66. The study was supported by the National Key Basic Research and Development Program of China (2017YFD0500104). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgments
Conflict of interests
The authors declare that they have no conflict of interest.
References
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Banergee K. Emerging viral infections with special reference to India. Indian J. Med. Res. 1996;103:177–200. [PubMed] [Google Scholar]
- Baron M.D., Holzer B. Nairobi sheep disease virus/Ganjam virus. Rev. Sci. Tech. Int. Epiz. 2015;34:411–417. doi: 10.20506/rst.34.2.2367. [DOI] [PubMed] [Google Scholar]
- Black W.C., Piesman J. Phylogeny of hard-and soft-tick taxa (Acari: Ixodida) based on mithochondrial 16S rDNA sequences. Proc. Natl. Acad. Sci. USA. 1994;91:10034–10038. doi: 10.1073/pnas.91.21.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandawate C.N., Rajagopalan P.K., Pavri K.M., Work T., H. Virus isolations from mosquitoes collected in North Arcot district, Madras state, and Chittoor district, Andhar Pradesh between November 1955 and October 1957. Indian J. Med. Res. 1969;57:1420–1426. [PubMed] [Google Scholar]
- Dandawate C.N., Work T., Webb H., Shah, K.V J.K. Isolation of Ganjam virus from a human case of febrile illness: a report of a laboratory infection and serological survey of human sera from three different states of India. Indian J. Med. Res. 1969;57:975–982. [PubMed] [Google Scholar]
- Dantas-Torres F., Chomel B.B., Otranto D. Ticks and tick-borne diseases: a One Health perspective. Trends Parasitol. 2012;28:437–446. doi: 10.1016/j.pt.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Dong Y., Xie M., Jiang Y., Xiao N., Du X., Zhang W., Tosser-Klopp G., Wang J., Yang S., Liang J., Chen W., Chen J., Zeng P., Hou Y., Bian C., Pan S., Li Y., Liu X., Wang W., Servin B., Sayre B., Zhu B., Sweeney D., Moore R., Nie W., Shen Y., Zhao R., Zhang G., Li J., Faraut T., Womack J., Zhang Y., Kijas J., Cockett N., Xu X., Zhao S., Wang J., Wang W. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus) Nat. Biotechnol. 2012;31:135–141. doi: 10.1038/nbt.2478. [DOI] [PubMed] [Google Scholar]
- Fang L.-Q., Liu K., Li X.-L., Liang S., Yang Y., Yao H.-W., Sun R.-X., Sun Y., Chen W.-J., Zuo S.-Q., Ma M.-J., Li H., Jiang J.-F., Liu W., Yang X.F., Gray G.C., Krause P.J., Cao W.-C. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect. Dis. 2015;15:1467–1479. doi: 10.1016/S1473-3099(15)00177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente J. dl, Villar M., Cabezas-Cruz A., Estrada-Pena A., Ayllon N., Alberdi P. Tick-host-pathogen interactions: conflict and cooperation. PLoS Pathog. 2016;12:e1005488. doi: 10.1371/journal.ppat.1005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter M. Zoonotic risks from small ruminants. Vet. Microbiol. 2015;181:53–65. doi: 10.1016/j.vetmic.2015.07.015. [DOI] [PubMed] [Google Scholar]
- Gong S., He B., Wang Z., Shang L., Wei F., Liu Q., Tu C. Nairobi sheep disease virus RNA in Ixodid ticks, China, 2013. Emerg. Infect. Dis. 2015;21:718–720. doi: 10.3201/eid2104.141602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulia-Nuss M., Nuss A.B., Meyer J.M., Sonenshine D.E., Roe R.M., Waterhouse R.M., Sattelle D.B., de la Fuente J., Ribeiro J.M., Megy K., Thimmapuram J., Miller J.R., Walenz B.P., Koren S., Hostetler J.B., Thiagarajan M., Joardar V.S., Hannick L.I., Bidwell S., Hammond M.P., Young S., Zeng Q., Abrudan J.L., Almeida F.C., Ayllón N., Bhide K., Bissinger B.W., Bonzon-Kulichenko E., Buckingham S.D., Caffrey D.R., Caimano M.J., Croset V., Driscoll T., Gilbert D., Gillespie J.J., Giraldo-Calderón G.I., Grabowski J.M., Jiang D., Khalil S.M.S., Kim D., Kocan K.M., Koči J., Kuhn R.J., Kurtti T.J., Lees K., Lang E.G., Kennedy R.C., Kwon H., Perera R., Qi Y., Radolf J.D., Sakamoto J.M., Sánchez-Gracia A., Severo M.S., Silverman N., Šimo L., Tojo M., Tornador C., Van Zee J.P., Vázquez J., Vieira F.G., Villar M., Wespiser A.R., Yang Y., Zhu J., Arensburger P., Pietrantonio P.V., Barker S.C., Shao R., Zdobnov E.M., Hauser F., Grimmelikhuijzen C.J.P., Park Y., Rozas J., Benton R., Pedra J.H.F., Nelson D.R., Unger M.F., Tubio J.M.C., Tu Z., Robertson H.M., Shumway M., Sutton G., Wortman J.R., Lawson D., Wikel S.K., Nene V.M., Fraser C.M., Collins F.H., Birren B., Nelson K.E., Caler E., Hill C.A. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 2016;7:10507. doi: 10.1038/ncomms10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajarizadeh B., Grebely J., Dore G.J. Epidemiology and natural history of HCV infection. Nat. Rev. Gastro Hepat. 2013;10:553–562. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- He B., Huang X., Zhang F., Tan W., Matthijnssens J., Qin S., Xu L., Zhao Z., Yang Le, Wang Q., Hu T., Bao X., Wu J., Tu C. Group A rotaviruses in Chinese bats: genetic composition, serology, and evidence for bat-to-human transmission and reassortment. J. Virol. 2017;91:e02493–02416. doi: 10.1128/JVI.02493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Li Z., Yang F., Zheng J., Feng Y., Guo H., Li Y., Wang Y., Su N., Zhang F., Fan Q., Tu C. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel Mammalian viruses. PLoS One. 2013;8:e61950. doi: 10.1371/journal.pone.0061950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Du Y., Hu X., Ma H., Wang H., You A., Kang K., Chen H., Zhang L., Liu G., Xu B. Epidemiological and etiological characteristics of fever, thrombocytopenia and leukopenia syndrome in Henan province, China, 2011–2012. PLoS One. 2014;9:e91166. doi: 10.1371/journal.pone.0091166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses (ICTV), 2018. Virus taxonomy: 2017 release, 〈https://talk.ictvonline.org/taxonomy〉.
- Jin C., Jiang H., Liang M., Han Y., Gu W., Zhang F., Zhu H., Wu W., Chen T., Li C., Zhang W., Zhang Q., Qu J., Wei Q., Qin C., Li D. SFTS virus infection in nonhuman primates. J. Infect. Dis. 2015;211:915–925. doi: 10.1093/infdis/jiu564. [DOI] [PubMed] [Google Scholar]
- Joshi M.V., Elankumaran S., Joshi G.D., Albert A., Padbidri V.S., Murali Manohar B. Post-epizootic survey of rift valley fever-like illness among sheep at Veerapuram, Chennai, Tamil Nadu. Indian J. Virol. 1998;14:155–157. [Google Scholar]
- Joshi M.V., Geevarghese G., Joshi G.D., Ghodke Y.S., Mourya D.T., Mishra A.C. Isolation of Ganjam virus from ticks collected off domestic animals around Pune, Maharashtra, India. J. Med. Entomol. 2005;42:204–206. doi: 10.1093/jmedent/42.2.204. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T.M., Creevey C.J., Pentony M.M., Naughton T.J., McLnerney J.O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Yi J., Kim G., Choi S., Jun K., Kim N., Choe P., Kim N., Lee J., Oh M. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg. Infect. Dis. 2013;19:1892–1894. doi: 10.3201/eid1911.130792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.-Y., Liu W., Bowden T.A., Cui N., Zhuang L., Liu K., Zhang Y.-Y., Cao W.-C., Pybus O.G. Evolutionary and molecular analysis of the emergent severe fever with thrombocytopenia syndrome virus. Epidemics. 2013;5:1–10. doi: 10.1016/j.epidem.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Li Z., Wang Z., He B., Wang S., Wei F., Tu C., Liu Q. The first molecular evidence of severe fever with thrombocytopenia syndrome virus in ticks in Jilin, Northeastern China. Ticks Tick. borne Dis. 2016;7:1280–1283. doi: 10.1016/j.ttbdis.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Liu J.W., Wen H.L., Fang L.Z., Zhang Z.T., He S.T., Xue Z.F., Ma D.Q., Zhang X.S., Wang T., Yu H., Zhang Y., Zhao L., Yu X.J. Prevalence of SFTSV among Asian house shrews and rodents, China, January-August 2013. Emerg. Infect. Dis. 2013;20:2126–2128. doi: 10.3201/eid2012.141013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Cui N., Fang L.-Q., Wang B.-J., Lu Q.-B., Peng W., Li H., Wang L.-Y., Liang S., Wang H.-Y., Zhang Y.-Y., Zhuang L., Yang H., Gray G.C., de Vlas S.J., Liu W., Cao W.-C. Epidemiologic features and environmental risk factors of severe fever with thrombocytopenia syndrome, Xinyang, China. PLoS Negl. Trop. Dis. 2014;8:e2820. doi: 10.1371/journal.pntd.0002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., He B., Huang S.-Y., Wei F., Zhu X.-Q. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. Lancet Infect. Dis. 2014;14:763–772. doi: 10.1016/S1473-3099(14)70718-2. [DOI] [PubMed] [Google Scholar]
- Liu W., Lu Q.-B., Cui N., Li H., Wang L.-Y., Liu K., Yang Z.-D., Wang B.-J., Wang H.-Y., Zhang Y.-Y., Zhuang L., Hu C.-Y., Yuan C., Fan X.-J., Wang Z., Zhang L., Zhang X.-A., Walker D.H., Cao W.-C. Case-fatality ratio and effectiveness of Ribavirin therapy among hospitalized patients in China who had severe fever with thrombocytopenia syndrome. Clin. Infect. Dis. 2013;57:1292–1299. doi: 10.1093/cid/cit530. [DOI] [PubMed] [Google Scholar]
- Marczinke B.I., Nichol S.T. Nairobi sheep disease virus, an important tick-borne pathogen of sheep and goats in Africa, is also present in Asia. Virology. 2002;303:146–151. doi: 10.1006/viro.2002.1514. [DOI] [PubMed] [Google Scholar]
- McMullan L.K., Folk S.M., Kelly A.J., MacNeil A., Goldsmith C.S., Metcalfe M.G., Batten B.C., Albarino C.G., Zaki S.R., Rollin P.E., Nicholson W.L., Nichol S.T. A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med. 2012;367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- Niu G., Li J., Liang M., Jiang X., Jiang M., Yin H., Wang Z., Li C., Zhang Q., Jin C., Wang X., Ding S., Xing Z., Wang S., Bi Z., Li D. Sever fever with thrombocytopenia syndrome virus among domesticated animals, China. Emerg. Infect. Dis. 2013;19:756–763. doi: 10.3201/eid1905.120245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera L.P., Peiris J.S.M., Weilgama D.J., Calisher C.H., Shope R.E. Nairobi sheep disease virus isolated from Haemaphysalis intermedia ticks collected in Sri Lanka. Ann. Trop. Med. Parasitol. 1996;90:91–93. doi: 10.1080/00034983.1996.11813031. [DOI] [PubMed] [Google Scholar]
- Scheel T.K.H., Kapoor A., Nishiuchi E., Brock K.V., Yu Y., Andrus L., Gu M., Renshaw R.W., Dubovi E.J., McDonough S.P., Van de Walle G.R., Lipkin W.I., Divers T.J., Tennant B.C., Rice C.M. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc. Natl. Acad. Sci. USA. 2015;112:2192–2197. doi: 10.1073/pnas.1500265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Hu S., Liu X., Yang J., Liu D., Wu L., Wang H., Hu Z., Deng F., Shen S. Migration, recombination, and reassortment are involved in the evolution of severe fever with thrombocytopenia syndrome bunyavirus. Infect. Genet. Evol. 2017;47:109–117. doi: 10.1016/j.meegid.2016.11.015. [DOI] [PubMed] [Google Scholar]
- Sudeep A.B., Jadi R.S., Mishra A.C. Ganjam virus. Indian J. Med. Res. 2009;130:514–519. [PubMed] [Google Scholar]
- Takahashi T., Maeda K., Suzuki T., Ishido A., Shigeoka T., Tominaga T., Kamei T., Honda M., Ninomiya D., Sakai T., Senba T., Kaneyuki S., Sakaguchi S., Satoh A., Hosokawa T., Kawabe Y., Kurihara S., Izumikawa K., Kohno S., Azuma T., Suemori K., Yasukawa M., Mizutani T., Omatsu T., Katayama Y., Miyahara M., Ijuin M., Doi K., Okuda M., Umeki K., Saito T., Fukushima K., Nakajima K., Yoshikawa T., Tani H., Fukushi S., Fukuma A., Ogata M., Shimojima M., Nakajima N., Nagata N., Katano H., Fukumoto H., Sato Y., Hasegawa H., Yamagishi T., Oishi K., Kurane I., Morikawa S., Saijo M. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J. Infect. Dis. 2014;209:816–827. doi: 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffan A., Jonassen C.M., De Battisti C., Schiavon E., Kofstad T., Capua I., Cattoli G. Genetic characterization of a new astrovirus detected in dogs suffering from diarrhoea. Vet. Microbiol. 2009;139:147–152. doi: 10.1016/j.vetmic.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Li X.-L., Liu M., Song X.-J., Zhang H., Wang Y.-B., Tian B.-P., Xing X.-S., Li S.-Y. Epidemiological characteristics and environmental risk factors of severe fever with thrombocytopenia syndrome in Hubei province, China, from 2011 to 2016. Front. Microbiol. 2017;8:387. doi: 10.3389/fmicb.2017.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Hu C., Zhang D., Tang S., Zhang Z., Kou Z., Fan 'Z., Bente D., Zeng C., Li T. Metagenomic profile of the viral communities in Rhipicephalus spp. ticks from Yunnan, China. PLoS One. 2015;10:e0121609. doi: 10.1371/journal.pone.0121609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Sun Y. Classification and identification of important ticks of China. In: Lu B., Wu Y., editors. Classification and Identification of Important Medical Insects of China. Henan Science and Technology Publishing House; Zhengzhou, China: 2003. pp. 652–713. (1 ed.) (1 ed.) [Google Scholar]
- Yang Le, Wu J., Hu T., Qin S., Deng B., Liu J., Zhang F., He B., Tu C. Genetic diversity of bat orthohepadnaviruses in China and a proposed new nomenclature. Infect. Genet Evol. 2018;63:135–143. doi: 10.1016/j.meegid.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.-J., Liang M.-F., Zhang S.-Y., Liu Y., Li J.-D., Sun Y.-L., Zhang L., Zhang Q.-F., Popov V.L., Li C., Qu J., Li Q., Zhang Y.-P., Hai R., Wu W., Wang Q., Zhan F.-X., Wang X.-J., Kan B., Wang S.-W., Wan K.-L., Jing H.-Q., Lu J.-X., Yin W.-W., Zhou H., Guan X.-H., Liu J.-F., Bi Z.-Q., Liu G.-H., Ren J., Wang H., Zhao Z., Song J.-D., He J.-R., Wan T., Zhang J.-S., Fu X.-P., Sun L.-N., Dong X.-P., Feng Z.-J., Yang W.-Z., Hong T., Zhang Y., Walker D.H., Wang Y., Li D.-X. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 2011;364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J., Wang Q., Cheng J., Hu B., Li J., Zhan F., Song Y., Guo D. Current status of severe fever with thrombocytopenia syndrome in China. Virol. Sin. 2017;32:51–62. doi: 10.1007/s12250-016-3931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Sun J., Yan J., Lv H., Chai C., Sun Y., Shao B., Jiang J., Chen Z., Kortekaas J., Zhang Y. Antibodies against severe fever with thrombocytopenia syndrome virus in healthy persons, China, 2013. Emerg. Infect. Dis. 2014;20:1355–1357. doi: 10.3201/eid2008.131796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z., Zhou D.-J., Qin X.-C., Tian J.-H., Xiong Y., Wang J.-B., Chen X.-P., Gao D.-Y., He Y.-W., Jin D., Sun Q., Guo W.-P., Wang W., Yu B., Li J., Dai Y.-A., Li W., Peng J.-S., Zhang G.-B., Zhang S., Chen X.-M., Wang Y., Li M.-H., Lu X., Ye C., de Jong M.D., Xu J. The ecology, genetic diversity, and phylogeny of Huaiyangshan virus in China. J. Virol. 2012;86:2864–2868. doi: 10.1128/JVI.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]